Using Whole Exome Sequencing to Identify Genetic Causes of Neurodevelopmental Disorders in a Cohort of 11 Patients: A Single Center Experience
Abstract
1. Introduction
2. Results
2.1. MN37
2.2. MN90
2.3. MN107
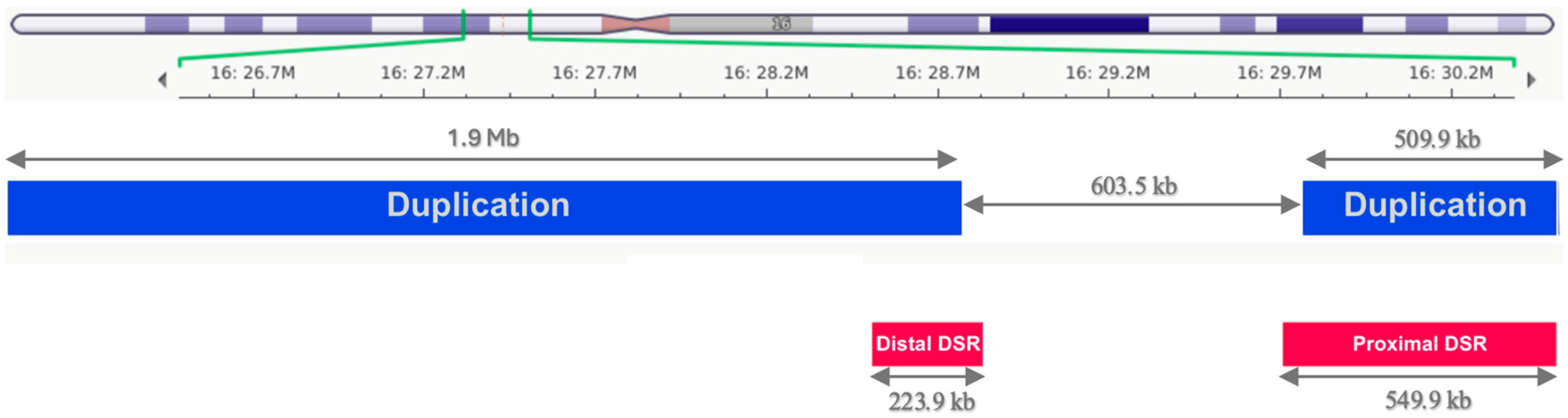
2.4. MN126
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Laboratory Methods
4.3. Bioinformatic Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NDD | Neurodevelopmental delay |
| WES | Whole exome sequencing |
| DSM-V | Diagnostic and Statistical Manual of Mental Disorders, 5th Edition |
| ASDs | autism spectrum disorders |
| GDD | global developmental delay |
| ID | intellectual disability |
| NHIS | national health insurance system |
| CNVs | copy number variations |
| MLPA | multiple ligation-dependent probe amplification |
| MM | Molecular Medicine |
| MUTH | Markusovszky University Teaching Hospital |
| NGC | National Genomic Center |
| SNV | single nucleotide variant |
| CDLS | Cornelia de Lange syndrome |
| ERN | The European Reference Network |
| ITHACA | Intellectual disability, TeleHealth, Autism, and Congenital Anomalies |
| CPG | clinical practice guideline |
| ADHD | Attention Deficit Hyperactivity Disorder |
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Publishing: Arlington, VA, USA, 2013. [Google Scholar]
- Postma, J.K.; Harrison, M.A.; Kutcher, S.; Webster, R.J.; Cloutier, M.; Bourque, D.K.; Yu, A.C.; Carter, M.T. The diagnostic yield of genetic and metabolic investigations in syndromic and nonsyndromic patients with autism spectrum disorder, global developmental delay, or intellectual disability from a dedicated neurodevelopmental disorders genetics clinic. Am. J. Med. Genet. Part A 2024, 194, e63791. [Google Scholar] [CrossRef] [PubMed]
- McRae, J.F.; Clayton, S.; Fitzgerald, T.W.; Kaplanis, J.; Prigmore, E.; Rajan, D.; Sifrim, A.; Aitken, S.; Akawi, N.; Alvi, M.; et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017, 542, 433–438. [Google Scholar]
- Fu, J.M.; Satterstrom, F.K.; Peng, M.; Brand, H.; Collins, R.L.; Dong, S.; Klei, L.; Stevens, C.R.; Cusick, C.; Babadi, M.; et al. Rare coding variation illuminates the allelic architecture, risk genes, cellular expression patterns and phenotypic context of autism. Nat. Genet. 2022, 54, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Dhindsa, R.S.; Weido, B.A.; Dhindsa, J.S.; Shetty, A.J.; Sands, C.F.; Petrovski, S.; Vitsios, D.; Zoghbi, A.W. Genome-wide prediction of dominant and recessive neurodevelopmental disorder-associated genes. Am. J. Hum. Genet. 2025, 112, 693–708. [Google Scholar] [CrossRef]
- Sandin, S.; Lichtenstein, P.; Kuja-Halkola, R.; Hultman, C.; Larsson, H.; Reichenberg, A. The heritability of autism spectrum disorder. JAMA 2017, 318, 1182–1184. [Google Scholar] [CrossRef]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of autism spectrum disorders: A meta-analysis of twin studies. J. Child. Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef]
- Reichenberg, A.; Cederlöf, M.; McMillan, A.; Trzaskowski, M.; Kapra, O.; Fruchter, E.; Ginat, K.; Davidson, M.; Weiser, M.; Larsson, H.; et al. Discontinuity in the genetic and environmental causes of the intellectual disability spectrum. Proc. Natl. Acad. Sci. USA 2016, 113, 1098–1103. [Google Scholar] [CrossRef]
- Savatt, J.M.; Myers, S.M. Genetic testing in neurodevelopmental disorders. Front. Pediatr. 2021, 9, 526779. [Google Scholar] [CrossRef] [PubMed]
- Brian, J.A.; Zwaigenbaum, L.; Ip, A. Standards of diagnostic assessment for autism spectrum disorder. Paediatr. Child Health 2019, 24, 444–460. [Google Scholar] [CrossRef]
- Carter, M.T.; Cloutier, M.; Tsampalieros, A.; Webster, R. Genetic and metabolic investigations for individuals with neurodevelopmental disorders: A survey of Canadian geneticists’ practices. Am. J. Med. Genet. A 2021, 185, 1757–1766. [Google Scholar] [CrossRef]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010, 86, 749–764. [Google Scholar] [CrossRef]
- Schaefer, G.B.; Mendelsohn, N.J. Professional Practice and Guidelines Committee. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet. Med. 2013, 15, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Tompa, M.; Urban, P.; Galik, B.; Gazdagh, G.; Kajtar, B.; Kiss, Z.; Gyenesei, A.; Kalman, B. A new diagnostic service for rare (neurologic) diseases in West-Hungary. Ideggyogy. Szle. 2025, 78, 9–10. [Google Scholar]
- Sinkó, G.; Dénes, E.; Farkas, G.; Tompa, M.; Kálmán, B. Globoid Cell Leukodystrophy (Krabbe Disease). Ideggyogy. Szle. 2025; in press. [Google Scholar]
- Nagy, Z.Á.; Bátor, G.; Vadvári, Á.; Tompa, M.; Tankó, M.L.; Sevcic, K.; Kálmán, B. Genetikai vizsgálattal igazolt Blau-szindróma ritka klinikai megjelenése. Orv. Hetil. 2025, 166, 623–630. [Google Scholar] [CrossRef]
- Bartalis, K.; Kálmán, B.; Kisely, M. Ectrodactyly-ectodermal dysplasia-clefting (EEC) syndrome. Case Rep. Orv. Hetil. 2023, 164, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.A.; Shen, Y.; Korn, J.M.; Arking, D.E.; Miller, D.T.; Fossdal, R.; Saemundsen, E.; Stefansson, H.; Ferreira, M.A.R.; Green, T.; et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008, 358, 667–675. [Google Scholar] [CrossRef]
- Rosenfeld, J.A.; Coppinger, J.; Bejjani, B.A.; Girirajan, S.; Eichler, E.E.; Shaffer, L.G.; Ballif, B.C. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J. Neurodev. Disord. 2010, 2, 26–38. [Google Scholar] [CrossRef]
- Cooper, G.M.; Coe, B.P.; Girirajan, S.; Rosenfeld, J.A.; Vu, T.H.; Baker, C.; Williams, C.; Stalker, H.; Hamid, R.; Hannig, V.; et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011, 43, 838–846, Correction in Nat. Genet. 2014, 46, 1040. [Google Scholar] [CrossRef]
- Bachmann-Gagescu, R.; Mefford, H.C.; Cowan, C.; Glew, G.M.; Hing, A.V.; Wallace, S.; Bader, P.I.; Hamati, A.; Reitnauer, P.J.; Smith, R.; et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet. Med. 2010, 12, 641–647. [Google Scholar] [CrossRef]
- Loviglio, M.N.; Leleu, M.; Männik, K.; Passeggeri, M.; Giannuzzi, G.; van der Werf, I.; Waszak, S.M.; Zazhytska, M.; Roberts-Caldeira, I.; Gheldof, N.; et al. Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Mol. Psychiatry 2017, 22, 836–849. [Google Scholar] [CrossRef]
- Sadler, B.; Haller, G.; Antunes, L.; Bledsoe, X.; Morcuende, J.; Giampietro, P.; Raggio, C.; Miller, N.; Kidane, Y.; Wise, C.A.; et al. Distal chromosome 16p11.2 duplications containing SH2B1 in patients with scoliosis. J. Med. Genet. 2019, 56, 427–433. [Google Scholar] [CrossRef]
- Parenti, I.; Diab, F.; Ruiz Gil, S.; Mulugeta, E.; Casa, V.; Berutti, R.; Brouwer, R.W.W.; Dupé, V.; Eckhold, J.; Graf, E.; et al. MAU2 and NIPBL variants impair the heterodimerization of the cohesin loader subunits and cause Cornelia de Lange syndrome. Cell Rep. 2020, 31, 107647. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.; Vissers, L.; de Vries, B.B.A. The genetics of intellectual disability. Brain Sci. 2023, 13, 231. [Google Scholar] [CrossRef]
- Haneveld, K.M.J.; Oerbekke, M.S.; Szakszon, K.; Cornel, M.C.; Gaasterland, C.M.W.; van Eeghen, A.M. Priority-setting criteria for clinical practice guideline development on rare genetic neurodevelopmental disorders: A Delphi study within the European Reference Network ITHACA. J. Clin. Epidemiol. 2025, 182, 111761. [Google Scholar] [CrossRef] [PubMed]
- Zablotsky, B.; Black, L.I.; Maenner, M.J.; Schieve, L.A.; Danielson, M.L.; Bitsko, R.H.; Blumberg, S.J.; Kogan, M.D.; Boyle, C.A. Prevalence and trends of developmental disabilities among children in the United States: 2009–2017. Pediatrics 2019, 144, e20190811. [Google Scholar] [CrossRef]
- Leblond, C.S.; Le, T.-L.; Malesys, S.; Cliquet, F.; Tabet, A.-C.; Delorme, R.; Rolland, T.; Bourgeron, T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol. Cell. Neurosci. 2021, 113, 103623. [Google Scholar] [CrossRef]
- Peng, X.; Jia, X.; Wang, H.; Chen, J.; Zhang, X.; Tan, S.; Duan, X.; Qiu, C.; Hu, M.; Hou, H.; et al. Disrupting Integrator complex subunit INTS6 causes neurodevelopmental disorders and impairs neurogenesis and synapse development. J. Clin. Investig. 2025, 18, e191729. [Google Scholar] [CrossRef]
- Bradbrook, S.M.; Graham, G.; Carter, M.T.; Kibaek, M.; Fagerberg, C.; Larsen, M.J.; Dawson, K.; Meuter, C.; Pepler, A.; Besnard, T.; et al. De novo truncating variants in ZNF865 cause a novel neurodevelopmental disorder. Am. J. Med. Genet. A 2025, 11, e64242. [Google Scholar] [CrossRef] [PubMed]
- Beyad, F.; Bonyadi, M.; Barzegar, M. Identification of a novel TRIT1 mutation in a consanguineous Iranian-Azeri-Turkish family with global developmental delay. Int. J. Dev. Neurosci. 2025, 85, e70053. [Google Scholar] [CrossRef]
- Archer, J.; Goh, S.; Miteff, C.; O’Donnell, S.; Park, K.; Goel, H. CRELD1-associated neurodevelopmental disorder: Three new individuals from unrelated families. Genes 2025, 16, 972. [Google Scholar] [CrossRef]
- Pruitt, A.; Gupta, A.R.; Hoffman, E.J. Molecular and genetic mechanisms in autism spectrum disorder. Ann. Neurol. 2025. Online ahead of print. [Google Scholar] [CrossRef]
- PerezGrovas-Saltijeral, A.; Stones, J.; Orji, O.C.; Shaker, H.; Knight, H.M. Modification of the RNA methylome in neurodevelopmental disorders. Curr. Opin. Genet. Dev. 2025, 92, 102330. [Google Scholar] [CrossRef]
- Jackson, A.; Thaker, N.; Blakes, A.; Rice, G.; Griffiths-Jones, S.; Balasubramanian, M.; Campbell, J.; Shannon, N.; Choi, J.; Hong, J.; et al. Analysis of R-loop forming regions identifies RNU2-2 and RNU5B-1 as neurodevelopmental disorder genes. Nat. Genet. 2025, 57, 1362–1366. [Google Scholar] [CrossRef]
- Holling, T.; von Kroge, S.; Hecher, L.; Amling, M.; Schinke, T.; Kutsche, K.; Oheim, R. Assessment and treatment of osteoporosis in a patient with a neurodevelopmental disorder caused by a RNU4-2 pathogenic variant (ReNU syndrome). JBMR Plus 2025, 9, ziaf084. [Google Scholar] [CrossRef]
- Garas, P.; Takacs, Z.K.; Balázs, J. Longitudinal suicide risk in children and adolescents with attention deficit and hyperactivity disorder: A systematic review and meta-analysis. Brain Behav. 2025, 15, e70618. [Google Scholar] [CrossRef] [PubMed]
- Ágrez, K.; Vakli, P.; Weiss, B.; Vidnyánszky, Z.; Bunford, N. Assessing the association between ADHD and brain maturation in late childhood and emotion regulation in early adolescence. Transl. Psychiatry 2025, 15, 185. [Google Scholar] [CrossRef] [PubMed]
- Zsigmond, A.; Till, Á.; Bene, J.; Czakó, M.; Mikó, A.; Hadzsiev, K. Case report of suspected gonadal mosaicism in FOXP1-related neurodevelopmental disorder. Int. J. Mol. Sci. 2024, 25, 5709. [Google Scholar] [CrossRef]
- Papp-Hertelendi, R.; Tényi, T.; Hadzsiev, K.; Hau, L.; Benyus, Z.; Csábi, G. First report on the association of SCN1A mutation, childhood schizophrenia and autism spectrum disorder without epilepsy. Psychiatry Res. 2018, 270, 1175–1176. [Google Scholar] [CrossRef]
- Levy, T.; Siper, P.M.; Lerman, B.; Halpern, D.; Zweifach, J.; Belani, P.; Thurm, A.; Kleefstra, T.; Berry-Kravis, E.; Buxbaum, J.D.; et al. DDX3X Syndrome: Summary of Findings and Recommendations for Evaluation and Care. Pediatr. Neurol. 2023, 138, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Devasahayam Arokia Balaya, R.; Kanekar, S.; Kumar, S.; Kandasamy, R.K. Role of DEAD/DEAH-box helicases in immunity, infection and cancers. Cell Commun. Signal. 2025, 23, 292. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef]
- Taschuk, F.; Cherry, S. DEAD-box helicases: Sensors, regulators, and effectors for antiviral defense. Viruses 2020, 12, 181. [Google Scholar] [CrossRef]
- Perčulija, V.; Ouyang, S. Diverse roles of DEAD/DEAH-box helicases in innate immunity and diseases. In Helicases from All Domains of Life; Academic Press: Cambridge, MA, USA, 2019; pp. 141–171. [Google Scholar] [CrossRef]
- Lennox, A.L.; Hoye, M.L.; Jiang, R.; Johnson-Kerner, B.L.; Suit, L.A.; Venkataramanan, S.; Sheehan, C.J.; Alsina, F.C.; Fregeau, B.; Aldinger, K.A.; et al. Pathogenic DDX3X mutations impair RNA metabolism and neurogenesis during fetal cortical development. Neuron 2020, 106, 404–420. [Google Scholar] [CrossRef]
- Snijders Blok, L.; Madsen, E.; Juusola, J.; Gilissen, C.; Baralle, D.; Reijnders, M.R.F.; Venselaar, H.; Helsmoortel, C.; Cho, M.T.; Hoischen, A.; et al. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 2015, 97, 343–352. [Google Scholar] [CrossRef]
- Tang, L.; Levy, T.; Guillory, S.; Halpern, D.; Zweifach, J.; Kiss, I.G.; Foss-Feig, J.H.; Frank, Y.; Lozano, R.; Belani, P.; et al. Prospective and detailed behavioral phenotyping in DDX3X syndrome. Mol. Autism 2021, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Mashayekhi, F.; Salehi, Z. Autism spectrum disorder genetics: A comprehensive review. Rev. Neurosci. 2025. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Eyring, K.W.; Liu, C.; Elhajjaoui, N.; Abuhanna, K.D.; Zhang, Y.; von Behren, Z.; Eskin, E.; Geschwind, D.H.; Luo, C. A single-cell atlas of DNA methylation in autism spectrum disorder reveals distinct regulatory and aging signatures. bioRxiv 2025. [Google Scholar] [CrossRef]
- De Franco, E.; Watson, R.A.; Weninger, W.J.; Wong, C.C.; Flanagan, S.E.; Caswell, R.; Green, A.; Tudor, C.; Lelliott, C.J.; Geyer, S.H.; et al. A specific CNOT1 mutation results in a novel syndrome of pancreatic agenesis and holoprosencephaly through impaired pancreatic and neurological development. Am. J. Hum. Genet. 2019, 104, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Lan, X.; Song, X.; Xu, W.; Zhang, Y.; Wang, S.; Xiao, M.; Yang, Y.; Zhang, H.; Wu, S. Clinical characteristics and identification of novel CNOT1 variants in three unrelated Chinese families with Vissers-Bodmer Syndrome. Heliyon 2024, 10, e26743. [Google Scholar] [CrossRef]
- Kruszka, P.; Berger, S.I.; Weiss, K.; Everson, J.L.; Martinez, A.F.; Hong, S.; Anyane-Yeboa, K.; Lipinski, R.J.; Muenke, M. A CCR4-NOT transcription complex, subunit 1, CNOT1, variant associated with holoprosencephaly. Am. J. Hum. Genet. 2019, 104, 990–993. [Google Scholar] [CrossRef]
- Vissers, L.E.L.M.; Kalvakuri, S.; de Boer, E.; Geuer, S.; Oud, M.; van Outersterp, I.; Kwint, M.; Witmond, M.; Kersten, S.; Polla, D.L.; et al. De novo variants in CNOT1, a central component of the CCR4-NOT complex involved in gene expression and RNA and protein stability, cause neurodevelopmental delay. Am. J. Hum. Genet. 2020, 107, 164–172. [Google Scholar] [CrossRef]
- Wu, T.; Chen, X.; Zhang, X. Vissers-Bodmer syndrome caused by a novel de novo CNOT1 frameshift variant. Am. J. Med. Genet. A 2024, 194, 363–367. [Google Scholar] [CrossRef]
- Shinawi, M.; Liu, P.; Kang, S.-H.L.; Shen, J.; Belmont, J.W.; Scott, D.A.; Probst, F.J.; Craigen, W.J.; Graham, B.H.; Pursley, A.; et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010, 47, 332–341. [Google Scholar] [CrossRef]
- Yasuhiko, Y.; Haraguchi, S.; Kitajima, S.; Takahashi, Y.; Kanno, J.; Saga, Y. Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proc. Natl. Acad. Sci. USA 2006, 103, 3651–3656. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, T.; Uchikawa, M.; Yoshida, M.; Bell, D.M.; Lovell-Badge, R.; Papaioannou, V.E.; Kondoh, H. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 2011, 470, 394–398. [Google Scholar] [CrossRef]
- Jacquemont, S.; Reymond, A.; Zufferey, F.; Harewood, L.; Walters, R.G.; Kutalik, Z.; Martinet, D.; Shen, Y.; Valsesia, A.; Beckmann, N.D.; et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 2011, 478, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Gillis, L.A.; McCallum, J.; Kaur, M.; DeScipio, C.; Yaeger, D.; Mariani, A.; Kline, A.D.; Li, H.; Devoto, M.; Jackson, L.G.; et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am. J. Hum. Genet. 2004, 75, 610–623. [Google Scholar] [CrossRef]
- Tonkin, E.T.; Wang, T.-J.; Lisgo, S.; Bamshad, M.J.; Strachan, T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004, 36, 636–641. [Google Scholar] [CrossRef]
- Kao, H.J.; Wang, E.H.F.; Yeh, E.C.; Chen, H.H.; Hsieh, F.J.; Ko, T.M.; Hwu, W.L.; Kwok, P.Y.; Lee, N.C. Identification of de novo chromosomal translocations disrupting NIPBL in a patient with Cornelia de Lange syndrome by full genome analysis. Mol. Genet. Genomic Med. 2025, 13, e70115. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, G.; Wagh, K.; Stavreva, D.A.; Jiménez-Panizo, A.; Kim, S.; Lion, M.; Alegre-Martí, A.; Rinaldi, L.; Johnson, T.A.; Gilson, E.; et al. Transcription factors form a ternary complex with NIPBL/MAU2 to localize cohesin at enhancers. Nucleic Acids Res. 2025, 53, gkaf415. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; Tei, S.; Izumi, K.; Krantz, I.D.; Bando, M.; Shirahige, K. A common molecular mechanism underlying Cornelia de Lange and CHOPS syndromes. Curr. Biol. 2025, 35, 1353–1363.e5. [Google Scholar] [CrossRef]
- Jahnke, P.; Xu, W.; Wülling, M.; Albrecht, M.; Gabriel, H.; Gillessen-Kaesbach, G.; Kaiser, F.J. The cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 2008, 36, 6450–6458. [Google Scholar] [CrossRef]
- van den Berg, D.L.C.; Azzarelli, R.; Oishi, K.; Martynoga, B.; Urbán, N.; Dekkers, D.H.W.; Demmers, J.A.; Guillemot, F. Nipbl interacts with Zfp609 and the integrator complex to regulate cortical neuron migration. Neuron 2017, 93, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Rigotti, L.; Rebellato, S.; Lettieri, A.; Castiglioni, S.; Mariani, M.; Totaro, S.; Saitta, C.; Gervasini, C.; Fazio, G.; Massa, V.; et al. Cohesins: Crossroad between Cornelia de Lange spectrum and cancer predisposition. Am. J. Med. Genet. A 2025, 197, e64076. [Google Scholar] [CrossRef] [PubMed]
- Durkie, M.; Cassidy, E.-J.; Berry, I.; Owens, M.; Turnbull, C.; Scott, R.H.; Taylor, R.W.; Deans, Z.C.; Ellard, S.; Baple, E.L.; et al. 2024, ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2024 (v1.2), Association for Clinical Genomic Science. Available online: https://www.genomicseducation.hee.nhs.uk/wp-content/uploads/2024/08/ACGS-2024_UK-practice-guidelines-for-variant-classification.pdf (accessed on 18 August 2025).
| Proband | MN37 | MN90 | MN107 | MN126 |
|---|---|---|---|---|
| Age of disease onset (years) | 1 | 1 | 1 | congenital |
| Age at study (years) | 10 | 12 | 7 | 16 |
| History of hypotonia | no | no | yes | no |
| Motor delay | yes | yes | yes | no |
| Speech delay | yes | yes | yes | yes |
| Somatic delay | no | no | low weight | no |
| Seizures | no | yes | no | yes |
| ID | yes | yes | yes | yes |
| ASD | no | yes | no | no |
| ADHD | no | no | yes | no |
| Behavior problems | yes | yes | yes | no |
| Dysmorphism | minor | minor | minor | moderate |
| Other | no | no | eczema | asymmetric breasts |
| MRI abnormality | narrow pons and brainstem, dilatation of occipital horns of lateral ventricules | normal anatomical variation | no | no |
| Positive family history of NDDs | yes, but remote | younger brother ADHD, 4 cousins on mother’s side have ASD | no | paternal aunt had ID |
| Previous genetic tests | none | 46XY kariotype, FMR1 normal alleles | 46XY kariotype | 46XX karyotype, array CGH planned, but not completed |
| Gene | DDX3X | CNOT1 | See details in Results | NIPBL |
| SNV | NM_001356.5: c.869C>A, NP_001347.3: p.S290* | NM_001265612.2: c.920delG, NP_001252541.1: p.G307Afs*32 | no | NM_133433.4: c.6839A>G, NP_597677.2: p.Gln2280Arg |
| CNV | no | no | 16p11.2 (16:29,690,418-30,200,285)x3 and 16p12.1-p11.2 (16:27,078,317-29,001,333)x3 | no |
| Variant classification | LP | LP | P | VUS |
| Novel | yes | yes | yes | yes |
| Established diagnosis | DDX3X syndrome | Vissers-Bodmer syndrome | chromosome 16p11.2 duplication syndrome | Cornelia de Lange syndrome |
| Genes | DDX3X | CNOT1 | NIPBL |
|---|---|---|---|
| Variants | NM_001356.5:c.869C>A NP_001347.3: p.S290* | NM_001265612.2:c.920delG NP_001252541.1:p.G307Afs*32 | NM_133433.4:c.6839A>G NP_597677.2:p.Gln2280Arg |
| MSA-SIFT | - | - | 1.00 |
| MSA-PolyPhen2 | - | - | 1.000 |
| PhyloP | 7.77 | 7.80 | 8.96 |
| GERP++ | 19.50 | 19.56 | 18.56 |
| Combined Annotation-Dependent Depletion (CADD) | 9.67 | 4.65 | 5.33 |
| Revel | - | - | 0.97 |
| Alpha Missense | - | - | 0.996 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tompa, M.; Sinko, G.; Mally, J.; Karteszi, J.; Kalman, B. Using Whole Exome Sequencing to Identify Genetic Causes of Neurodevelopmental Disorders in a Cohort of 11 Patients: A Single Center Experience. Int. J. Mol. Sci. 2025, 26, 10176. https://doi.org/10.3390/ijms262010176
Tompa M, Sinko G, Mally J, Karteszi J, Kalman B. Using Whole Exome Sequencing to Identify Genetic Causes of Neurodevelopmental Disorders in a Cohort of 11 Patients: A Single Center Experience. International Journal of Molecular Sciences. 2025; 26(20):10176. https://doi.org/10.3390/ijms262010176
Chicago/Turabian StyleTompa, Marton, Gabriella Sinko, Judit Mally, Judit Karteszi, and Bernadette Kalman. 2025. "Using Whole Exome Sequencing to Identify Genetic Causes of Neurodevelopmental Disorders in a Cohort of 11 Patients: A Single Center Experience" International Journal of Molecular Sciences 26, no. 20: 10176. https://doi.org/10.3390/ijms262010176
APA StyleTompa, M., Sinko, G., Mally, J., Karteszi, J., & Kalman, B. (2025). Using Whole Exome Sequencing to Identify Genetic Causes of Neurodevelopmental Disorders in a Cohort of 11 Patients: A Single Center Experience. International Journal of Molecular Sciences, 26(20), 10176. https://doi.org/10.3390/ijms262010176






