Enhancing Local Functional Structure Features to Improve Drug–Target Interaction Prediction
Abstract
1. Introduction
2. Results and Discussion
2.1. Performance Evaluation
2.2. Ablation Studies
2.3. Case Studies
2.3.1. DTI Prediction as a Guide for Molecular Simulation
2.3.2. Interpretable Prediction of Functional Structures
3. Materials and Methods
3.1. Evaluation Metrics and Implementation
3.2. Datasets
3.3. Method
3.3.1. Structure-Enhanced Drug Feature Encoder
3.3.2. Structure-Enhanced Protein Feature Encoder
3.3.3. Gated Cross-Attention Module
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ciccotti, G.; Dellago, C.; Ferrario, M.; Hernández, E.R.; Tuckerman, M.E. Molecular Simulations: Past, Present, and Future (a Topical Issue in EPJB). Eur. Phys. J. B 2022, 95, 3, Correction to Eur. Phys. J. B 2022, 95, 17. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, J.; Meenu, M.; Xu, B. Anti-Obesity Mechanisms Elucidation of Essential Oil Components from Artemisiae Argyi Folium (Aiye) by the Integration of GC-MS, Network Pharmacology, and Molecular Docking. Int. J. Food Prop. 2025, 28, 2452445. [Google Scholar] [CrossRef]
- Yu, Y.; Xu, S.; He, R.; Liang, G. Application of molecular simulation methods in food science: Status and prospects. J. Agric. Food Chem. 2023, 71, 2684–2703. [Google Scholar] [CrossRef]
- Son, A.; Kim, W.; Park, J.; Lee, W.; Lee, Y.; Choi, S.; Kim, H. Utilizing Molecular Dynamics Simulations, Machine Learning, Cryo-EM, and NMR Spectroscopy to Predict and Validate Protein Dynamics. Int. J. Mol. Sci. 2024, 25, 9725. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Wei, Z. Molecular Simulation for Food Protein–Ligand Interactions: A Comprehensive Review on Principles, Current Applications, and Emerging Trends. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13280. [Google Scholar] [CrossRef] [PubMed]
- Müller, K.-R.; Mika, S.; Tsuda, K.; Schölkopf, K. An Introduction to Kernel-Based Learning Algorithms. In Handbook of Neural Network Signal Processing, 1st ed.; CRC Press: Boca Raton, FL, USA, 2002; ISBN 978-1-315-22041-3. [Google Scholar]
- Prašnikar, E.; Ljubič, M.; Perdih, A.; Borišek, J. Machine Learning Heralding a New Development Phase in Molecular Dynamics Simulations. Artif. Intell. Rev. 2024, 57, 102. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Pahikkala, T.; Airola, A.; Pietilä, S.; Shakyawar, S.; Szwajda, A.; Tang, J.; Aittokallio, T. Toward More Realistic Drug-Target Interaction Predictions. Brief. Bioinform. 2015, 16, 325–337. [Google Scholar] [CrossRef]
- Noé, F.; Tkatchenko, A.; Müller, K.-R.; Clementi, C. Machine Learning for Molecular Simulation. Annu. Rev. Phys. Chem. 2020, 71, 361–390. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, M. Navigating the Landscape of Enzyme Design: From Molecular Simulations to Machine Learning. Chem. Soc. Rev. 2024, 53, 8202–8239. [Google Scholar] [CrossRef]
- Askr, H.; Elgeldawi, E.; Aboul Ella, H.; Elshaier, Y.A.M.M.; Gomaa, M.M.; Hassanien, A.E. Deep Learning in Drug Discovery: An Integrative Review and Future Challenges. Artif. Intell. Rev. 2023, 56, 5975–6037. [Google Scholar] [CrossRef]
- Huang, K.; Xiao, C.; Glass, L.M.; Sun, J. MolTrans: Molecular Interaction Transformer for Drug–Target Interaction Prediction. Bioinformatics 2021, 37, 830–836. [Google Scholar] [CrossRef] [PubMed]
- Bian, J.; Zhang, X.; Zhang, X.; Xu, D.; Wang, G. MCANet: Shared-Weight-Based MultiheadCrossAttention Network for Drug–Target Interaction Prediction. Brief. Bioinform. 2023, 24, bbad082. [Google Scholar] [CrossRef]
- Bai, P.; Miljković, F.; John, B.; Lu, H. Interpretable Bilinear Attention Network with Domain Adaptation Improves Drug–Target Prediction. Nat. Mach. Intell. 2023, 5, 126–136. [Google Scholar] [CrossRef]
- Feng, B.-M.; Zhang, Y.-Y.; Niu, N.-W.-J.; Zheng, H.-Y.; Wang, J.-L.; Feng, W.-F. DeFuseDTI: Interpretable Drug Target Interaction Prediction Model with Dual-Branch Encoder and Multiview Fusion. Future Gener. Comput. Syst. 2024, 161, 239–247. [Google Scholar] [CrossRef]
- Ning, Q.; Wang, Y.; Zhao, Y.; Sun, J.; Jiang, L.; Wang, K.; Yin, M. DMHGNN: Double Multi-View Heterogeneous Graph Neural Network Framework for Drug-Target Interaction Prediction. Artif. Intell. Med. 2025, 159, 103023. [Google Scholar] [CrossRef]
- Li, J.; Bi, X.; Ma, W.; Jiang, H.; Liu, S.; Lu, Y.; Wei, Z.; Zhang, S. MHAN-DTA: A Multiscale Hybrid Attention Network for Drug-Target Affinity Prediction. IEEE J. Biomed. Health Inform. 2024, 1–12. [Google Scholar] [CrossRef]
- Liang, X.; Lai, G.; Yu, J.; Lin, T.; Wang, C.; Wang, W. Herbal Ingredient-Target Interaction Prediction via Multi-Modal Learning. Inf. Sci. 2025, 711, 122115. [Google Scholar] [CrossRef]
- Feng, B.-M.; Zhang, Y.-Y.; Zhou, X.-C.; Wang, J.-L.; Feng, Y.-F. MolLoG: A Molecular Level Interpretability Model Bridging Local to Global for Predicting Drug Target Interactions. J. Chem. Inf. Model. 2024, 64, 4348–4358. [Google Scholar]
- Liu, Y.; Qiu, L.; Li, A.; Fei, R.; Li, J.; Wu, F.-X. Prediction of miRNA Family Based on Class-Incremental Learning. In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024; pp. 197–200. [Google Scholar]
- Gao, Y.; Li, F.; Meng, F.; Ge, D.; Ren, Q.; Shang, J. Spatial Domains Identification Based on Multi-View Contrastive Learning in Spatial Transcriptomics. In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024; pp. 521–527. [Google Scholar]
- Hu, H.; Wang, X.; Zhang, Y.; Chen, Q.; Guan, Q. A Comprehensive Survey on Contrastive Learning. Neurocomputing 2024, 610, 128645. [Google Scholar] [CrossRef]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How Powerful Are Graph Neural Networks? arXiv 2019. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015. [Google Scholar] [CrossRef]
- Lee, I.; Keum, J.; Nam, H. DeepConv-DTI: Prediction of Drug-Target Interactions via Deep Learning with Convolution on Protein Sequences. PLoS Comput. Biol. 2019, 15, e1007129. [Google Scholar] [CrossRef]
- Nguyen, T.; Le, H.; Quinn, T.P.; Nguyen, T.; Le, T.D.; Venkatesh, S. GraphDTA: Predicting Drug–Target Binding Affinity with Graph Neural Networks. Bioinformatics 2021, 37, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, H.; Cui, P.; Li, S.; Wang, H.; Wu, Z. NFSA-DTI: A Novel Drug–Target Interaction Prediction Model Using Neural Fingerprint and Self-Attention Mechanism. Int. J. Mol. Sci. 2024, 25, 11818. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Q.; Zhang, C.; Feng, B.; Shang, J.; Zhang, L. IHDFN-DTI: Interpretable Hybrid Deep Feature Fusion Network for Drug–Target Interaction Prediction. Interdiscip. Sci. Comput. Life Sci. 2025, 1–15. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2017. [Google Scholar] [CrossRef]
- Randeni, N.; Luo, J.; Wu, Y.; Xu, B. Elucidating the Anti-Diabetic Mechanisms of Mushroom Chaga (Inonotus obliquus) by Integrating LC-MS, Network Pharmacology, Molecular Docking, and Bioinformatics. Int. J. Mol. Sci. 2025, 26, 5202. [Google Scholar] [CrossRef]
- Davis, L.L.; Behl, S.; Lee, D.; Zeng, H.; Skubiak, T.; Weaver, S.; Hefting, N.; Larsen, K.G.; Hobart, M. Brexpiprazole and Sertraline Combination Treatment in Posttraumatic Stress Disorder: A Phase 3 Randomized Clinical Trial. JAMA Psychiatry 2025, 82, 218–227. [Google Scholar] [CrossRef]
- Halavaty, A.S.; Kim, Y.; Minasov, G.; Shuvalova, L.; Dubrovska, I.; Winsor, J.; Zhou, M.; Onopriyenko, O.; Skarina, T.; Papazisi, L.; et al. Structural Characterization and Comparison of Three Acyl-Carrier-Protein Synthases from Pathogenic Bacteria. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Burley, S.K.; Berman, H.M.; Kleywegt, G.J.; Markley, J.L.; Nakamura, H.; Velankar, S. Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography: Methods and Protocols; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Springer: New York, NY, USA, 2017; pp. 627–641. ISBN 978-1-4939-7000-1. [Google Scholar]
- Li, J.; Zhai, X.; Liu, J.; Lam, C.K.; Meng, W.; Wang, Y.; Li, S.; Wang, Y.; Li, K. Integrated Causal Inference Modeling Uncovers Novel Causal Factors and Potential Therapeutic Targets of Qingjin Yiqi Granules for Chronic Fatigue Syndrome. Acupunct. Herb. Med. 2024, 4, 122. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Zhao, D.; Yu, X.; Shen, X.; Zhou, Y.; Wang, S.; Qiu, Y.; Chen, Y.; Zhu, F. TTD: Therapeutic Target Database Describing Target Druggability Information. Nucleic Acids Res. 2024, 52, D1465–D1477. [Google Scholar] [CrossRef] [PubMed]
- Ghanizadeh, A. Sertraline-Associated Hair Loss. J. Drugs Dermatol. 2008, 7, 693–694. [Google Scholar]
- Fishback, J.A.; Robson, M.J.; Xu, Y.-T.; Matsumoto, R.R. Sigma Receptors: Potential Targets for a New Class of Antidepressant Drug. Pharmacol. Ther. 2010, 127, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, L.; Andersen, J.; Thomsen, M.; Hansen, S.M.R.; Zhao, X.; Sandelin, A.; Strømgaard, K.; Kristensen, A.S. Interaction of Antidepressants with the Serotonin and Norepinephrine Transporters: Mutational Studies of the S1 Substrate Binding Pocket. J. Biol. Chem. 2012, 287, 43694–43707. [Google Scholar] [CrossRef]
- Daws, L.C. Unfaithful Neurotransmitter Transporters: Focus on Serotonin Uptake and Implications for Antidepressant Efficacy. Pharmacol. Ther. 2009, 121, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cui, J.; Xu, T.; Xu, Y.; Long, M.; Li, J.; Liu, M.; Yang, T.; Du, Y.; Xu, Q. Advances in the Preparation, Characterization, and Biological Functions of Chitosan Oligosaccharide Derivatives: A Review. Carbohydr. Polym. 2024, 332, 121914. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.A.; DeVeaux, A.L.; Juliano, B.R.; Photenhauer, A.L.; Boulinguiez, M.; Bornschein, R.E.; Wawrzak, Z.; Ruotolo, B.T.; Terrapon, N.; Koropatkin, N.M. BoGH13ASus from Bacteroides Ovatus Represents a Novel α-Amylase Used for Bacteroides Starch Breakdown in the Human Gut. Cell. Mol. Life Sci. 2023, 80, 232. [Google Scholar] [CrossRef]
- Nithin, C.; Kmiecik, S.; Błaszczyk, R.; Nowicka, J.; Tuszyńska, I. Comparative Analysis of RNA 3D Structure Prediction Methods: Towards Enhanced Modeling of RNA–Ligand Interactions. Nucleic Acids Res. 2024, 52, 7465–7486. [Google Scholar] [CrossRef]
- Ludwiczak, O.; Antczak, M.; Szachniuk, M. Assessing Interface Accuracy in Macromolecular Complexes. PLoS ONE 2025, 20, e0319917. [Google Scholar] [CrossRef] [PubMed]
- Bai, P.; Miljković, F.; Ge, Y.; Greene, N.; John, B.; Lu, H. Hierarchical Clustering Split for Low-Bias Evaluation of Drug-Target Interaction Prediction. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 641–644. [Google Scholar]
- Zitnik, M.; Agrawal, M.; Leskovec, J. Modeling Polypharmacy Side Effects with Graph Convolutional Networks. Bioinformatics 2018, 34, i457–i466. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sun, J.; Guan, J.; Zheng, J.; Zhou, S. Improving Compound–Protein Interaction Prediction by Building up Highly Credible Negative Samples. Bioinformatics 2015, 31, i221–i229. [Google Scholar] [CrossRef]
- Davis, M.I.; Hunt, J.P.; Herrgard, S.; Ciceri, P.; Wodicka, L.M.; Pallares, G.; Hocker, M.; Treiber, D.K.; Zarrinkar, P.P. Comprehensive Analysis of Kinase Inhibitor Selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]

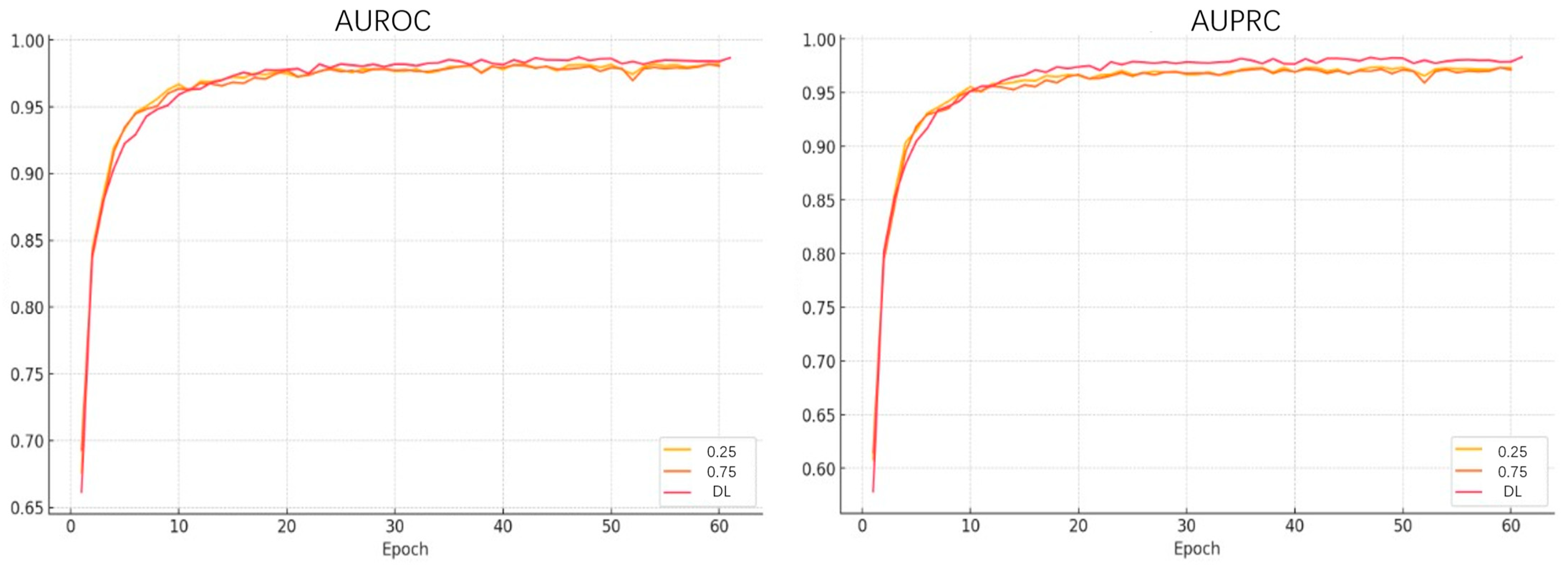


| Method | AUROC | AUPRC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| BindingDB | |||||
| DeepConv-DTI | 0.944 ± 0.004 | 0.925 ± 0.005 | 0.882 ± 0.007 | 0.873 ± 0.018 | 0.884 ± 0.009 |
| GraphDTA | 0.950 ± 0.003 | 0.934 ± 0.002 | 0.888 ± 0.005 | 0.882 ± 0.012 | 0.887 ± 0.008 |
| MolTrans | 0.952 ± 0.002 | 0.933 ± 0.004 | 0.887 ± 0.006 | 0.884 ± 0.019 | 0.883 ± 0.011 |
| DrugBAN | 0.956 ± 0.003 | 0.943 ± 0.003 | 0.897 ± 0.003 | 0.890 ± 0.015 | 0.896 ± 0.008 |
| NFSA-DTI | 0.951 ± 0.003 | 0.933 ± 0.004 | 0.883 ± 0.004 | 0.892 ± 0.008 | 0.908 ± 0.012 |
| IHDFN-DTI | 0.955 ± 0.002 | 0.939 ± 0.003 | 0.893 ± 0.003 | 0.884 ± 0.012 | 0.912 ± 0.009 |
| LoF-DTI | 0.963 ± 0.005 | 0.947 ± 0.005 | 0.902 ± 0.002 | 0.896 ± 0.015 | 0.918 ± 0.007 |
| BioSNAP | |||||
| DeepConv-DTI | 0.886 ± 0.006 | 0.890 ± 0.006 | 0.805 ± 0.009 | 0.760 ± 0.029 | 0.851 ± 0.011 |
| GraphDTA | 0.887 ± 0.008 | 0.890 ± 0.007 | 0.800 ± 0.007 | 0.745 ± 0.032 | 0.854 ± 0.025 |
| MolTrans | 0.890 ± 0.006 | 0.891 ± 0.005 | 0.804 ± 0.003 | 0.755 ± 0.021 | 0.846 ± 0.022 |
| DrugBAN | 0.903 ± 0.005 | 0.900 ± 0.004 | 0.836 ± 0.009 | 0.825 ± 0.018 | 0.849 ± 0.013 |
| NFSA-DTI | 0.897 ± 0.004 | 0.895 ± 0.008 | 0.832 ± 0.010 | 0.807 ± 0.015 | 0.844 ± 0.011 |
| IHDFN-DTI | 0.903 ± 0.005 | 0.908 ± 0.006 | 0.835 ± 0.007 | 0.815 ± 0.022 | 0.862 ± 0.008 |
| LoF-DTI | 0.905 ± 0.003 | 0.904 ± 0.002 | 0.841 ± 0.005 | 0.812 ± 0.020 | 0.872 ± 0.014 |
| DAVIS | |||||
| DeepConvDTI | 0.884 ± 0.008 | 0.299 ± 0.039 | 0.774 ± 0.012 | 0.754 ± 0.040 | 0.876 ± 0.013 |
| DeepDTA | 0.880 ± 0.007 | 0.301 ± 0.044 | 0.773 ± 0.010 | 0.765 ± 0.045 | 0.880 ± 0.024 |
| MolTrans | 0.892 ± 0.004 | 0.371 ± 0.031 | 0.779 ± 0.017 | 0.781 ± 0.023 | 0:878 ± 0.012 |
| DrugBAN | 0.892 ± 0.005 | 0.333 ± 0.039 | 0.770 ± 0.015 | 0.751 ± 0.024 | 0.869 ± 0.011 |
| NFSA-DTI | 0.884 ± 0.008 | 0.329 ± 0.028 | 0.774 ± 0.012 | 0.754 ± 0.030 | 0.866 ± 0.013 |
| IHDFN-DTI | 0.876 ± 0.005 | 0.348 ± 0.032 | 0.778 ± 0.010 | 0.778 ± 0.013 | 0.874 ± 0.007 |
| LoF-DTI | 0.894 ± 0.005 | 0.354 ± 0.023 | 0.782 ± 0.015 | 0.782 ± 0.015 | 0.882 ± 0.005 |
| Human | |||||
| DeepConvDTI | 0.975 ± 0.002 | 0.969 ± 0.003 | 0.941 ± 0.002 | 0.915 ± 0.008 | 0.934 ± 0.015 |
| DeepDTA | 0.975 ± 0.002 | 0.969 ± 0.003 | 0.941 ± 0.002 | 0.915 ± 0.008 | 0.934 ± 0.015 |
| MolTrans | 0.973 ± 0.003 | 0.968 ± 0.003 | 0.943 ± 0.003 | 0.918 ± 0.007 | 0.936 ± 0.013 |
| DrugBAN | 0.981 ± 0.004 | 0.974 ± 0.006 | 0.938 ± 0.005 | 0.927 ± 0.011 | 0.938 ± 0.018 |
| NFSA-DTI | 0.980 ± 0.002 | 0.966 ± 0.005 | 0.943 ± 0.005 | 0.930 ± 0.007 | 0.947 ± 0.014 |
| IHDFN-DTI | 0.983 ± 0.004 | 0.980 ± 0.003 | 0.945 ± 0.002 | 0.938 ± 0.009 | 0.953 ± 0.006 |
| LoF-DTI | 0.985 ± 0.004 | 0.977 ± 0.002 | 0.948 ± 0.007 | 0.944 ± 0.012 | 0.953 ± 0.008 |
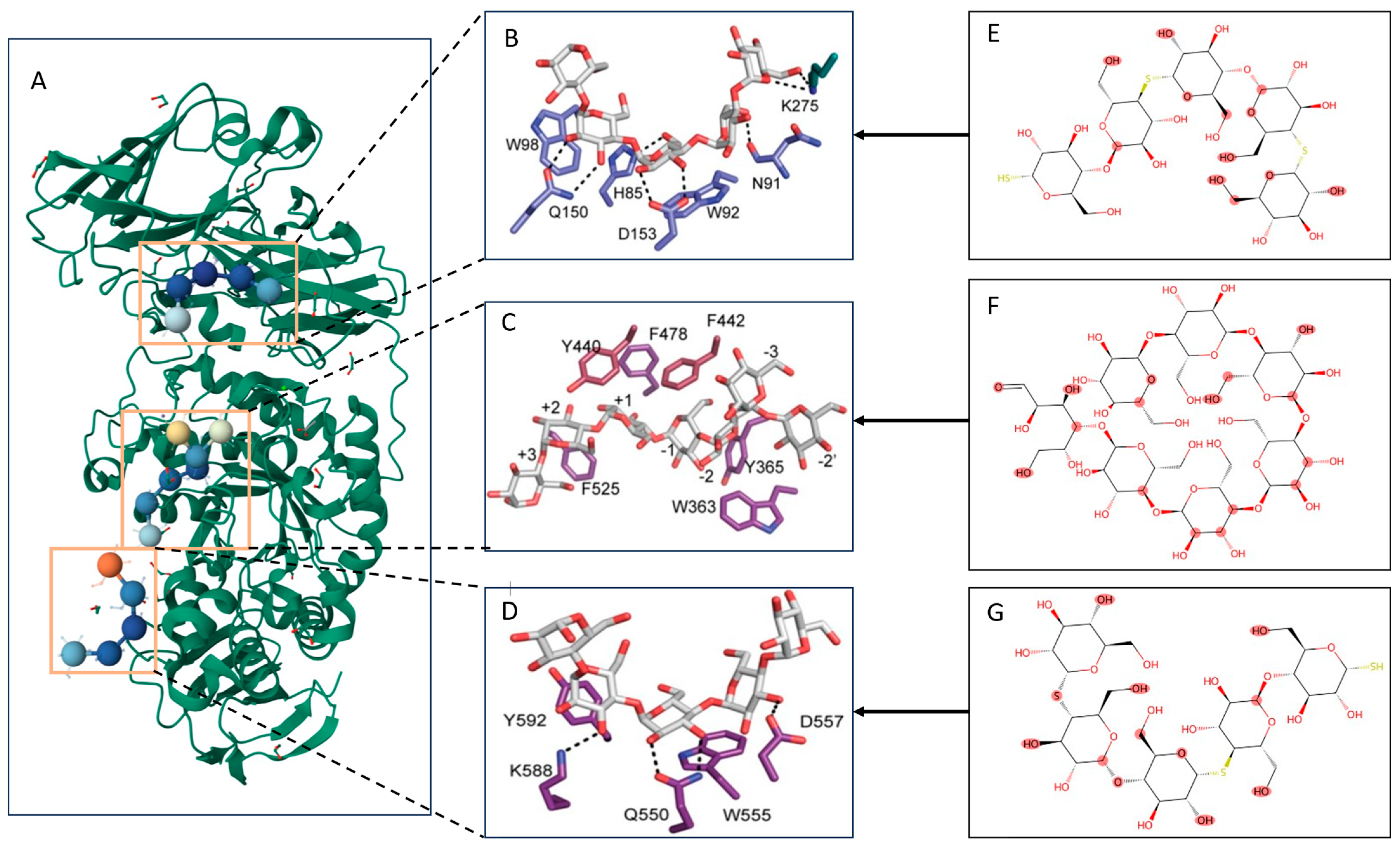
| Drug | Target | Prontein | Compound |
|---|---|---|---|
 | P31645 [37] |  | COA |
| Q01950 [38] | A3P | ||
| Q99720 [39] | MRD | ||
| P23975 [40] | MPD | ||
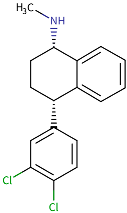
| Sertralin | P08684 [41] | 3QMN | ACT |
| Hyperparameter | Setting |
|---|---|
| Optimizer | Adam |
| Learning rate | 1 × 10−5 |
| MAX_Epoch | 100 |
| BATCH_SIZE | 64 |
| Number of residual blocks | 2 |
| GIN layers | 4 |
| CNN kernel size | [3, 6, 9] |
| Heads of attention | 4 |
| Attention pooling size | 3 |
| Dataset | #Drugs | #Proteins | #Interactions |
|---|---|---|---|
| BindingDB | 14,643 | 2623 | 49,200 |
| BioSNAP | 4510 | 2181 | 27,465 |
| Human | 2726 | 2001 | 6728 |
| DAVIS | 72 | 382 | 11,885 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, B.; Du, H.; Tong, H.H.Y.; Wang, X.; Li, K. Enhancing Local Functional Structure Features to Improve Drug–Target Interaction Prediction. Int. J. Mol. Sci. 2025, 26, 10194. https://doi.org/10.3390/ijms262010194
Feng B, Du H, Tong HHY, Wang X, Li K. Enhancing Local Functional Structure Features to Improve Drug–Target Interaction Prediction. International Journal of Molecular Sciences. 2025; 26(20):10194. https://doi.org/10.3390/ijms262010194
Chicago/Turabian StyleFeng, Baoming, Haofan Du, Henry H. Y. Tong, Xu Wang, and Kefeng Li. 2025. "Enhancing Local Functional Structure Features to Improve Drug–Target Interaction Prediction" International Journal of Molecular Sciences 26, no. 20: 10194. https://doi.org/10.3390/ijms262010194
APA StyleFeng, B., Du, H., Tong, H. H. Y., Wang, X., & Li, K. (2025). Enhancing Local Functional Structure Features to Improve Drug–Target Interaction Prediction. International Journal of Molecular Sciences, 26(20), 10194. https://doi.org/10.3390/ijms262010194








