High Proteolytic and Collagenolytic Activity in an Environmental Vibrio Isolate: Insights into Tissue-Degrading Virulence Factors
Abstract
1. Introduction
2. Results and Discussion
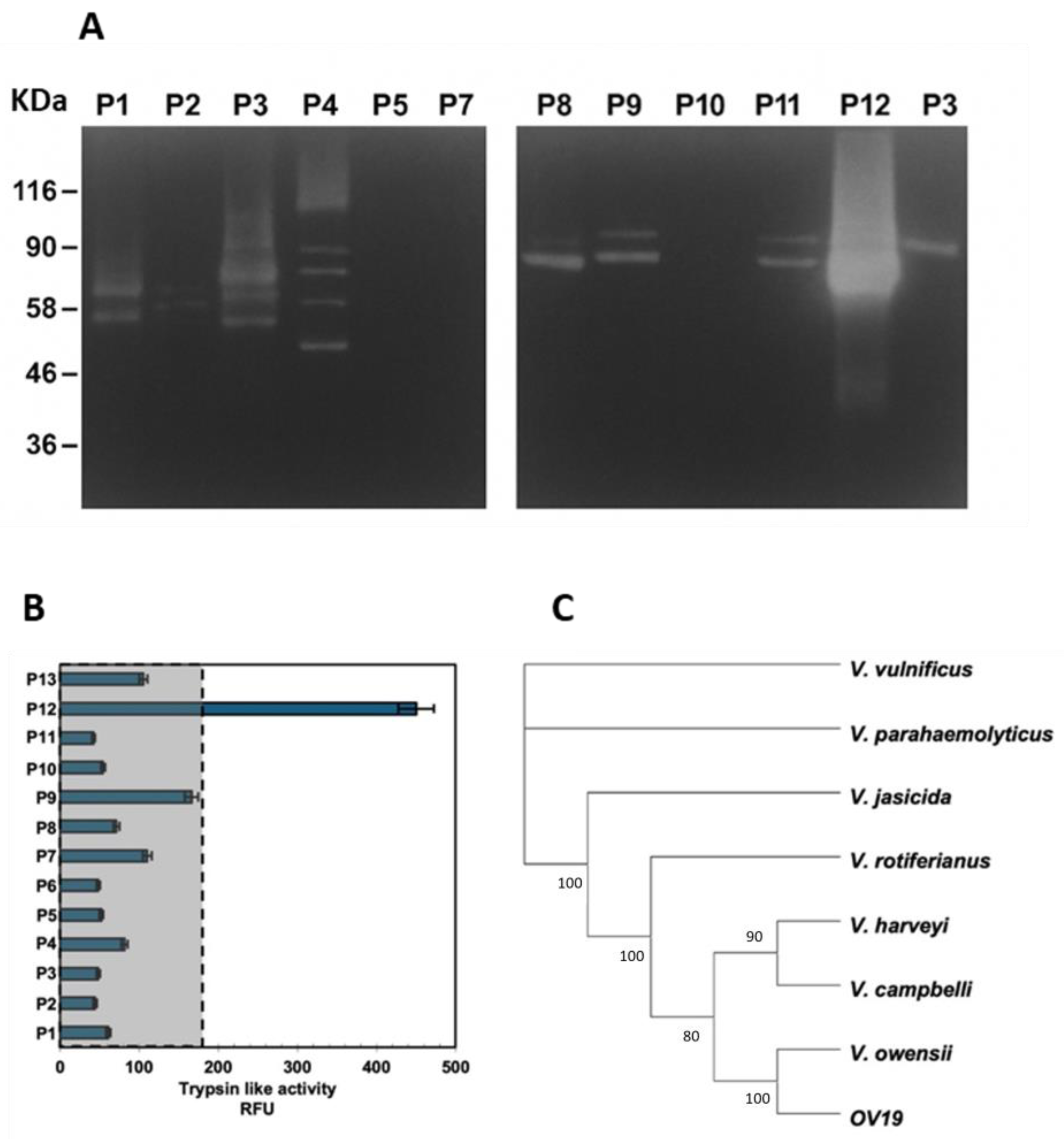
2.1. Bacterial Isolation, Analyses of Enzymes Secreted from Vibrio Isolates and Phylogenetic Analyses
2.2. Molecular Identification of OV19
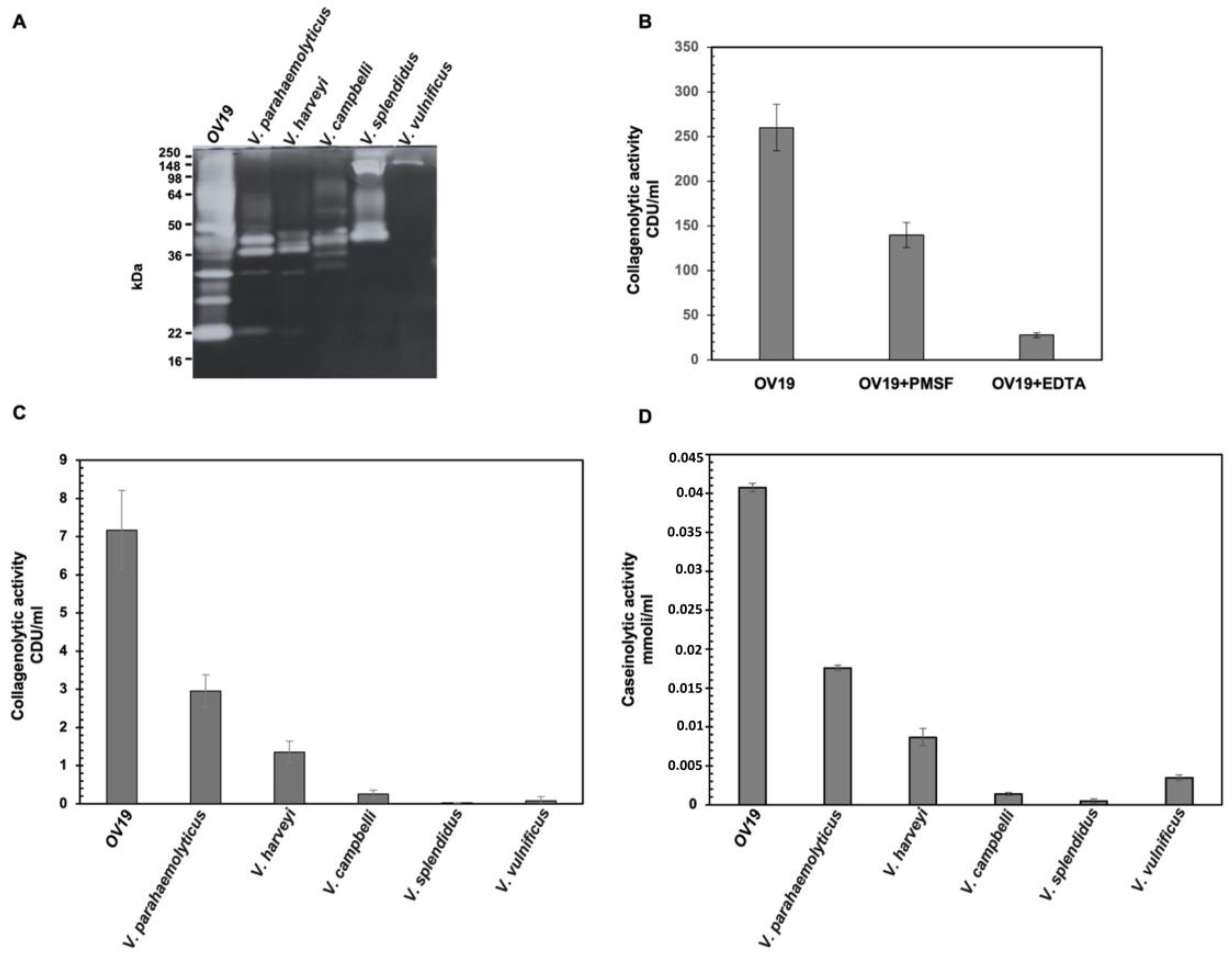
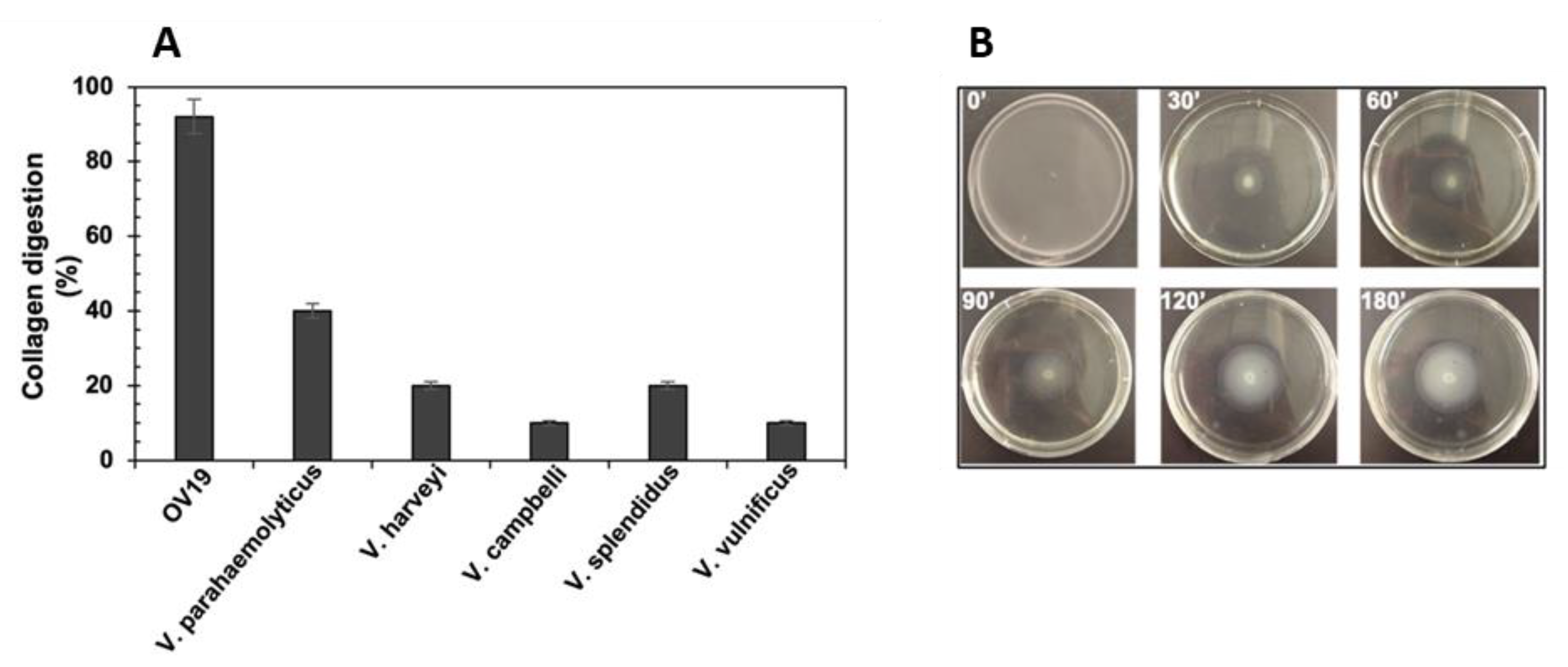
2.3. Comparison of Collagenolytic Activity of OV19 with Vibrio Type-Strains
3. Materials and Methods
3.1. Specimens’ Collection, Bacterial Isolation and Cultivation
3.2. Molecular and Bioinformatic Analyses
3.3. Screening of Protease-Producing Bacteria
3.4. SDS-PAGE and Zymography
3.5. Proteases and Collagenase Activity
3.6. Collagen Degradation Test
3.7. Swarming Motility Test
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2021, 4, 1–19, Erratum in Nat. Rev. Dis. Primers 2021, 7, 15. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-Cholera Vibrios: The microbial barometer of climate change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Eisen, J.A.; Nelson, W.C.; Clayton, R.A.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Umayam, L.; et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000, 406, 477–483. [Google Scholar] [CrossRef]
- Okada, K.; Iida, T.; Kita-Tsukamoto, K.; Honda, T. Vibrios commonly possess two chromosomes. J. Bacteriol. 2005, 187, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Waldor, M.K. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004, 155, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Pant, A.; Bag, S.; Saha, B.; Verma, J.; Kumar, P.; Banerjee, S.; Kumar, B.; Kumar, Y.; Desigamani, A.; Maiti, S.; et al. Molecular insights into the genome dynamics and interactions between core and acquired genomes of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2020, 117, 23762–23773. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Thompson, C.C.; Vicente, A.C.P.; Souza, R.C.; Vasconcelos, A.T.R.; Vesth, T.; Alves, N.; Ussery, D.W.; Iida, T.; Thompson, F.L. Genomic taxonomy of vibrios. BMC Evol. Biol. 2009, 9, 258. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New insights into a deadly opportunistic pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef]
- Vezzulli, L.; Colwell, R.R.; Pruzzo, C. Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 2013, 65, 817–825. [Google Scholar] [CrossRef]
- Ina-Salwany, M.Y.; Al-Saari, N.; Mohamad, A.; Mursidi, F.A.; Mohd-Aris, A.; Amal, M.N.A.; Kasai, H.; Mino, S.; Sawabe, T.; Zamri-Saad, M. Vibriosis in fish: A review on disease development and prevention. J. Aquat. Anim. Health 2019, 31, 3–22. [Google Scholar] [CrossRef]
- Dubert, J.; Barja, J.L.; Romalde, J.L. New insights into pathogenic vibrios affecting bivalves in Hatcheries: Present and Future Prospects. Front. Microbiol. 2017, 8, 762. [Google Scholar] [CrossRef]
- Sanches-Fernandes, G.M.M.; Sá-Correia, I.; Costa, R. Vibriosis outbreaks in aquaculture: Addressing Environmental and Public Health Concerns and Preventive Therapies Using Gilthead Seabream Farming as a Model System. Front. Microbiol. 2022, 13, 904815. [Google Scholar] [CrossRef] [PubMed]
- Orlić, K.; Kapetanović, D.; Kazazić, S.; Smrzlić, I.V.; Barac, F.; Nerlović, V.; Buha, T.; Bolotin, J.; Kožul, V.; Bobanović-Ćolić, S.; et al. Diversity, virulence and antibiotic resistance of Vibrio harveyi clade species associated with bivalve aquaculture within marine protected areas. Aquaculture 2025, 594, 741392. [Google Scholar] [CrossRef]
- Lu, K.; Li, Y.; Chen, R.; Yang, H.; Wang, Y.; Xiong, W.; Xu, F.; Yuan, Q.; Liang, H.; Xiao, X.; et al. Pathogenic mechanism of Vibrio vulnificus infection. Future Microbiol. 2023, 18, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Nicosia, A.; Salamone, M.; Tagliavia, M. Targeting virulence genes expression in Vibrio vulnificus. Int. J. Mol. Sci. 2022, 23, 15278. [Google Scholar] [CrossRef]
- Miyoshi, S. Extracellular proteolytic enzymes produced by human pathogenic Vibrio species. Front. Microbiol. 2013, 4, 339. [Google Scholar] [CrossRef]
- Wang, R.; Zhong, Y.; Gu, X.; Yuan, J.; Saeed, A.F.; Wang, S. The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 2015, 6, 144, Erratum in Front. Microbiol. 2015, 6, 437. [Google Scholar] [CrossRef]
- Huang, J.; Zeng, B.; Liu, D.; Wu, R.; Zhang, J.; Liao, B.; He, H.; Bian, F. Classification and structural insight into vibriolysin-like proteases of Vibrio pathogenicity. Microb. Pathog. 2018, 117, 335–340. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a fish pathogen: Virulence factors, diagnosis and prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Huang, L.; Fu, Y.; Sun, W.; Brzostek, J. Editorial: Vibrio virulence regulation and Host Interactions. Front. Cell. Infect. Microbiol. 2021, 11, 793464. [Google Scholar] [CrossRef]
- Salamone, M.; Nicosia, A.; Ghersi, G.; Tagliavia, M. Vibrio proteases for biomedical applications: Modulating the proteolytic secretome of V. alginolyticus and V. parahaemolyticus for improved enzymes production. Microorganisms 2019, 7, 387. [Google Scholar] [CrossRef]
- Archer, E.J.; Baker-Austin, C.; Osborn, T.J.; Jones, N.R.; Martínez-Urtaza, J.; Trinanes, J.; Oliver, J.D.; González, F.J.C.; Lake, I.R. Climate warming and increasing Vibrio vulnificus infections in North America. Sci. Rep. 2023, 13, 3893. [Google Scholar] [CrossRef]
- Kim, Y.R.; Lee, S.E.; Kook, H.; Yeom, J.A.; Na, H.S.; Kim, S.Y.; Chung, S.S.; Choy, H.E.; Rhee, J.H. Vibrio vulnificus RTX toxin kills host cells only after contact of the bacteria with host cells. Cell. Microbiol. 2008, 10, 848–862. [Google Scholar] [CrossRef]
- Canellas, A.L.B.; Marques, M.d.O.N.; Lopes, M.V.; Lage, A.; Klautau, M.; Muricy, G.; de Oliveira, B.F.R.; Laport, M.S. Functional and genomic insights into the biotechnological potential of Vibrio spp. Isolated from deeply polluted and pristine environments. Curr. Microbiol. 2024, 82, 36. [Google Scholar] [CrossRef]
- Tagliavia, M.; Salamone, M.; Bennici, C.; Quatrini, P.; Cuttitta, A. A modified culture medium for improved isolation of marine vibrios. MicrobiologyOpen 2019, 8, e00835. [Google Scholar] [CrossRef]
- Canellas, A.L.B.; Faria, A.R.; Dias, G.R.; Teixeira, L.M.; Laport, M.S. Polyphasic identification of Vibrio species from aquatic sources using mass spectrometry, housekeeping gene sequencing and whole genome analysis. Sci. Rep. 2024, 14, 26250. [Google Scholar] [CrossRef] [PubMed]
- Thompson, F.L.; Gevers, D.; Thompson, C.C.; Dawyndt, P.; Naser, S.; Hoste, B.; Munn, C.B.; Swings, J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 2005, 71, 5107–5115. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Macián, M.C.; Arahal, D.R.; Garay, E.; Pujalte, M.J. Multilocus sequence analysis of the central clade of the genus Vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol. 2010, 60, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 2007, 189, 7932–7936. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, H.; Su, Y.; Liu, S.; Xu, L.; Guo, Z.; Wu, J.; Cheng, C.; Feng, J. Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genom. 2019, 20, 761. [Google Scholar] [CrossRef]
- Le Roux, F.; Blokesch, M. Eco-evolutionary dynamics linked to Horizontal Gene Transfer in Vibrios. Annu. Rev. Microbiol. 2018, 72, 89–110. [Google Scholar] [CrossRef]
- Xu, M.; Xu, M.; Tu, Q. Comparative evaluation of Vibrio delineation methodologies in post-genomic era. Environ. Microbiol. Rep. 2021, 13, 209–217. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Cao, H.Y.; Ding, H.T.; Su, H.N.; Liu, S.C.; Liu, G.; Zhang, X.; Li, C.Y.; Peng, M.; et al. Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis. Nat. Commun. 2022, 13, 566. [Google Scholar] [CrossRef]
- Pozo, F.; Borbor, M.; Solórzano, R.; Sonnenholzner, S.; Bayot, B. Optimized swarming motility assay to identify anti-virulence products against Vibrio parahaemolyticus, a pathogen of farmed shrimp. MethodsX 2024, 12, 102622. [Google Scholar] [CrossRef] [PubMed]
- Tagliavia, M.; Catania, V.; Dell’Omo, G.; Massa, B. High-performance PCR for alleles discrimination of chromo-helicase-DNA binding protein (CHD1) gene in bird sexing. Biology 2023, 12, 300. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Salamone, M.; Nicosia, A.; Bennici, C.; Quatrini, P.; Catania, V.; Mazzola, S.; Ghersi, G.; Cuttitta, A. Comprehensive Analysis of a Vibrio parahaemolyticus Strain Extracellular Serine Protease VpSP37. PLoS ONE 2015, 10, e0126349. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Salamone, M.; Seidita, G.; Cuttitta, A.; Rigogliuso, S.; Mazzola, S.; Bertuzzi, F.; Ghersi, G. A new method to value efficiency of enzyme blends for pancreatic tissue digestion. Transplant. Proc. 2010, 42, 2043–2048, Erratum in Transplant. Proc. 2010, 42, 3898–3900. [Google Scholar] [CrossRef]
- Salamone, M.; Saladino, S.; Pampalone, M.; Campora, S.; Ghersi, G. Tissue dissociation and primary cells isolation using recombinant collagenases Class I and II. Chem. Eng. Trans. 2014, 38, 247–252. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salamone, M.; Nicosia, A.; Ghersi, G.; Cuttitta, A.; Quatrini, P.; Tagliavia, M. High Proteolytic and Collagenolytic Activity in an Environmental Vibrio Isolate: Insights into Tissue-Degrading Virulence Factors. Int. J. Mol. Sci. 2025, 26, 10153. https://doi.org/10.3390/ijms262010153
Salamone M, Nicosia A, Ghersi G, Cuttitta A, Quatrini P, Tagliavia M. High Proteolytic and Collagenolytic Activity in an Environmental Vibrio Isolate: Insights into Tissue-Degrading Virulence Factors. International Journal of Molecular Sciences. 2025; 26(20):10153. https://doi.org/10.3390/ijms262010153
Chicago/Turabian StyleSalamone, Monica, Aldo Nicosia, Giulio Ghersi, Angela Cuttitta, Paola Quatrini, and Marcello Tagliavia. 2025. "High Proteolytic and Collagenolytic Activity in an Environmental Vibrio Isolate: Insights into Tissue-Degrading Virulence Factors" International Journal of Molecular Sciences 26, no. 20: 10153. https://doi.org/10.3390/ijms262010153
APA StyleSalamone, M., Nicosia, A., Ghersi, G., Cuttitta, A., Quatrini, P., & Tagliavia, M. (2025). High Proteolytic and Collagenolytic Activity in an Environmental Vibrio Isolate: Insights into Tissue-Degrading Virulence Factors. International Journal of Molecular Sciences, 26(20), 10153. https://doi.org/10.3390/ijms262010153







