Topical Probiotics as a Novel Approach in the Treatment of Chronic Dermatoses Associated with Skin Dysbiosis: A Narrative Review
Abstract
1. Introduction
2. The Skin Microbiome and Its Role in Inflammatory Skin Disease
2.1. The Healthy Skin Microbiome: Composition, Function, Environmental Influence
2.2. Microbiome Alterations in Atopic Dermatitis (AD)
2.2.1. The Role of Staphylococcus aureus in AD
2.2.2. Non-S. aureus-Related Microbial Changes in AD
2.3. Microbiome Alterations in Psoriasis
3. Topical Probiotics in the Treatment of Chronic Dermatoses
3.1. Mechanistic Insights into Topical Probiotics: Immunomodulation, Pathogen Inhibition, and Barrier Support
3.2. Experimental Validation of Immunomodulation of Probiotics Applied Topically
3.3. Competitive Inhibition and Biofilm Disruption
3.4. Barrier Restoration and Decreased Transepidermal Water Loss (TEWL)
3.5. Probiotic Metabolite Production and Dermatological Impact
3.6. Systemic Immunomodulation via the Gut–Skin Axis: The Role of Oral Probiotics
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gilchrest, B.A. Skin Aging 2003: Recent Advances and Current Concepts. Cutis 2003, 72, 5–10. [Google Scholar]
- Chung, J.H.; Seo, J.Y.; Choi, H.R.; Lee, M.K.; Youn, C.S.; Rhie, G.; Cho, K.H.; Kim, K.H.; Park, K.C.; Eun, H.C. Modulation of Skin Collagen Metabolism in Aged and Photoaged Human Skin In Vivo. J. Investig. Dermatol. 2001, 117, 1218–1224. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Dréno, B.; Dagnelie, M.A.; Khammari, A.; Corvec, S. The Skin Microbiome: A New Actor in Inflammatory Acne. Am. J. Clin. Dermatol. 2020, 21, 18–24. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal Shifts in the Skin Microbiome Associated with Disease Flares and Treatment in Children with Atopic Dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Šuler Baglama, Š.; Trčko, K. Skin and Gut Microbiota Dysbiosis in Autoimmune and Inflammatory Skin Diseases. Acta Dermatovenerol. Alp. Pannonica Adriat. 2022, 31, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Borrego-Ruiz, A.; Borrego, J.J. Nutritional and Microbial Strategies for Treating Acne, Alopecia, and Atopic Dermatitis. Nutrients 2024, 16, 3559. [Google Scholar] [CrossRef] [PubMed]
- Rozas, M.; Hart de Ruijter, A.; Fabrega, M.J.; Zorgani, A.; Guell, M.; Paetzold, B.; Brillet, F. From Dysbiosis to Healthy Skin: Major Contributions of Cutibacterium acnes to Skin Homeostasis. Microorganisms 2021, 9, 628. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, C.V.; Antiga, E.; Lulli, M. Oral and Topical Probiotics and Postbiotics in Skincare and Dermatological Therapy: A Concise Review. Microorganisms 2023, 11, 1420. [Google Scholar] [CrossRef]
- da Silva Vale, A.; de Melo Pereira, G.V.; de Oliveira, A.C.; de Carvalho Neto, D.P.; Herrmann, L.W.; Karp, S.G.; Soccol, V.T.; Soccol, C.R. Production, Formulation, and Application of Postbiotics in the Treatment of Skin Conditions. Fermentation 2023, 9, 264. [Google Scholar] [CrossRef]
- Gallo, R.L. Human Skin Is the Largest Epithelial Surface for Interaction with Microbes. J. Investig. Dermatol. 2017, 137, 1213–1214. [Google Scholar] [CrossRef] [PubMed]
- Cundell, A.M. Microbial Ecology of the Human Skin. Microb. Ecol. 2018, 76, 113–120. [Google Scholar] [CrossRef]
- Murillo, N.; Raoult, D. Skin Microbiota: Overview and Role in the Skin Diseases Acne Vulgaris and Rosacea. Future Microbiol. 2013, 8, 209–222. [Google Scholar] [CrossRef]
- Andersen, B.M. Prevention and Control of Infections in Hospitals; Springer International Publishing: Cham, Switzerland, 2019; ISBN 978-3-319-99920-3. [Google Scholar]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S.; Zheng, Q.; Hodkinson, B.P.; SanMiguel, A.J.; Minot, S.; Bushman, F.D.; Grice, E.A. The Human Skin Double-Stranded DNA Virome: Topographical and Temporal Diversity, Genetic Enrichment, and Dynamic Associations with the Host Microbiome. mBio 2015, 6. [Google Scholar] [CrossRef]
- Jacob, S.; VanDaele, M.A.; Brown, J.N. Treatment of Demodex-associated Inflammatory Skin Conditions: A Systematic Review. Dermatol. Ther. 2019, 32. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Murillo, N.; Aubert, J.; Raoult, D. Microbiota of Demodex Mites from Rosacea Patients and Controls. Microb. Pathog. 2014, 71–72, 37–40. [Google Scholar] [CrossRef]
- Kloos, W.E.; Musselwhite, M.S. Distribution and Persistence of Staphylococcus and Micrococcus Species and Other Aerobic Bacteria on Human Skin. Appl. Microbiol. 1975, 30, 381–395. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida Albicans Cell-Type Switching and Functional Plasticity in the Mammalian Host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Somerville, D.A. The normal flora of the skin in different age groups. Br. J. Dermatol. 1969, 81, 248–258. [Google Scholar] [CrossRef]
- Fluhr, J.W.; Kao, J.; Ahn, S.K.; Feingold, K.R.; Elias, P.M.; Jain, M. Generation of Free Fatty Acids from Phospholipids Regulates Stratum Corneum Acidification and Integrity. J. Investig. Dermatol. 2001, 117, 44–51. [Google Scholar] [CrossRef]
- Roth, R.R.; James, W.D. Microbial Ecology of the Skin. Annu. Rev. Microbiol. 1988, 42, 441–464. [Google Scholar] [CrossRef]
- Kong, H.H. Skin Microbiome: Genomics-Based Insights into the Diversity and Role of Skin Microbes. Trends Mol. Med. 2011, 17, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The Interpersonal and Intrapersonal Diversity of Human-Associated Microbiota in Key Body Sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, I.J.; Wright, E.M.; Tagg, J.R.; Jain, R.; Hale, J.D.F. Skin Microbiome—The Next Frontier for Probiotic Intervention. Probiotics Antimicrob. Proteins 2022, 14, 630–647. [Google Scholar] [CrossRef]
- Meisel, J.S.; Sfyroera, G.; Bartow-McKenney, C.; Gimblet, C.; Bugayev, J.; Horwinski, J.; Kim, B.; Brestoff, J.R.; Tyldsley, A.S.; Zheng, Q.; et al. Commensal Microbiota Modulate Gene Expression in the Skin. Microbiome 2018, 6, 20. [Google Scholar] [CrossRef]
- Williams, M.R.; Costa, S.K.; Zaramela, L.S.; Khalil, S.; Todd, D.A.; Winter, H.L.; Sanford, J.A.; O’Neill, A.M.; Liggins, M.C.; Nakatsuji, T.; et al. Quorum Sensing between Bacterial Species on the Skin Protects against Epidermal Injury in Atopic Dermatitis. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Eng, R.M.; Seite, S. The Role of Cutaneous Microbiota Harmony in Maintaining a Functional Skin Barrier. J. Drugs Dermatol. 2017, 16, 12–18. [Google Scholar] [CrossRef]
- Pausan, M.R.; Csorba, C.; Singer, G.; Till, H.; Schöpf, V.; Santigli, E.; Klug, B.; Högenauer, C.; Blohs, M.; Moissl-Eichinger, C. Exploring the Archaeome: Detection of Archaeal Signatures in the Human Body. Front. Microbiol. 2019, 10, 2796. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A Diversity Profile of the Human Skin Microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.H.; Andersson, B.; Clavel, T.; Common, J.E.; Jackson, S.A.; Olson, N.D.; Segre, J.A.; Traidl-Hoffmann, C. Performing Skin Microbiome Research: A Method to the Madness. J. Investig. Dermatol. 2017, 137, 561–568. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from Human Skin Commensal Bacteria Protect against Staphylococcus aureus and Are Deficient in Atopic Dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human Skin Microbiota Is a Rich Source of Bacteriocin-Producing Staphylococci That Kill Human Pathogens. FEMS Microbiol. Ecol. 2019, 95, fiy241. [Google Scholar] [CrossRef]
- Gaitanis, G.; Tsiouri, G.; Spyridonos, P.; Stefos, T.; Stamatas, G.N.; Velegraki, A.; Bassukas, I.D. Variation of Cultured Skin Microbiota in Mothers and Their Infants during the First Year Postpartum. Pediatr. Dermatol. 2019, 36, 460–465. [Google Scholar] [CrossRef]
- Sanford, J.A.; Gallo, R.L. Functions of the Skin Microbiota in Health and Disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef]
- Maheswary, T.; Nurul, A.A.; Fauzi, M.B. The Insights of Microbes’ Roles in Wound Healing: A Comprehensive Review. Pharmaceutics 2021, 13, 981. [Google Scholar] [CrossRef] [PubMed]
- Wanke, I.; Steffen, H.; Christ, C.; Krismer, B.; Götz, F.; Peschel, A.; Schaller, M.; Schittek, B. Skin Commensals Amplify the Innate Immune Response to Pathogens by Activation of Distinct Signaling Pathways. J. Investig. Dermatol. 2011, 131, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in Healthy Skin, Update for Dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef]
- Friedrich, A.; Paz, M.; Leoni, J.; González Maglio, D. Message in a Bottle: Dialog between Intestine and Skin Modulated by Probiotics. Int. J. Mol. Sci. 2017, 18, 1067. [Google Scholar] [CrossRef]
- Park, Y.J.; Lee, H.K. The Role of Skin and Orogenital Microbiota in Protective Immunity and Chronic Immune-Mediated Inflammatory Disease. Front. Immunol. 2018, 8, 1955. [Google Scholar] [CrossRef]
- Coates, M.; Blanchard, S.; MacLeod, A.S. Innate Antimicrobial Immunity in the Skin: A Protective Barrier against Bacteria, Viruses, and Fungi. PLoS Pathog. 2018, 14, e1007353. [Google Scholar] [CrossRef]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.-J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C.; et al. Commensal–Dendritic-Cell Interaction Specifies a Unique Protective Skin Immune Signature. Nature 2015, 520, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Canesso, M.C.C.; Vieira, A.T.; Castro, T.B.R.; Schirmer, B.G.A.; Cisalpino, D.; Martins, F.S.; Rachid, M.A.; Nicoli, J.R.; Teixeira, M.M.; Barcelos, L.S. Skin Wound Healing Is Accelerated and Scarless in the Absence of Commensal Microbiota. J. Immunol. 2014, 193, 5171–5180. [Google Scholar] [CrossRef] [PubMed]
- Linehan, J.L.; Harrison, O.J.; Han, S.-J.; Byrd, A.L.; Vujkovic-Cvijin, I.; Villarino, A.V.; Sen, S.K.; Shaik, J.; Smelkinson, M.; Tamoutounour, S.; et al. Non-Classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell 2018, 172, 784–796. [Google Scholar] [CrossRef] [PubMed]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in Exploring and Manipulating the Human Skin Microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Z.; Zhang, H.; Guo, Y.; Yao, Z. Update on the Pathogenesis and Therapy of Atopic Dermatitis. Clin. Rev. Allergy Immunol. 2021, 61, 324–338. [Google Scholar] [CrossRef]
- Garmhausen, D.; Hagemann, T.; Bieber, T.; Dimitriou, I.; Fimmers, R.; Diepgen, T.; Novak, N. Characterization of Different Courses of Atopic Dermatitis in Adolescent and Adult Patients. Allergy 2013, 68, 498–506. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus aureus Toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Joo, H.-S.; Chatterjee, S.S.; Otto, M. Phenol-Soluble Modulins—Critical Determinants of Staphylococcal Virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus Colonization of the Skin and Antimicrobial Peptides. Expert. Rev. Dermatol. 2010, 5, 183–195. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and Signal Transduction in the Regulation of Staphylococcal Virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef]
- Godakova, S.A.; Noskov, A.N.; Vinogradova, I.D.; Ugriumova, G.A.; Solovyev, A.I.; Esmagambetov, I.B.; Tukhvatulin, A.I.; Logunov, D.Y.; Naroditsky, B.S.; Shcheblyakov, D.V.; et al. Camelid VHHs Fused to Human Fc Fragments Provide Long Term Protection Against Botulinum Neurotoxin A in Mice. Toxins 2019, 11, 464. [Google Scholar] [CrossRef]
- Altunbulakli, C.; Reiger, M.; Neumann, A.U.; Garzorz-Stark, N.; Fleming, M.; Huelpuesch, C.; Castro-Giner, F.; Eyerich, K.; Akdis, C.A.; Traidl-Hoffmann, C. Relations between Epidermal Barrier Dysregulation and Staphylococcus Species–Dominated Microbiome Dysbiosis in Patients with Atopic Dermatitis. J. Allergy Clin. Immunol. 2018, 142, 1643–1647.e12. [Google Scholar] [CrossRef]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-Host Interplay in Atopic Dermatitis and Psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef]
- Chng, K.R.; Tay, A.S.L.; Li, C.; Ng, A.H.Q.; Wang, J.; Suri, B.K.; Matta, S.A.; McGovern, N.; Janela, B.; Wong, X.F.C.C.; et al. Whole Metagenome Profiling Reveals Skin Microbiome-Dependent Susceptibility to Atopic Dermatitis Flare. Nat. Microbiol. 2016, 1, 16106. [Google Scholar] [CrossRef]
- Koh, L.F.; Ong, R.Y.; Common, J.E. Skin Microbiome of Atopic Dermatitis. Allergol. Int. 2022, 71, 31–39. [Google Scholar] [CrossRef]
- Rauer, L.; Reiger, M.; Bhattacharyya, M.; Brunner, P.M.; Krueger, J.G.; Guttman-Yassky, E.; Traidl-Hoffmann, C.; Neumann, A.U. Skin Microbiome and Its Association with Host Cofactors in Determining Atopic Dermatitis Severity. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 772–782. [Google Scholar] [CrossRef]
- Simpson, E.L.; Villarreal, M.; Jepson, B.; Rafaels, N.; David, G.; Hanifin, J.; Taylor, P.; Boguniewicz, M.; Yoshida, T.; De Benedetto, A.; et al. Patients with Atopic Dermatitis Colonized with Staphylococcus aureus Have a Distinct Phenotype and Endotype. J. Investig. Dermatol. 2018, 138, 2224–2233. [Google Scholar] [CrossRef] [PubMed]
- Hülpüsch, C.; Tremmel, K.; Hammel, G.; Bhattacharyya, M.; de Tomassi, A.; Nussbaumer, T.; Neumann, A.U.; Reiger, M.; Traidl-Hoffmann, C. Skin PH–Dependent Staphylococcus aureus Abundance as Predictor for Increasing Atopic Dermatitis Severity. Allergy 2020, 75, 2888–2898. [Google Scholar] [CrossRef]
- Tauber, M.; Balica, S.; Hsu, C.-Y.; Jean-Decoster, C.; Lauze, C.; Redoules, D.; Viodé, C.; Schmitt, A.-M.; Serre, G.; Simon, M.; et al. Staphylococcus aureus Density on Lesional and Nonlesional Skin Is Strongly Associated with Disease Severity in Atopic Dermatitis. J. Allergy Clin. Immunol. 2016, 137, 1272–1274.e3. [Google Scholar] [CrossRef]
- Cho, S.-H.; Strickland, I.; Tomkinson, A.; Fehringer, A.P.; Gelfand, E.W.; Leung, D.Y.M. Preferential Binding of Staphylococcus aureus to Skin Sites of Th2-Mediated Inflammation in a Murine Model. J. Investig. Dermatol. 2001, 116, 658–663. [Google Scholar] [CrossRef]
- Miajlovic, H.; Fallon, P.G.; Irvine, A.D.; Foster, T.J. Effect of Filaggrin Breakdown Products on Growth of and Protein Expression by Staphylococcus aureus. J. Allergy Clin. Immunol. 2010, 126, 1184–1190.e3. [Google Scholar] [CrossRef]
- Cabanillas, B.; Novak, N. Atopic Dermatitis and Filaggrin. Curr. Opin. Immunol. 2016, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.Y.M.; Guttman-Yassky, E. Deciphering the Complexities of Atopic Dermatitis: Shifting Paradigms in Treatment Approaches. J. Allergy Clin. Immunol. 2014, 134, 769–779. [Google Scholar] [CrossRef]
- Salamzade, R.; Swaney, M.H.; Kalan, L.R. Comparative Genomic and Metagenomic Investigations of the Corynebacterium tuberculostearicum Species Complex Reveals Potential Mechanisms Underlying Associations To Skin Health and Disease. Microbiol. Spectr. 2023, 11, e03578-22. [Google Scholar] [CrossRef]
- Edslev, S.M.; Olesen, C.M.; Nørreslet, L.B.; Ingham, A.C.; Iversen, S.; Lilje, B.; Clausen, M.-L.; Jensen, J.S.; Stegger, M.; Agner, T.; et al. Staphylococcal Communities on Skin Are Associated with Atopic Dermatitis and Disease Severity. Microorganisms 2021, 9, 432. [Google Scholar] [CrossRef]
- Bjerre, R.D.; Holm, J.B.; Palleja, A.; Sølberg, J.; Skov, L.; Johansen, J.D. Skin Dysbiosis in the Microbiome in Atopic Dermatitis Is Site-Specific and Involves Bacteria, Fungus and Virus. BMC Microbiol. 2021, 21, 256. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Cheon, H.I.; Hur, M.S.; Kim, M.J.; Jung, W.H.; Lee, Y.W.; Choe, Y.B.; Ahn, K.J. Analysis of the Skin Mycobiome in Adult Patients with Atopic Dermatitis. Exp. Dermatol. 2018, 27, 366–373. [Google Scholar] [CrossRef]
- Zhang, E.; Tanaka, T.; Tajima, M.; Tsuboi, R.; Nishikawa, A.; Sugita, T. Characterization of the Skin Fungal Microbiota in Patients with Atopic Dermatitis and in Healthy Subjects. Microbiol. Immunol. 2011, 55, 625–632. [Google Scholar] [CrossRef]
- Selander, C.; Zargari, A.; Möllby, R.; Rasool, O.; Scheynius, A. Higher PH Level, Corresponding to That on the Skin of Patients with Atopic Eczema, Stimulates the Release of Malassezia sympodialis Allergens. Allergy 2006, 61, 1002–1008. [Google Scholar] [CrossRef]
- Matricardi, P.M.; Kleine-Tebbe, J.; Hoffmann, H.J.; Valenta, R.; Hilger, C.; Hofmaier, S.; Aalberse, R.C.; Agache, I.; Asero, R.; Ballmer-Weber, B.; et al. EAACI Molecular Allergology User’s Guide. Pediatr. Allergy Immunol. 2016, 27, 1–250. [Google Scholar] [CrossRef]
- Glatz, M.; Buchner, M.; Bartenwerffer, W.; Schmid-Grendelmeier, P.; Worm, M.; Hedderich, J.; Fölster-Holst, R. Malassezia spp.-Specific Immunoglobulin E Level Is a Marker for Severity of Atopic Dermatitis in Adults. Acta Derm. Venereol. 2015, 95, 191–196. [Google Scholar] [CrossRef]
- Selander, C.; Engblom, C.; Nilsson, G.; Scheynius, A.; Andersson, C.L. TLR2/MyD88-Dependent and -Independent Activation of Mast Cell IgE Responses by the Skin Commensal Yeast Malassezia sympodialis. J. Immunol. 2009, 182, 4208–4216. [Google Scholar] [CrossRef]
- Hiragun, T.; Ishii, K.; Hiragun, M.; Suzuki, H.; Kan, T.; Mihara, S.; Yanase, Y.; Bartels, J.; Schröder, J.-M.; Hide, M. Fungal Protein MGL_1304 in Sweat Is an Allergen for Atopic Dermatitis Patients. J. Allergy Clin. Immunol. 2013, 132, 608–615.e4. [Google Scholar] [CrossRef]
- Sparber, F.; De Gregorio, C.; Steckholzer, S.; Ferreira, F.M.; Dolowschiak, T.; Ruchti, F.; Kirchner, F.R.; Mertens, S.; Prinz, I.; Joller, N.; et al. The Skin Commensal Yeast Malassezia Triggers a Type 17 Response That Coordinates Anti-Fungal Immunity and Exacerbates Skin Inflammation. Cell Host Microbe 2019, 25, 389–403.e6. [Google Scholar] [CrossRef] [PubMed]
- Bitschar, K.; Sauer, B.; Focken, J.; Dehmer, H.; Moos, S.; Konnerth, M.; Schilling, N.A.; Grond, S.; Kalbacher, H.; Kurschus, F.C.; et al. Lugdunin Amplifies Innate Immune Responses in the Skin in Synergy with Host- and Microbiota-Derived Factors. Nat. Commun. 2019, 10, 2730. [Google Scholar] [CrossRef] [PubMed]
- Suwarsa, O.; Hazari, M.N.; Dharmadji, H.P.; Dwiyana, R.F.; Effendi, R.M.R.A.; Hidayah, R.M.N.; Avriyanti, E.; Gunawan, H.; Sutedja, E. A Pilot Study: Composition and Diversity of 16S RRNA Based Skin Bacterial Microbiome in Indonesian Atopic Dermatitis Population. Clin. Cosmet. Investig. Dermatol. 2021, 14, 1737–1744. [Google Scholar] [CrossRef]
- Schmid, B.; Künstner, A.; Fähnrich, A.; Bersuch, E.; Schmid-Grendelmeier, P.; Busch, H.; Glatz, M.; Bosshard, P.P. Dysbiosis of Skin Microbiota with Increased Fungal Diversity Is Associated with Severity of Disease in Atopic Dermatitis. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1811–1819. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Suh, D.H.; Lee, S.; Kim, H.S.; Cho, S.H.; Woo, Y.R. Associations Between Skin Microbiome and Metabolome in the Pathogenesis of Atopic Dermatitis Patients With Scalp Involvement. Allergy Asthma Immunol. Res. 2024, 16, 668. [Google Scholar] [CrossRef]
- Sugumaran, D.; Yong, A.C.H.; Stanslas, J. Advances in Psoriasis Research: From Pathogenesis to Therapeutics. Life Sci. 2024, 355, 122991. [Google Scholar] [CrossRef]
- Vičić, M.; Kaštelan, M.; Brajac, I.; Sotošek, V.; Massari, L.P. Current Concepts of Psoriasis Immunopathogenesis. Int. J. Mol. Sci. 2021, 22, 11574. [Google Scholar] [CrossRef]
- Assarsson, M.; Söderman, J.; Dienus, O.; Seifert, O. Significant Differences in the Bacterial Microbiome of the Pharynx and Skin in Patients with Psoriasis Compared with Healthy Controls. Acta Derm. Venereol. 2020, 100, adv00273. [Google Scholar] [CrossRef]
- Wei, J.; Zhu, J.; Xu, H.; Zhou, D.; Elder, J.T.; Tsoi, L.C.; Patrick, M.T.; Li, Y. Alcohol Consumption and Smoking in Relation to Psoriasis: A Mendelian Randomization Study. Br. J. Dermatol. 2022, 187, 684–691. [Google Scholar] [CrossRef]
- Zou, X.; Zou, X.; Gao, L.; Zhao, H. Gut Microbiota and Psoriasis: Pathogenesis, Targeted Therapy, and Future Directions. Front. Cell Infect. Microbiol. 2024, 14, 1430586. [Google Scholar] [CrossRef]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, Y.; Yabe, R.; Chung, S.-H.; Murayama, M.A.; Yoshida, K.; Matsuo, K.; Kubo, S.; Saijo, S.; Nakamura, Y.; Matsue, H.; et al. IL-36α from Skin-Resident Cells Plays an Important Role in the Pathogenesis of Imiquimod-Induced Psoriasiform Dermatitis by Forming a Local Autoamplification Loop. J. Immunol. 2018, 201, 167–182. [Google Scholar] [CrossRef]
- Valdimarsson, H.; Baker, B.S.; Jónsdóttir, I.; Powles, A.; Fry, L. Psoriasis: A T-Cell-Mediated Autoimmune Disease Induced by Streptococcal Superantigens? Immunol. Today 1995, 16, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.Y.; Travers, J.B.; Giorno, R.; Norris, D.A.; Skinner, R.; Aelion, J.; Kazemi, L.V.; Kim, M.H.; Trumble, A.E.; Kotb, M. Evidence for a Streptococcal Superantigen-Driven Process in Acute Guttate Psoriasis. J. Clin. Investig. 1995, 96, 2106–2112. [Google Scholar] [CrossRef]
- Gao, Z.; Tseng, C.; Strober, B.E.; Pei, Z.; Blaser, M.J. Substantial Alterations of the Cutaneous Bacterial Biota in Psoriatic Lesions. PLoS ONE 2008, 3, e2719. [Google Scholar] [CrossRef]
- Fahlén, A.; Engstrand, L.; Baker, B.S.; Powles, A.; Fry, L. Comparison of Bacterial Microbiota in Skin Biopsies from Normal and Psoriatic Skin. Arch. Dermatol. Res. 2012, 304, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Alekseyenko, A.V.; Perez-Perez, G.I.; De Souza, A.; Strober, B.; Gao, Z.; Bihan, M.; Li, K.; Methé, B.A.; Blaser, M.J. Community Differentiation of the Cutaneous Microbiota in Psoriasis. Microbiome 2013, 1, 31. [Google Scholar] [CrossRef]
- Drago, L.; De Grandi, R.; Altomare, G.; Pigatto, P.; Rossi, O.; Toscano, M. Skin Microbiota of First Cousins Affected by Psoriasis and Atopic Dermatitis. Clin. Mol. Allergy 2016, 14, 2. [Google Scholar] [CrossRef]
- Tett, A.; Pasolli, E.; Farina, S.; Truong, D.T.; Asnicar, F.; Zolfo, M.; Beghini, F.; Armanini, F.; Jousson, O.; De Sanctis, V.; et al. Unexplored Diversity and Strain-Level Structure of the Skin Microbiome Associated with Psoriasis. NPJ Biofilms Microbiomes 2017, 3, 14. [Google Scholar] [CrossRef]
- Chang, H.-W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the Cutaneous Microbiome in Psoriasis and Potential Role in Th17 Polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, M.; Duvetorp, A.; Dienus, O.; Söderman, J.; Seifert, O. Significant Changes in the Skin Microbiome in Patients with Chronic Plaque Psoriasis after Treatment with Narrowband Ultraviolet B. Acta Derm. Venereol. 2018, 98, 428–436. [Google Scholar] [CrossRef]
- Stehlikova, Z.; Kostovcik, M.; Kostovcikova, K.; Kverka, M.; Juzlova, K.; Rob, F.; Hercogova, J.; Bohac, P.; Pinto, Y.; Uzan, A.; et al. Dysbiosis of Skin Microbiota in Psoriatic Patients: Co-Occurrence of Fungal and Bacterial Communities. Front. Microbiol. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Kayıran, M.A.; Sahin, E.; Koçoğlu, E.; Sezerman, O.U.; Gürel, M.S.; Karadağ, A.S. Is Cutaneous Microbiota a Player in Disease Pathogenesis? Comparison of Cutaneous Microbiota in Psoriasis and Seborrheic Dermatitis with Scalp Involvement. Indian. J. Dermatol. Venereol. Leprol. 2022, 88, 738. [Google Scholar] [CrossRef]
- Ruuskanen, M.O.; Vats, D.; Potbhare, R.; RaviKumar, A.; Munukka, E.; Ashma, R.; Lahti, L. Towards Standardized and Reproducible Research in Skin Microbiomes. Environ. Microbiol. 2022, 24, 3840–3860. [Google Scholar] [CrossRef]
- Perugini, P.; Grignani, C.; Condrò, G.; van der Hoeven, H.; Ratti, A.; Mondelli, A.; Colpani, A.; Bleve, M. Skin Microbiota: Setting up a Protocol to Evaluate a Correlation between the Microbial Flora and Skin Parameters. Biomedicines 2023, 11, 966. [Google Scholar] [CrossRef]
- Quan, C.; Chen, X.-Y.; Li, X.; Xue, F.; Chen, L.-H.; Liu, N.; Wang, B.; Wang, L.-Q.; Wang, X.-P.; Yang, H.; et al. Psoriatic Lesions Are Characterized by Higher Bacterial Load and Imbalance between Cutibacterium and Corynebacterium. J. Am. Acad. Dermatol. 2020, 82, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506. [Google Scholar] [CrossRef]
- Yu, Y.; Dunaway, S.; Champer, J.; Kim, J.; Alikhan, A. Changing Our Microbiome: Probiotics in Dermatology. Br. J. Dermatol. 2020, 182, 39–46. [Google Scholar] [CrossRef]
- Al-Ghazzewi, F.H.; Tester, R.F. Impact of Prebiotics and Probiotics on Skin Health. Benef. Microbes 2014, 5, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Båtsman, A.; Salminen, S. Probiotics for the Skin: A New Area of Potential Application? Lett. Appl. Microbiol. 2003, 36, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Lopes, E.G.; Moreira, D.A.; Gullón, P.; Gullón, B.; Cardelle-Cobas, A.; Tavaria, F.K. Topical Application of Probiotics in Skin: Adhesion, Antimicrobial and Antibiofilm in Vitro Assays. J. Appl. Microbiol. 2017, 122, 450–461. [Google Scholar] [CrossRef]
- Schön, M.P. Adaptive and Innate Immunity in Psoriasis and Other Inflammatory Disorders. Front. Immunol. 2019, 10, 1764. [Google Scholar] [CrossRef]
- Guéniche, A.; Benyacoub, J.; Buetler, T.M.; Smola, H.; Blum, S. Supplementation with Oral Probiotic Bacteria Maintains Cutaneous Immune Homeostasis after UV Exposure. Eur. J. Dermatol. 2006, 16, 511–517. [Google Scholar]
- Chae, M.; Kim, B.J.; Na, J.; Kim, S.Y.; Lee, J.O.; Kim, Y.J.; Lee, E.; Cho, D.; Roh, J.; Kim, W. Antimicrobial Activity of Lactiplantibacillus plantarum APsulloc 331261 and APsulloc 331266 against Pathogenic Skin Microbiota. Front Biosci 2021, 13, 237–248. [Google Scholar] [CrossRef]
- Negi, A.; Kuo, C.W.; Hazam, P.K.; Yeh, J.C.; Lin, W.C.; Lou, Y.C.; Yu, C.Y.; Yu, T.L.; Lu, T.M.; Chen, J.Y. Disruption of MRSA Biofilm and Virulence by Deep-Sea Probiotics: Impacts on Energy Metabolism and Host Antimicrobial Peptides. Probiotics Antimicrob. Proteins 2025, 17, 2394–2416. [Google Scholar] [CrossRef]
- Myles, I.A.; Earland, N.J.; Anderson, E.D.; Moore, I.N.; Kieh, M.D.; Williams, K.W.; Saleem, A.; Fontecilla, N.M.; Welch, P.A.; Darnell, D.A.; et al. First-in-Human Topical Microbiome Transplantation with Roseomonas Mucosa for Atopic Dermatitis. JCI Insight 2018, 3, e120608. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Gallo, R.L.; Shafiq, F.; Tong, Y.; Chun, K.; Butcher, A.M.; Cheng, J.Y.; Hata, T.R. Use of Autologous Bacteriotherapy to Treat Staphylococcus aureus in Patients with Atopic Dermatitis: A Randomized Double-Blind Clinical Trial. JAMA Dermatol. 2021, 157, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Feng, C.; Zhang, T.; Martínez-Ríos, V.; Martorell, P.; Tortajada, M.; Cheng, S.; Cheng, S.; Duan, Z. Effects of a Lotion Containing Probiotic Ferment Lysate as the Main Functional Ingredient on Enhancing Skin Barrier: A Randomized, Self-Control Study. Sci. Rep. 2023, 13, 16879. [Google Scholar] [CrossRef] [PubMed]
- Prince, T.; McBain, A.J.; O’Neill, C.A. Lactobacillus Reuteri Protects Epidermal Keratinocytes from Staphylococcus aureus-Induced Cell Death by Competitive Exclusion. Appl. Environ. Microbiol. 2012, 78, 5119–5126. [Google Scholar] [CrossRef]
- Park, J.-Y.; Lee, J.Y.; Hong, S.; Heo, H.; Lee, H.; Kim, Y.G.; Kim, B.-K.; Choi, S.-I.; Lee, J. Limosilactobacillus fermentum MG5368 and Lactiplantibacillus plantarum MG989 Regulates Skin Health in UVB-Induced HaCaT Cells and Hairless Mice Model. Nutrients 2024, 16, 4083. [Google Scholar] [CrossRef]
- Souak, D.; Barreau, M.; Courtois, A.; André, V.; Duclairoir Poc, C.; Feuilloley, M.G.J.; Gault, M. Challenging Cosmetic Innovation: The Skin Microbiota and Probiotics Protect the Skin from UV-Induced Damage. Microorganisms 2021, 9, 936. [Google Scholar] [CrossRef]
- Jeong, J.H.; Lee, C.Y.; Chung, D.K. Probiotic Lactic Acid Bacteria and Skin Health. Crit. Rev. Food Sci. Nutr. 2016, 56, 2331–2337. [Google Scholar] [CrossRef] [PubMed]
- Callewaert, C.; Ravard Helffer, K.; Lebaron, P. Skin Microbiome and Its Interplay with the Environment. Am. J. Clin. Dermatol. 2020, 21, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Rosignoli, C.; Thibaut de Ménonville, S.; Orfila, D.; Béal, M.; Bertino, B.; Aubert, J.; Mercenier, A.; Piwnica, D. A Topical Treatment Containing Heat-Treated Lactobacillus Johnsonii NCC 533 Reduces Staphylococcus aureus Adhesion and Induces Antimicrobial Peptide Expression in an in Vitro Reconstructed Human Epidermis Model. Exp. Dermatol. 2018, 27, 358–365. [Google Scholar] [CrossRef]
- Wang, Y.; Moon, A.; Huang, J.; Sun, Y.; Qiu, H.J. Antiviral Effects and Underlying Mechanisms of Probiotics as Promising Antivirals. Front. Cell Infect. Microbiol. 2022, 12, 928050. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Fischer, T.C.; Wohlrab, J.; Barnard, J.; Alió, A.B. Study of the Efficacy, Tolerability, and Safety of 2 Fixed-Dose Combination Gels in the Management of Acne Vulgaris. Cutis 2009, 84, 223–229. [Google Scholar]
- Lebeer, S.; Oerlemans, E.F.M.; Claes, I.; Henkens, T.; Delanghe, L.; Wuyts, S.; Spacova, I.; van den Broek, M.F.L.; Tuyaerts, I.; Wittouck, S.; et al. Selective Targeting of Skin Pathobionts and Inflammation with Topically Applied Lactobacilli. Cell Rep. Med. 2022, 3, 100521. [Google Scholar] [CrossRef]
- Gueniche, A.; Knaudt, B.; Schuck, E.; Volz, T.; Bastien, P.; Martin, R.; Röcken, M.; Breton, L.; Biedermann, T. Effects of Nonpathogenic Gram-Negative Bacterium Vitreoscilla Filiformis Lysate on Atopic Dermatitis: A Prospective, Randomized, Double-Blind, Placebo-Controlled Clinical Study. Br. J. Dermatol. 2008, 159, 1357–1363. [Google Scholar] [CrossRef]
- Guéniche, A.; Hennino, A.; Goujon, C.; Dahel, K.; Bastien, P.; Martin, R.; Jourdain, R.; Breton, L. Improvement of Atopic Dermatitis Skin Symptoms by Vitreoscilla Filiformis Bacterial Extract. Eur. J. Dermatol. 2006, 16, 380–384. [Google Scholar] [PubMed]
- Nguyen, A.T.; Kim, M.; Kim, Y.E.; Kim, H.; Lee, S.; Lee, Y.; Kim, K.Y. MSF Enhances Human Antimicrobial Peptide β-Defensin (HBD2 and HBD3) Expression and Attenuates Inflammation via the NF- κ B and P38 Signaling Pathways. Molecules 2023, 28, 2744. [Google Scholar] [CrossRef]
- Chen, Y.H.; Wu, C.S.; Chao, Y.H.; Lin, C.C.; Tsai, H.Y.; Li, Y.R.; Chen, Y.Z.; Tsai, W.H.; Chen, Y.K. Lactobacillus pentosus GMNL-77 Inhibits Skin Lesions in Imiquimod-Induced Psoriasis-like Mice. J. Food Drug Anal. 2017, 25, 559–566. [Google Scholar] [CrossRef]
- Asadullah, K.; Sterry, W.; Stephanek, K.; Jasulaitis, D.; Leupold, M.; Audring, H.; Volk, H.D.; Döcke, W.D. IL-10 Is a Key Cytokine in Psoriasis: Proof of Principle by IL-10 Therapy: A New Therapeutic Approach. J. Clin. Investig. 1998, 101, 783–794. [Google Scholar] [CrossRef]
- Tsai, W.H.; Chou, C.H.; Chiang, Y.J.; Lin, C.G.; Lee, C.H. Regulatory Effects of Lactobacillus plantarum-GMNL6 on Human Skin Health by Improving Skin Microbiome. Int. J. Med. Sci. 2021, 18, 1114–1120. [Google Scholar] [CrossRef]
- Gueniche, A.; Philippe, D.; Bastien, P.; Reuteler, G.; Blum, S.; Castiel-Higounenc, I.; Breton, L.; Benyacoub, J. Randomised Double-Blind Placebo-Controlled Study of the Effect of Lactobacillus paracasei NCC 2461 on Skin Reactivity. Benef. Microbes 2014, 5, 137–145. [Google Scholar] [CrossRef]
- Gudadappanavar, A.; Hombal, P.; Timashetti, S.; Javali, S. Influence of Lactobacillus acidophilus and Lactobacillus plantarum on Wound Healing in Male Wistar Rats—An Experimental Study. Int. J. Appl. Basic. Med. Res. 2017, 7, 233–238. [Google Scholar] [CrossRef]
- Saha, U.B.; Saroj, S.D. Lactic Acid Bacteria: Prominent Player in the Fight against Human Pathogens. Expert Rev. Anti -Infect. Ther. 2022, 20, 1435–1453. [Google Scholar] [CrossRef]
- Öhnstedt, E.; Tomenius, H.L.; Frank, P.; Roos, S.; Vågesjö, E.; Phillipson, M. Accelerated Wound Healing in Minipigs by On-Site Production and Delivery of CXCL12 by Transformed Lactic Acid Bacteria. Pharmaceutics 2022, 14, 229. [Google Scholar] [CrossRef]
- Li, Y.; Tang, X.; Liu, Y.; Chen, X.; Wang, L.; Su, Y.; He, W.; Li, J.; Huang, Q.; Wu, A. Advanced Wound Healing with Chitosan Hydrogels Incorporating Metabolites from Whale-Derived Lactiplantibacillus plantarum HJ-S2. Front. Mater. 2025, 12, 1573222. [Google Scholar] [CrossRef]
- Peral, M.C.; Rachid, M.M.; Gobbato, N.M.; Huaman Martinez, M.A.; Valdez, J.C. Interleukin-8 Production by Polymorphonuclear Leukocytes from Patients with Chronic Infected Leg Ulcers Treated with Lactobacillus plantarum. Clin. Microbiol. Infect. 2010, 16, 281–286. [Google Scholar] [CrossRef]
- Peral, M.C.; Huaman Martinez, M.A.; Valdez, J.C. Bacteriotherapy with Lactobacillus plantarum in Burns. Int. Wound J. 2009, 6, 73–81. [Google Scholar] [CrossRef]
- Valdéz, J.C.; Peral, M.C.; Rachid, M.; Santana, M.; Perdigón, G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in Vitro and in Infected Burns: The Potential Use of Probiotics in Wound Treatment. Clin. Microbiol. Infect. 2005, 11, 472–479. [Google Scholar] [CrossRef]
- Hong, Y.F.; Lee, H.Y.; Jung, B.J.; Jang, S.; Chung, D.K.; Kim, H. Lipoteichoic Acid Isolated from Lactobacillus plantarum Down-Regulates UV-Induced MMP-1 Expression and up-Regulates Type I Procollagen through the Inhibition of Reactive Oxygen Species Generation. Mol. Immunol. 2015, 67, 248–255. [Google Scholar] [CrossRef]
- Lee, K.; Kim, H.J.; Kim, S.A.; Park, S.D.; Shim, J.J.; Lee, J.L. Exopolysaccharide from Lactobacillus plantarum Hy7714 Protects against Skin Aging through Skin–Gut Axis Communication. Molecules 2021, 26, 1651. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Lee, M.; Oh, H.; Seo, W.; Kim, G.S.; Ban, O.H.; Shin, M.; Jung, Y.H.; Yang, J. Enhanced Ceramides Production by Lactobacillus rhamnosus IDCC 3201 and Its Proposed Mechanism. Appl. Biol. Chem. 2021, 64, 50. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Feng, X.; Patel, R.; Patel, S.; Paranavitana, C. Antimicrobial and Antibiofilm Potential of Biosurfactants Isolated from Lactobacilli against Multi-Drug-Resistant Pathogens. BMC Microbiol. 2014, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xiao, Y.; Zhang, X.; Zhu, Z.; Zhang, H.; Wei, J.; Zhao, Z.; Li, J.; Chen, T. Probiotics Suppress LL37 Generated Rosacea-like Skin Inflammation by Modulating the TLR2/MyD88/NF-ΚB Signaling Pathway. Food Funct. 2024, 15, 8916–8934. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Yoon, W.; Lee, S.Y.; Shin, H.S.; Lim, M.Y.; Nam, Y.D.; Yoo, Y. Effects of Lactobacillus pentosus in Children with Allergen-Sensitized Atopic Dermatitis. J. Korean Med. Sci. 2020, 35, 1145984. [Google Scholar] [CrossRef]
- Groeger, D.; O’Mahony, L.; Murphy, E.F.; Bourke, J.F.; Dinan, T.G.; Kiely, B.; Shanahan, F.; Quigley, E.M.M. Bifidobacterium Infantis 35624 Modulates Host Inflammatory Processes beyond the Gut. Gut Microbes 2013, 4, 325–339. [Google Scholar] [CrossRef]
- Jiraskova Zakostelska, Z.; Reiss, Z.; Tlaskalova-Hogenova, H.; Rob, F. Paradoxical Reactions to Anti-TNFα and Anti-IL-17 Treatment in Psoriasis Patients: Are Skin and/or Gut Microbiota Involved? Dermatol. Ther. 2023, 13, 911–933. [Google Scholar] [CrossRef]
- Buhaș, M.C.; Candrea, R.; Gavrilaș, L.I.; Miere, D.; Tătaru, A.; Boca, A.; Cătinean, A. Transforming Psoriasis Care: Probiotics and Prebiotics as Novel Therapeutic Approaches. Int. J. Mol. Sci. 2023, 24, 11225. [Google Scholar] [CrossRef]
- Habeebuddin, M.; Karnati, R.K.; Shiroorkar, P.N.; Nagaraja, S.; Asdaq, S.M.B.; Anwer, M.K.; Fattepur, S. Topical Probiotics: More Than a Skin Deep. Pharmaceutics 2022, 14, 557. [Google Scholar] [CrossRef]
- Ray Mohapatra, A.; Harikrishnan, A.; Lakshmanan, D.; Jeevaratnam, K. Targeting Staphylococcus aureus and Its Biofilms with Novel Antibacterial Compounds Produced by Lactiplantibacillus plantarum SJ33. Arch. Microbiol. 2022, 204, 20. [Google Scholar] [CrossRef]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.M. Staphylococcus Epidermidis in the Human Skin Microbiome Mediates Fermentation to Inhibit the Growth of Propionibacterium Acnes: Implications of Probiotics in Acne Vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef]
- Kober, M.M.; Bowe, W.P. The Effect of Probiotics on Immune Regulation, Acne, and Photoaging. Int. J. Womens. Dermatol. 2015, 1, 85–89. [Google Scholar] [CrossRef]
- Mintoff, D.; Borg, I.; Pace, N.P. The Clinical Relevance of the Microbiome in Hidradenitis Suppurativa: A Systematic Review. Vaccines 2021, 9, 1076. [Google Scholar] [CrossRef]
- Frew, J.W.; Hawkes, J.E.; Krueger, J.G. Topical, Systemic and Biologic Therapies in Hidradenitis Suppurativa: Pathogenic Insights by Examining Therapeutic Mechanisms. Ther. Adv. Chronic. Dis. 2019, 10, 2040622319830646. [Google Scholar] [CrossRef]
- Dessinioti, C.; Katsambas, A. The Microbiome and Acne: Perspectives for Treatment. Dermatol. Ther. 2024, 14, 31–44. [Google Scholar] [CrossRef]
- Sánchez-Pellicer, P.; Eguren-Michelena, C.; García-Gavín, J.; Llamas-Velasco, M.; Navarro-Moratalla, L.; Núñez-Delegido, E.; Agüera-Santos, J.; Navarro-López, V. Rosacea, Microbiome and Probiotics: The Gut-Skin Axis. Front. Microbiol. 2024, 14, 1323644. [Google Scholar] [CrossRef]
- Daou, H.; Paradiso, M.; Hennessy, K.; Seminario-Vidal, L. Rosacea and the Microbiome: A Systematic Review. Dermatol. Ther. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Liegenfeld, S.C.; Stenzel, S.; Rembe, J.-D.; Dittmer, M.; Ramos, P.; Stuermer, E.K. Pathogenic and Non-Pathogenic Microbes in the Wound Microbiome—How to Flip the Switch. Microbiol. Res. 2025, 16, 39. [Google Scholar] [CrossRef]
- Burns, E.M.; Ahmed, H.; Isedeh, P.N.; Kohli, I.; Van Der Pol, W.; Shaheen, A.; Muzaffar, A.F.; Al-Sadek, C.; Foy, T.M.; Abdelgawwad, M.S.; et al. Ultraviolet Radiation, Both UVA and UVB, Influences the Composition of the Skin Microbiome. Exp. Dermatol. 2019, 28, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.J.; Navarro, C.; Durán, P.; Galan-Freyle, N.J.; Parra Hernández, L.A.; Pacheco-Londoño, L.C.; Castelanich, D.; Bermúdez, V.; Chacin, M. Antioxidants in Photoaging: From Molecular Insights to Clinical Applications. Int. J. Mol. Sci. 2024, 25, 2403. [Google Scholar] [CrossRef] [PubMed]
- Świerczewska, Z.; Lewandowski, M.; Surowiecka, A.; Barańska-Rybak, W. Microbiome in Hidradenitis Suppurativa—What We Know and Where We Are Heading. Int. J. Mol. Sci. 2022, 23, 11280. [Google Scholar] [CrossRef] [PubMed]

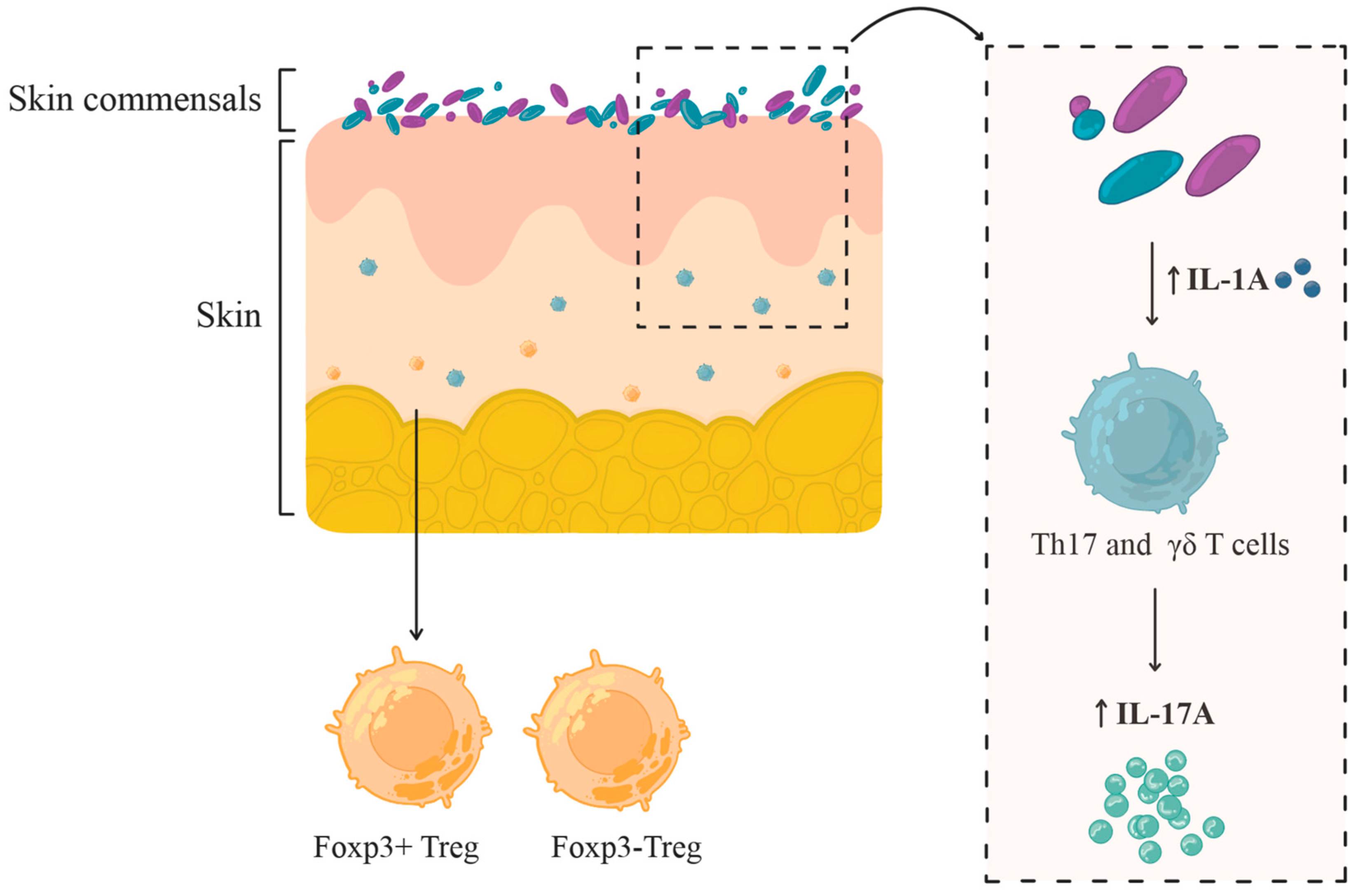
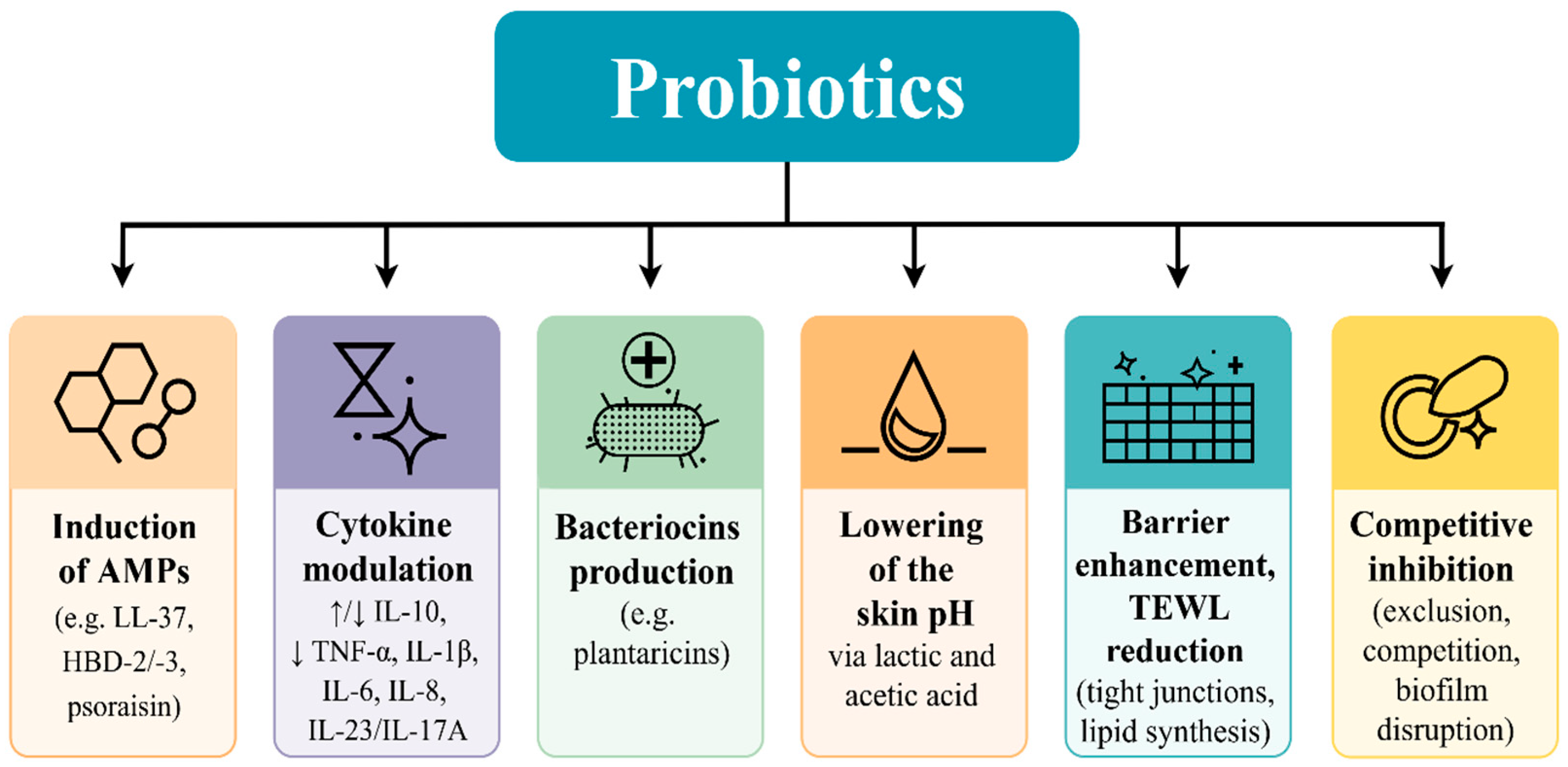
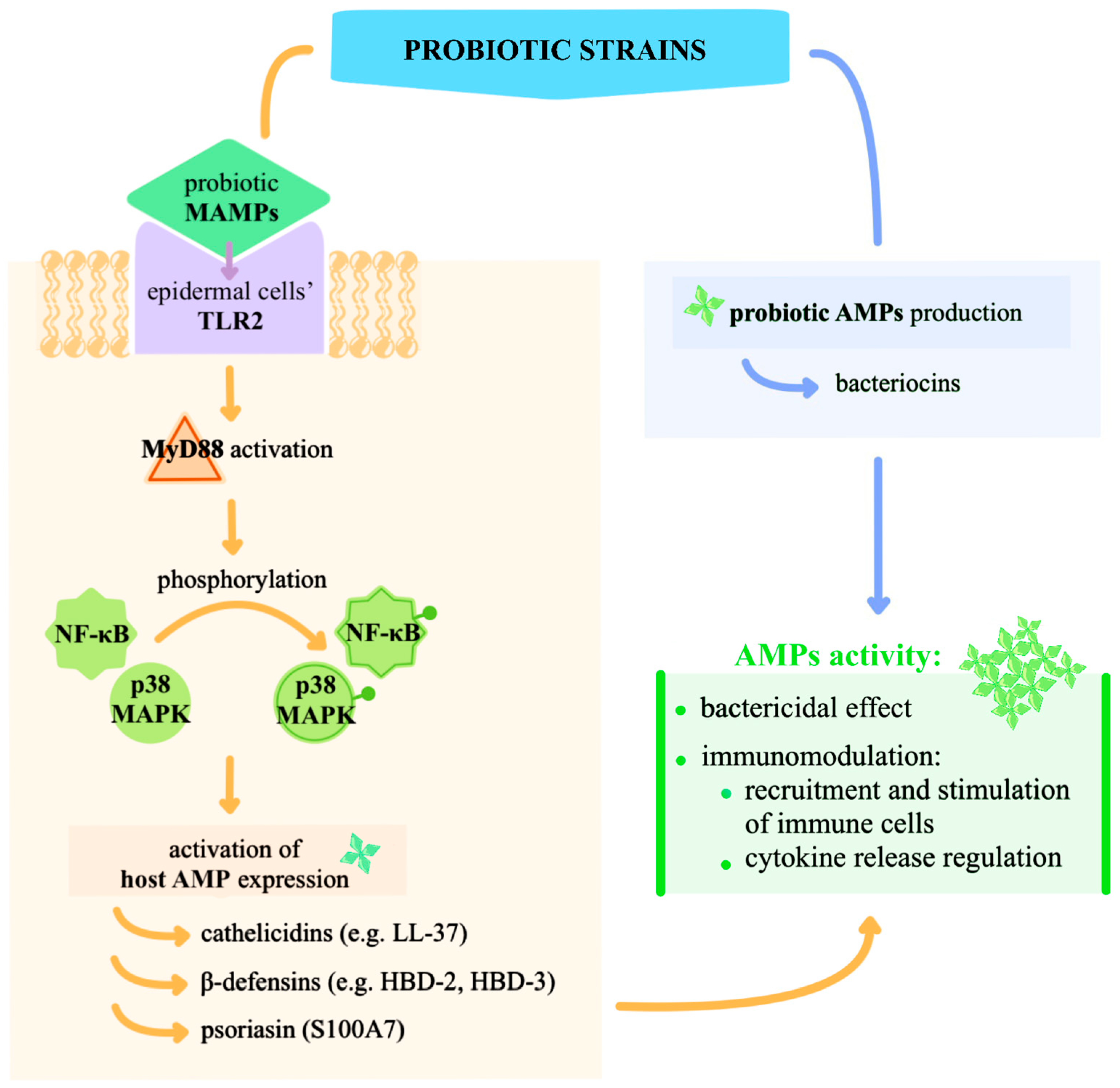

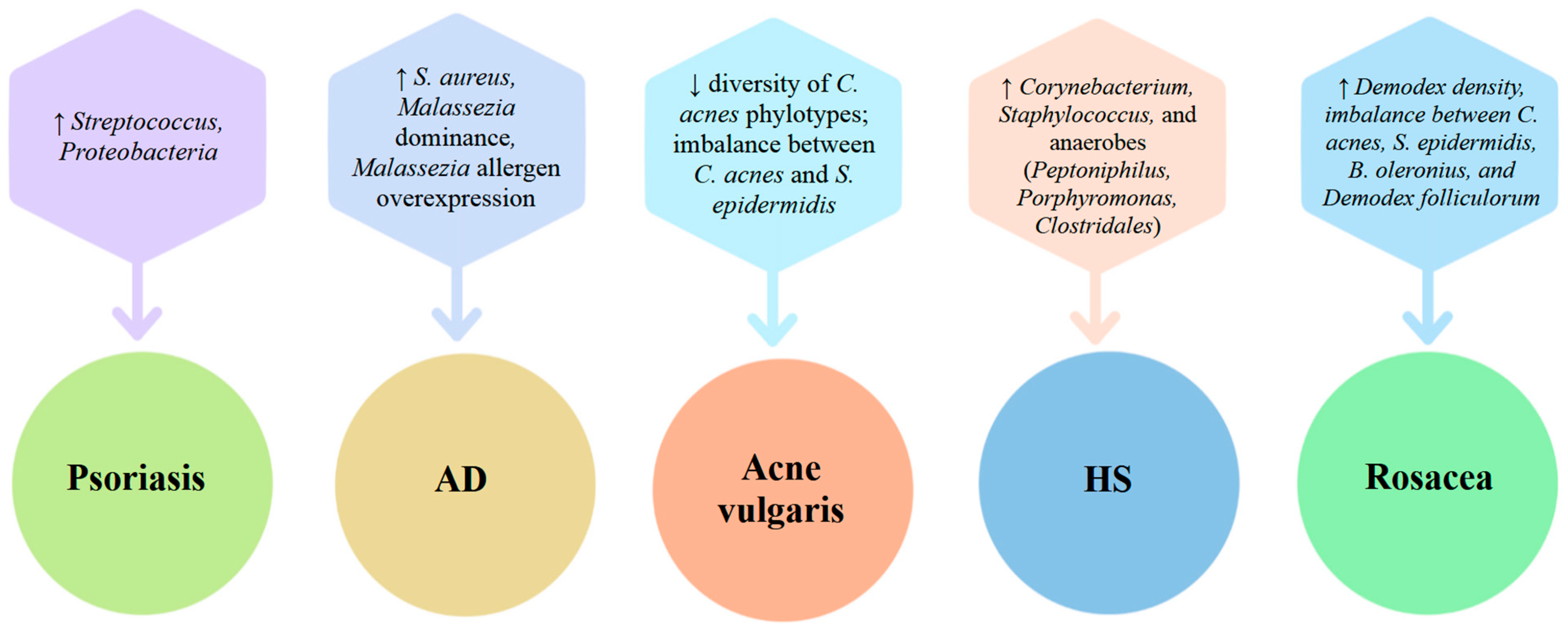
| Skin Site Type | Physiological Features | Representative Anatomical Locations | Dominant Bacterial Taxa |
|---|---|---|---|
| Sebaceous Sites [3,6,23,24] | Low moisture, high lipid content; acidic due to free fatty acids | Glabella, alar crease, external auditory canal, back, upper chest, face | Cutibacterium (Propionibacteriaceae), Staphylococcaceae, Corynebacteriaceae |
| Occiput | Staphylococcaceae, Corynebacteriaceae, Proteobacteria | ||
| Moist Sites [3,6,25] | High humidity and temperature; presence of glands and folds | Axillary vault, antecubital fossa, popliteal fossa, plantar heel | Proteobacteria, Staphylococcaceae, Bacteroidetes |
| Inguinal crease, umbilicus, gluteal crease | Corynebacteriaceae, Staphylococcaceae | ||
| Umbilicus | Corynebacteriaceae | ||
| Toe web space | Corynebacteriaceae, Staphylococcaceae, Cyanobacteria | ||
| Dry Sites [3,6,19] | Lower humidity; high microbial diversity but low temporal stability | Volar forearm, hypothenar palm, interdigital web space, plantar heel | Proteobacteria, Streptococcaceae, Actinobacteria (various), Bacteroidetes |
| Commensal Species | Cytokine Induction | Immune Cell Activation & Localization |
|---|---|---|
| Staphylococcus epidermidis | ↑ IL-17A | Induces Th17 and CD8+ T cells localized in the epidermis; CD8+ T cells produce IL-17A/IFN-γ and enhance barrier immunity |
| Cutibacterium acnes, Staphylococcus aureus | ↑ IL-17A, ↑ IFN-γ | Expand skin-resident IL-17A+ and IFN-γ T cells, but no CD8+ T cells response comparable to Staphylococcus epidermidis |
| Author (Year) | Study Group | Sample Type | Origin of Skin Samples | Bacteria/Fungi Alterations in AD |
|---|---|---|---|---|
| Zhang et al., 2011 (study about skin fungal microbiota) [73] | 9 patients with AD (3 each with mild, moderate and severe disease) | Scale samples collected using 7 cm × 9 cm OpSite strips (Smith & Nephew, Hull, UK) (Sugita method); each site sampled three times. | Scale samples collected from facial lesional sites (patients) and non-lesional skin (controls) | Malassezia—Mild/moderate AD: M. restricta > M. globosa. Severe AD: ratio M. restricta: M. globosa ≈ 1. Non-Malassezia yeasts: more diverse in AD (13.0 ± 3.0 spp.) vs. healthy (8.0 ± 1.9 spp.). |
| Fyhrquist et al., 2019 [58] | AD (n = 91); Controls (n = 126) | Skin samples collected using a sterile 2.5 cm ring filled with 1.5 mL PBS; skin scraped with glass rod (10× left, 10× left), no prior cleaning. | Skin samples collected from upper/lower back, posterior thigh, or buttocks | ↑ S. aureus (not in all lesions → possible endotypes) Loss of anaerobes (e.g., Lactobacillus, Finegoldia) → switch to aerobic metabolism S. aureus negatively correlates with S. epidermidis and Corynebacterium spp. |
| Edslev et al., 2021 (Staphylococcus comparison) [70] | AD (n = 94); Controls (n = 92) | Skin swab prepared using eSwabs (Copan, Brescia, Italy) | AD: lesional and non-lesional samples collected from the volar forearm and the cubital crease; Control: the antecubital crease | Severity of AD was associated with alterations in the Staphylococcus community. ↑ S. aureus, S. capitis, S. lugdunensis → directly correlated with disease severity. ↓ S. hominis → inversely correlated with disease severity; reduced abundance compared with healthy skin. |
| Suwarsa et al., 2021 [81] | AD (n = 12), 9 with mild disease and 3 moderate disease; Controls (n = 4) | Skin swab, sterile, pre-moistened swab rubbed for 20 s | Samples collected from volar forearm (cubital fossa) | Moderate AD—Dominance of Firmicutes, Bacilli, Bacillales Staphylococcaceae, Staphylococcus; highest abundance of S. aureus; reduced microbial diversity. Mild AD—Dominance of Proteobacteria, Gammaproteobacteria, Pseudomonadales, Moraxellaceae, Acinetobacter. |
| Schmid et al., 2022 [82] | AD (n = 16); Controls (n = 16) | Skin swab—flocked swabs (Floqswabs/eSwabs, COPAN, Brescia, Italy) pre-soaked in 0.9% NaCl (0.9%, Braun, Sempach, Switzerland); rubbed repeatedly over 4–8 cm2 of skin. | Skin swabs collected from antecubital crease, dorsal neck, glabella and vertex | ↑ S. aureus, ↓ Cutibacterium spp. Severe AD: Malassezia predominant, but ↑ non-Malassezia fungi (e.g., Candida, Debaryomyces); ↓ M. restricta, ↓ M. sympodialis, ↑ M. furfur compared to healthy individuals and mild-to-moderate AD. |
| Kim et al., 2024 [83] | AD (n = 20); Controls (n = 16) | Skin samples—swabs (TransportsystemTM 108C; Copan Diagnostics Inc., Murrieta, CA, USA) and tape strips (Cuderm Corporation, Dallas, TX, USA) | AD: lesional scalp and non-lesional scalp (at least 4 cm from the lesional skin) | ↑ Staphylococcus spp. and Kocuria spp. ↓ Cutibacterium and Lawsonella |
| Author (Year) | Study Group | Sample Type | Origin of Skin Samples | Bacteria Increased | Bacteria Decreased |
|---|---|---|---|---|---|
| Gao et al., 2008 [93] | Psoriasis patients (n = 6) | Skin biopsy: - Unaffected skin: 1 sample - Psoriatic lesions: ≥2 samples | Forearm/finger/elbow/shoulder/back/abdomen/leg/knee/arm | Firmicutes, Streptococcus | Actinobacteria, Propionibacterium, Proteobacteria |
| Fahlen et al., 2012 [94] | Psoriasis (n = 10), Controls (n = 12) | Skin biopsy: - Psoriasis: 2 mm biopsies from plaques - Control: 2 × 2 mm biopsies from excised lesions | Psoriasis: 4 trunk (3 back, 1 flank), 6 limbs (3 arm, 3 leg)Control: 8 trunk (6 back, 1 abdomen, 1 chest), 4 limbs (3 arms, 1 leg), 1 neck | Proteobacteria, Streptococcus | Propionibacteria, Staphylococcus |
| Alekseyenko et al., 2013 [95] | Psoriasis (n = 54), Controls (n = 37) | Skin swab (2 × 2 cm area, cotton pledget soaked in 0.15 M NaCl + 0.1% Tween 20): - Psoriasis: Lesion (plaque) and unaffected (contralateral) - Control | Psoriasis: face/scalp/back/abdomen/shoulder/arm/elbow/forearm/leg/thigh/knee/shin/foot/Control: 4 standardized sites per person (scalp, abdomen, inner elbow, kneecap) | Corynebacterium, Propionibacterium, Staphylococcus, Streptococcus | Cupriavidus, Flavisolibacter, Methylobacterium, Schlegelella |
| Drago et al., 2016 [96] | Psoriasis (n = 1), Controls (n = 1) | Skin biopsy (2 cm2 via curettage): - Psoriasis: 2 lesional and 2 non-lesional samples - Control: 2 samples | Psoriasis and control: the area behind the left ear | Proteobacteria, Bacteroidetes, Streptococcus, Rhodobacteraceae, Campylobacteraceae, and Moraxellaceae | Staphylococcus, Propionibacteriaceae |
| Tett et al., 2017 [97] | Psoriasis (n = 28) | Skin swab (based on the protocol validated and adopted by the HMPC, sterile cotton-tipped swabs (VWR, Milan, Italy) were moistened with SCF-1 buffer * | Psoriasis: the olecranon skin area and the retroauricular crease (behind the ear) from left and left body site | Staphylococcus, Novel/uncultured taxa (Anaerococcus spp., related Chromobacteriaceae/Neisseriaceae, novel Malassezia) | Overall microbial diversity ↓ |
| Chang et al., 2018 [98] | Psoriasis (n = 28), Controls (n = 26) | Skin swab (individually packed, sterile Epicentre Catch-All swabs): - Psoriasis: lesional + non-lesional samples - Control | Control and psoriasis non-lesional: 6 standardized sites per person (scalp, trunk, axilla, arm, leg, gluteal fold) Psoriasis lesional: only from sites with visible plaques among the 6 | Staphylococcus aureus, Proteobacteria | Staphylococcus epidermidis, Cutibacterium acnes, Actinobacter |
| Assarsson et al., 2018 [99] | Psoriasis (n = 26) | Skin swab (4 × 4 cm area, flocked swab soaked in 1 mL liquid Amies [ESwab™, Copan, Brescia, Italy]): Psoriasis - Lesional: target plaque - Non-lesional: adjacent site ≥ 10 cm from lesion | All samples from dry micro-environments | Firmicutes | Staphylococcus |
| Stehlikova et al., 2019 [100] | Psoriasis (n = 34), Controls (n = 25) | Skin swab (2 × 2 cm, FLOQSwabs™ COPAN Diagnostics Inc., United States, SCF-1 buffer *); Skin scraping (2 × 2 cm, scalpel, SCF-1 buffer *); Skin biopsy (2 mm punch, dry stored);—Psoriasis: lesional + non-lesional samples - Control | Control and psoriasis non-lesional: samples from dorsal (back) or olecranon (elbow) skin areas | Brevibacterium, Kocuria palustris, Gordonia | Staphylococcus; Propionibacterium compared to healthy skin on elbow |
| Assarsson et al., 2020 [86] | Psoriasis (n = 39), Controls (n = 70) | Skin swab (4 × 4 cm; using a flocked swab pre-moistened with 1 mL of liquid Amies medium (ESwab™, Copan Diagnostics Inc., Murrieta, CA, USA) | Control: pharynx and elbow skin; Psorasis: pharynx, lesional skin of elbow, adjacent non-lesional skin (≥10 cm from lesion) | Corynebacterium, 4 genera correlated with severity—Capnocytophaga, Leptotrichia, Abiotrophia and Tanne-rella | Streptococcus gordonii, Cutibacterium, Prevotella |
| Kayıran et al., 2022 [101] | Psoriasis (n = 10), Controls (n = 10) | Skin swab (rubbing swabs soaked sterile in DNA/RNA Shield™, Zymo Research, Irvine, CA, USA | Control: scalp Psoriasis: lesional and non-lesional hairy scalp | Staphylococcus, Streptococcus, Aquabacterium, Neisseria, Azospirillum, Mycobacterium, Finegoldia, Haemophilus, Ezakiella | Propionibacterium |
| Metabolite Class | Producing Strain Examples | Mechanisms of Action and Demonstrated Effects | Experimental Model In Vitro | Experimental Model In Vivo | Reference Number |
|---|---|---|---|---|---|
| LTA | Lactobacillus plantarum K8 | Inhibits MMP-1, suppresses ERK/JNK/AP-1/NF-κB, reduces ROS, increases type I procollagen | UVB-irradiated human dermal fibroblasts | - | [140] |
| Organic acids (lactic, acetic) | Lactobacillus plantarum, L. fermentum | Lowers skin pH, inhibits S. aureus and C. acnes, reduces oxidative stress, suppresses inflammatory mediators, promotes barrier function | HaCaT keratinocytes | UVB-stressed mouse skin | [118] |
| Plantaricins (bacteriocins) | Lactiplantibacillus plantarum (APsulloc 331261/266) | Suppresses S. aureus, C. acnes, Malassezia spp.; inhibits biofilm formation, destabilizes membrane integrity | Agar diffusion, co-culture, gene profiling | - | [112] |
| Peptidoglycan fragments | L. plantarum-GMNL6 | Stimulates collagen synthesis, upregulates SPTSSA, inhibits C. acnes and S. aureus biofilms | Skin models | observational clinical data in humans | [131] |
| Lipoteichoic acid & SCFAs | L. plantarum, L. casei | Enhance tight junction proteins, reduce TEWL, modulate cutaneous immune signaling via TLR2/NF-κB | Keratinocyte models | mouse skin assays | [128] |
| EPS | Lactobacillus casei, L. rhamnosus | Antioxidant, improves moisture retention, enhances barrier regeneration | Topical gel formulations | clinical skin hydration evaluations | [141,142] |
| Biosurfactants | L. plantarum, L. jensenii | Prevent adhesion of S. aureus, reduce biofilm persistence, modulate surface tension | Surface adhesion tests, microplate assays | - | [143] |
| Disease/Indication | Key Microbiome Alterations in Pathogenesis | Key Mechanism(s) of Topical Probiotics | Effects on Disease Course/Treatment |
|---|---|---|---|
| Psoriasis | ↓ microbial diversity; ↑ Streptococcus and other Firmicutes; variable Proteobacteria enrichment (strain-level differences); ↓ commensal Actinobacteria (Cutibacterium, S. epidermidis) * [58,86,90,93,94,95,96,98,99,104] | Immune modulation: dampening IL-1β/TNFα cascade; restoring barrier-microbiome balance; inhibition and enhancement of gene expression [147,148] | Reduced lesion inflammation; reduced TEWL, improved barrier; potentially decreased need for topical steroids [148,149] |
| Atopic Dermatitis (AD) | ↓ microbial diversity; overgrowth of S. aureus and opportunistic Staphylococci; suppression of S. epidermidis and Corynebacterium; fungal dysbiosis (Malassezia dominance, Malassezia allergen overexpression) [5,57,58,72,73,74,75] | Recolonization with commensals (e.g., S. epidermidis, Vitreoscilla filiformis, Roseomonas mucosa): S. aureus inhibition; upregulates AMPs (cathelicidin); modulates TLR2-mediated innate immune responses; reduces integrin-mediated infiltration, increases TNFAIP3/A20 expression [126,127]. | Improvement in eczema severity; decreased S. aureus colonization; itch reduction and barrier restoration, enhancement in cutaneous homeostasis [114,115] |
| Acne vulgaris | ↓ microbial diversity; ↓ diversity of C. acnes phylotypes; imbalance between C. acnes and S. epidermidis [4,155] | Strain-specific inhibition of C. acnes by succinic acid/fermentation by S. epidermidis or L. plantarum; anti-inflammatory IL-8 modulation; boosting AMP expression, destabilizing MRSA biofilms [125,150]. | Reduction in pustules/inflammatory lesions; reduced bacterial load; diminished IL-8 and cytokine-driven inflammation [150,151,152] |
| Hidradenitis suppurativa (HS) | Dysbiosis with overgrowth of Corynebacterium, Staphylococcus, and anaerobes (Peptoniphilus, Porphyromonas, Clostridales);↓ commensals (e.g., Cutibacterium);↓ niche heterogeneity [161] | Competitive exclusion of pathogenic flora; SCFA-mediated suppression of inflammation; AMP induction [153,154] | Potential reduction in abscess formation and inflammation; microbiome normalization; improved wound healing [153,154] |
| Rosacea | Increased Demodex density; ↑ TLR2 overexpression; ↑ AMPs (e.g., cathelicidins); dysbiosis of cutaneus microbiota (imbalance between C. acnes, S. epidermidis, B. oleronius, and Demodex folliculorum) [156,157] | Downregulation of TLR2–NF-κB–IL-8 pathway; reduction in LL-37 and ROS [144] | Reduced erythema, papules, and sensitivity; restoration of immune balance and microbial diversity [144,149] |
| Photoaging/Skin Aging | UV exposure alters skin microbiome diversity and metabolic capacity; decreased antioxidant pathways, increased MMP activity [159,160] | Reduction in oxidative stress, inhibition of MMP and AP-1/NF-κB pathways; transcriptional suppression, immune homeostasis and collagen synthesis support [118]. | Reduced wrinkle formation, improved elasticity and hydration; prevention of UV-induced ECM degradation [119,120]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowicka, D.; Kucharczyk, E.; Pawłuszkiewicz, K.; Korgiel, M.; Busłowicz, T.; Ponikowska, M. Topical Probiotics as a Novel Approach in the Treatment of Chronic Dermatoses Associated with Skin Dysbiosis: A Narrative Review. Int. J. Mol. Sci. 2025, 26, 10195. https://doi.org/10.3390/ijms262010195
Nowicka D, Kucharczyk E, Pawłuszkiewicz K, Korgiel M, Busłowicz T, Ponikowska M. Topical Probiotics as a Novel Approach in the Treatment of Chronic Dermatoses Associated with Skin Dysbiosis: A Narrative Review. International Journal of Molecular Sciences. 2025; 26(20):10195. https://doi.org/10.3390/ijms262010195
Chicago/Turabian StyleNowicka, Danuta, Emilia Kucharczyk, Karolina Pawłuszkiewicz, Matylda Korgiel, Tomasz Busłowicz, and Małgorzata Ponikowska. 2025. "Topical Probiotics as a Novel Approach in the Treatment of Chronic Dermatoses Associated with Skin Dysbiosis: A Narrative Review" International Journal of Molecular Sciences 26, no. 20: 10195. https://doi.org/10.3390/ijms262010195
APA StyleNowicka, D., Kucharczyk, E., Pawłuszkiewicz, K., Korgiel, M., Busłowicz, T., & Ponikowska, M. (2025). Topical Probiotics as a Novel Approach in the Treatment of Chronic Dermatoses Associated with Skin Dysbiosis: A Narrative Review. International Journal of Molecular Sciences, 26(20), 10195. https://doi.org/10.3390/ijms262010195






