Microbiome and Long COVID-19: Current Evidence and Insights
Abstract
1. Introduction
2. Gut Microbiome Alterations in Various Phases of Long COVID
2.1. Altered Gut Microbiome During COVID-19 Infection
2.2. Lasting Changes in Microbial Disruption in Long COVID
2.3. Trends in Taxonomic Shifts in Gut Microbiota
2.4. Oral and Respiratory Microbiomes
2.5. Unique Microbial Clusters Associated with Long COVID Symptoms
2.6. Methodological and Clinical Heterogeneity in COVID-19 Microbiome Studies
3. Proposed Mechanisms Linking Microbiome to Long COVID
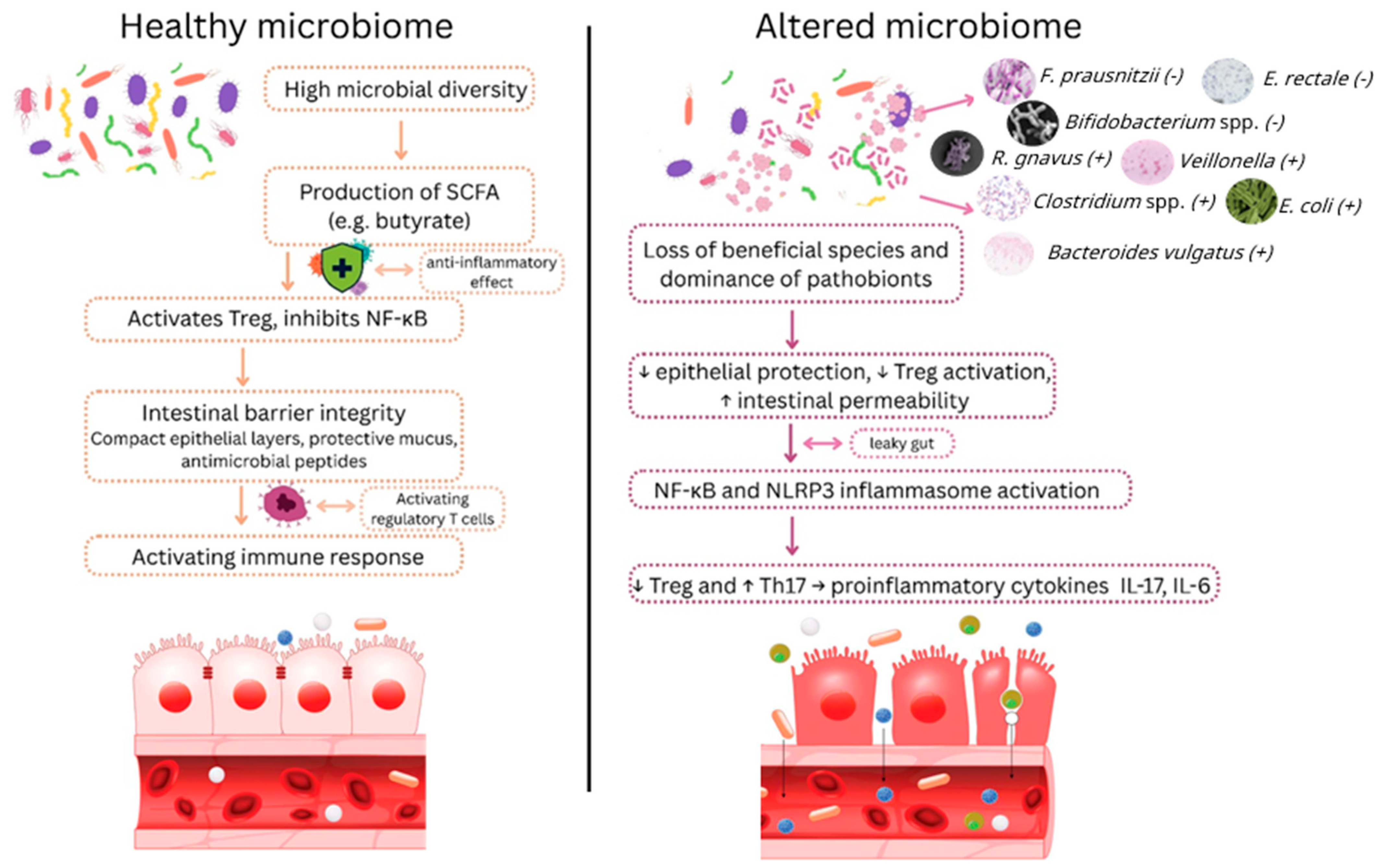
3.1. Gut–Immune Axis and Inflammation
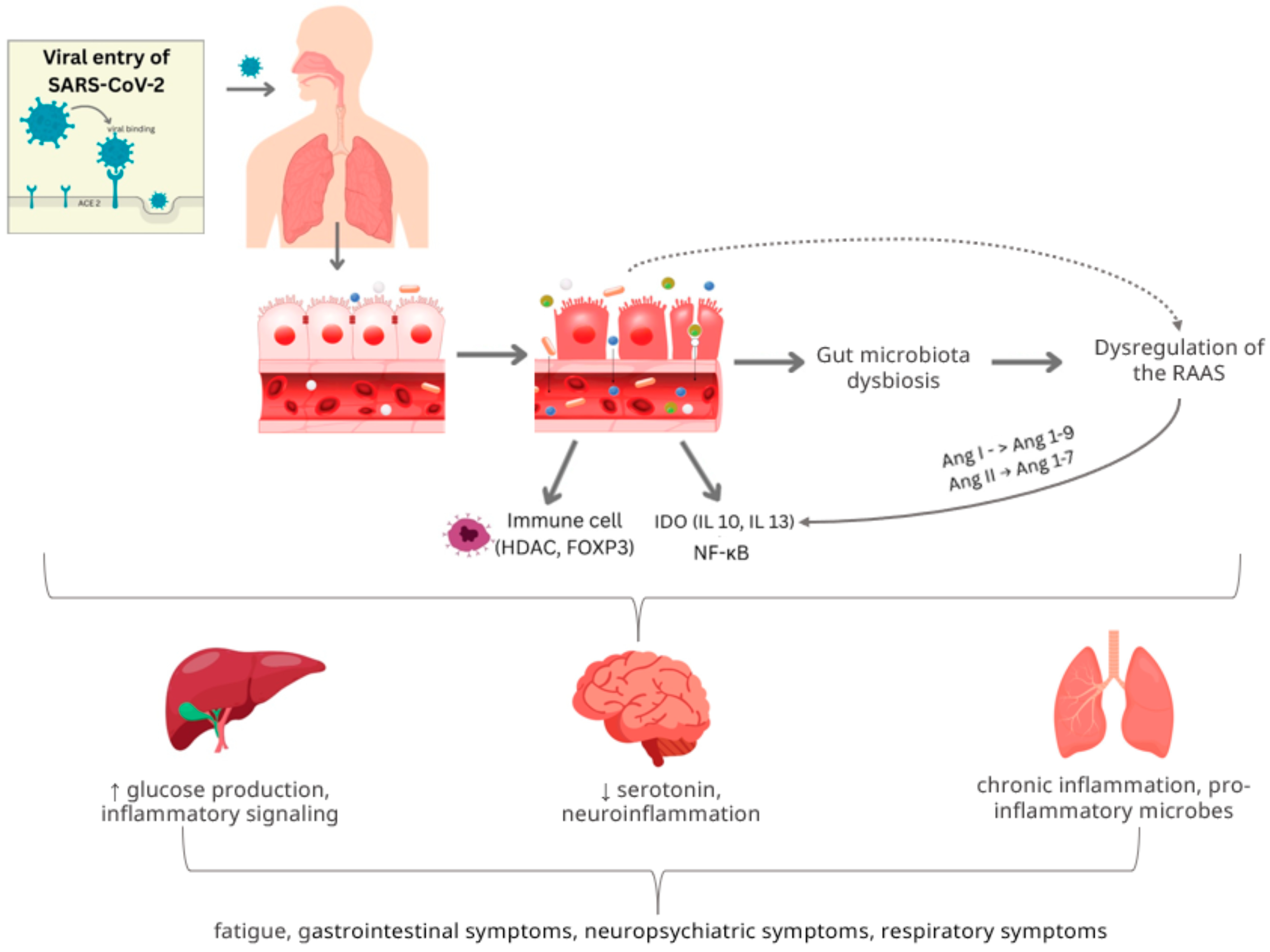
3.2. ACE2 and Gut Barrier Dysfunction
3.3. Microbiota-Gut–Brain Axis
3.4. Viral Persistence, Immune Priming and Autoantibody Formation
3.5. The Oral–Lung Aspiration Axis
3.6. Experimental Evidence: Causal Role of Microbiota
4. Microbial Biomarkers of Long COVID
4.1. Reduced SCFA Producers
4.2. Enriched Pathobionts
4.3. Microbiome Diversity Index
4.4. Specific Species or Functions
4.5. Combined Microbiome Signatures
5. Microbiome-Targeted Diagnostics and Therapeutics
5.1. Diagnostic Avenues
5.2. Prebiotics and Diet
5.3. Probiotics and Synbiotics
5.4. Microbiota Transplantation
6. Consensus and Controversies
6.1. Consensus Points
6.2. Controversies and Open Questions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 5-HT | 5-hydroxytryptamine |
| 5-HTP | 5-hydroxytryptophan |
| ACE2 | Angiotensin-converting enzyme 2 |
| Ang | Angiotensin |
| C-Trp | C-glycosyltrytophan |
| CT | Computed tomography |
| EGF | Epidermal growth factor |
| FEV1 | Forced expiratory volume |
| FGF | Fibroblast growth factor |
| FMT | Fecal microbiota transplantation |
| FOXP3 | Forkhead box P3 |
| FVC | Forced vital capacity |
| HDAC | Histone deacetylase |
| HSP | Heat-shock protein |
| IAA | Indole-3-acetic acid |
| ILA | Indole-3-lactic acid |
| IFN | Interferon |
| IDO | Indoleamine 2,3-dioxygenase |
| IPA | Indole-3-propionate |
| LBP | Lipopolysaccharide binding protein |
| LPS | Lipopolysaccharide |
| MAIT | Mucosal-associated invariant T |
| MDC | Macrophage-derived chemokine |
| ME/CFS | Myalgic encephalomyelitis/chronic fatigue syndrome |
| MIP-1b | Macrophage inflammatory protein-1 beta |
| MPIF-1 | Myeloid progenitor inhibitory factor 1 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLRP3 | NOD-, LRR-, and pyrin domain-containing protein 3 |
| PASC | Post-acute sequelae of COVID-19 |
| RAAS | Renin–angiotensin–aldosterone system |
| ROS | Reactive oxygen species |
| SCFA | Short-chain fatty acid |
| Th17 | T helper 17 cells |
| TLR | Toll-like receptor |
| TNF | Tumor necrosis factor |
| Treg | Regulatory T cell |
References
- Parums, D.V. Long COVID or Post-Acute Sequelae of SARS-CoV-2 Infection (PASC) and the Urgent Need to Identify Diagnostic Biomarkers and Risk Factors. Med. Sci. Monit. 2024, 30, e946512. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146, Erratum in Nat. Rev. Microbiol. 2023, 21, 408. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 698169. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Santacruz, C.; Tyrkalska, S.D.; Candel, S. The Microbiota in Long COVID. Int. J. Mol. Sci. 2024, 25, 1330. [Google Scholar] [CrossRef]
- Martín Giménez, V.M.; Modrego, J.; Gómez-Garre, D.; Manucha, W.; de las Heras, N. Gut Microbiota Dysbiosis in COVID-19: Modulation and Approaches for Prevention and Therapy. Int. J. Mol. Sci. 2023, 24, 12249. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Comba, I.Y.; Mars, R.A.T.; Yang, L.; Dumais, M.; Chen, J.; Van Gorp, T.M.; Harrington, J.J.; Sinnwell, J.P.; Johnson, S.; Holland, L.A.; et al. Gut Microbiome Signatures During Acute Infection Predict Long COVID. bioRxiv 2024. preprint. [Google Scholar] [CrossRef]
- Upadhyay, V.; Suryawanshi, R.K.; Tasoff, P.; McCavitt-Malvido, M.; Kumar, R.G.; Murray, V.W.; Noecker, C.; Bisanz, J.E.; Hswen, Y.; Ha, C.W.Y.; et al. Mild SARS-CoV-2 infection results in long-lasting microbiota instability. mBio 2023, 14, e0088923. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, K.Y.; Meng, T.Q.; Ye, Z.; Guo, S.M.; Li, Z.M.; Xiong, C.L.; Yin, Y.; Li, H.G.; Zhou, L.Q. Gut Microbiota May Not Be Fully Restored in Recovered COVID-19 Patients After 3-Month Recovery. Front. Nutr. 2021, 8, 638825. [Google Scholar] [CrossRef]
- Zhang, D.; Zhou, Y.; Ma, Y.; Chen, P.; Tang, J.; Yang, B.; Li, H.; Liang, M.; Xue, Y.; Liu, Y.; et al. Gut Microbiota Dysbiosis Correlates With Long COVID-19 at One-Year After Discharge. J. Korean Med. Sci. 2023, 38, e120. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2021, 12, 796025. [Google Scholar] [CrossRef]
- Chen, Y.; Gu, S.; Chen, Y.; Lu, H.; Shi, D.; Guo, J.; Wu, W.R.; Yang, Y.; Li, Y.; Xu, K.J.; et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut 2022, 71, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Ni, J.; Guo, W.; Ding, C.; Wang, F.; Wu, Y.; Zhao, Y.; Zhu, L.; Xu, K.; Chen, Y. Two-year follow-up of gut microbiota alterations in patients after COVID-19: From the perspective of gut enterotype. Microbiol. Spectr. 2025, 13, e02774-24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef]
- Krasaewes, K.; Chaiwarith, R.; Chattipakorn, N.; Chattipakorn, S.C. Profiles of gut microbiota associated with clinical outcomes in patients with different stages of SARS-CoV-2 infection. Life Sci. 2023, 332, 122136. [Google Scholar] [CrossRef]
- Carneiro, V.L.; Littlefield, K.M.; Watson, R.; Palmer, B.E.; Lozupone, C. Inflammation-associated gut microbiome in postacute sequelae of SARS-CoV-2 points towards new therapeutic targets. Gut 2024, 73, 376–378. [Google Scholar] [CrossRef]
- Moreno-Corona, N.C.; Lopez-Ortega, O.; Perez-Martinez, C.A.; Martinez-Castillo, M.; De Jesus-Gonzalez, L.A.; Leon-Reyes, G.; Leon-Juarez, M. Dynamics of the Microbiota and Its Relationship with Post-COVID-19 Syndrome. Int. J. Mol. Sci. 2023, 24, 14822. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.T.; Khan, H.; Khalid, A.; Mahmood, S.F.; Nasir, N.; Khanum, I.; de Siqueira, I.; Van Voorhis, W. Chronic inflammation in post-acute sequelae of COVID-19 modulates gut microbiome: A review of literature on COVID-19 sequelae and gut dysbiosis. Mol. Med. 2025, 31, 22. [Google Scholar] [CrossRef] [PubMed]
- Ailioaie, L.M.; Ailioaie, C.; Litscher, G. Infection, Dysbiosis and Inflammation Interplay in the COVID Era in Children. Int. J. Mol. Sci. 2023, 24, 10874. [Google Scholar] [CrossRef] [PubMed]
- Scheithauer, T.P.M.; Montijn, R.C.; Mieremet, A. Mieremet, Gut microbe-host interactions in post-COVID syndrome: A debilitating or restorative partnership? Gut Microbes 2024, 16, 2402544. [Google Scholar] [CrossRef]
- Hazan, S.; Stollman, N.; Bozkurt, H.S.; Dave, S.; Papoutsis, A.J.; Daniels, J.; Barrows, B.D.; Quigley, E.M.; Borody, T.J. Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity. BMJ Open Gastroenterol. 2022, 9, e000871, Erratum in BMJ Open Gastroenterol. 2025, 12, e000871corr1. [Google Scholar] [CrossRef]
- Raj, S.T.; Bruce, A.W.; Anbalagan, M.; Srinivasan, H.; Chinnappan, S.; Rajagopal, M.; Khanna, K.; Chandramoorthy, H.C.; Mani, R.R. COVID-19 influenced gut dysbiosis, post-acute sequelae, immune regulation, and therapeutic regimens. Front. Cell Infect. Microbiol. 2024, 14, 1384939. [Google Scholar] [CrossRef]
- Giannos, P.; Prokopidis, K. Gut dysbiosis and long COVID-19: Feeling gutted. J. Med. Virol. 2022, 94, 2917–2918. [Google Scholar] [CrossRef]
- Liu, Y.; Chan, M.T.V.; Chan, F.K.L.; Wu, W.K.K.; Ng, S.C.; Zhang, L. Lower gut abundance of Eubacterium rectale is linked to COVID-19 mortality. Front. Cell Infect. Microbiol. 2023, 13, 1249069. [Google Scholar] [CrossRef]
- Zhang, F.; Lau, R.I.; Liu, Q.; Su, Q.; Chan, F.K.L.; Ng, S.C. Gut microbiota in COVID-19: Key microbial changes, potential mechanisms and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 323–337, Erratum in Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 195. [Google Scholar] [CrossRef]
- Blankestijn, J.M.; Baalbaki, N.; Beijers, R.; Cornelissen, M.E.B.; Wiersinga, W.J.; Abdel-Aziz, M.I.; Maitland-van der Zee, A.H.; Consortium, P.O. Exploring Heterogeneity of Fecal Microbiome in Long COVID Patients at 3 to 6 Months After Infection. Int. J. Mol. Sci. 2025, 26, 1781. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Lau, R.I.; Liu, Q.; Li, M.K.T.; Yan Mak, J.W.; Lu, W.; Lau, I.S.F.; Lau, L.H.S.; Yeung, G.T.Y.; Cheung, C.P.; et al. The gut microbiome associates with phenotypic manifestations of post-acute COVID-19 syndrome. Cell Host Microbe 2024, 32, 651–660.e654. [Google Scholar] [CrossRef]
- Lau, R.I.; Su, Q.; Ching, J.Y.L.; Lui, R.N.; Chan, T.T.; Wong, M.T.L.; Lau, L.H.S.; Wing, Y.K.; Chan, R.N.Y.; Kwok, H.Y.H.; et al. Fecal Microbiota Transplantation for Sleep Disturbance in Post-acute COVID-19 Syndrome. Clin. Gastroenterol. Hepatol. 2024, 22, 2487–2496.e2486. [Google Scholar] [CrossRef]
- Lau, R.I.; Su, Q.; Lau, I.S.F.; Ching, J.Y.L.; Wong, M.C.S.; Lau, L.H.S.; Tun, H.M.; Mok, C.K.P.; Chau, S.W.H.; Tse, Y.K.; et al. A synbiotic preparation (SIM01) for post-acute COVID-19 syndrome in Hong Kong (RECOVERY): A randomised, double-blind, placebo-controlled trial. Lancet Infect. Dis. 2024, 24, 256–265. [Google Scholar] [CrossRef]
- Haran, J.P.; Bradley, E.; Zeamer, A.L.; Cincotta, L.; Salive, M.C.; Dutta, P.; Mutaawe, S.; Anya, O.; Meza-Segura, M.; Moormann, A.M.; et al. Inflammation-type dysbiosis of the oral microbiome associates with the duration of COVID-19 symptoms and long COVID. JCI Insight 2021, 6, e152346. [Google Scholar] [CrossRef]
- Bhanu, P.; Buchke, S.; Hemandhar-Kumar, N.; Varsha, P.; Kiran, S.K.R.; Vikneswaran, G.; Alva, A.; Basavaraj, G.S.; Kumar, J. Comparative metagenomic analysis of the oral microbiome in COVID-19 patients and healthy individuals. Sci. Rep. 2025, 15, 10303. [Google Scholar] [CrossRef]
- Xu, J.; Wu, D.; Yang, J.; Zhao, Y.; Liu, X.; Chang, Y.; Tang, Y.; Sun, F.; Zhao, Y. Adult Outpatients with Long COVID Infected with SARS-CoV-2 Omicron Variant. Part 1: Oral Microbiota Alterations. Am. J. Med. 2025, 138, 732–741.e2. [Google Scholar] [CrossRef] [PubMed]
- Paine, S.K.; Rout, U.K.; Bhattacharyya, C.; Parai, D.; Alam, M.; Nanda, R.R.; Tripathi, D.; Choudhury, P.; Kundu, C.N.; Pati, S.; et al. Temporal dynamics of oropharyngeal microbiome among SARS-CoV-2 patients reveals continued dysbiosis even after Viral Clearance. NPJ Biofilms Microbiomes 2022, 8, 67. [Google Scholar] [CrossRef] [PubMed]
- Drozdzik, A. Covid-19 and SARS-CoV-2 infection in periodontology: A narrative review. J. Periodontal Res. 2022, 57, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Ancona, G.; Alagna, L.; Alteri, C.; Palomba, E.; Tonizzo, A.; Pastena, A.; Muscatello, A.; Gori, A.; Bandera, A. Gut and airway microbiota dysbiosis and their role in COVID-19 and long-COVID. Front. Immunol. 2023, 14, 1080043. [Google Scholar] [CrossRef]
- Tan, L.; Zhong, M.M.; Liu, Q.; Chen, Y.; Zhao, Y.Q.; Zhao, J.; Dusenge, M.A.; Feng, Y.; Ye, Q.; Hu, J.; et al. Potential interaction between the oral microbiota and COVID-19: A meta-analysis and bioinformatics prediction. Front. Cell Infect. Microbiol. 2023, 13, 1193340. [Google Scholar] [CrossRef]
- Kageyama, S.; Takeshita, T.; Furuta, M.; Tomioka, M.; Asakawa, M.; Suma, S.; Takeuchi, K.; Shibata, Y.; Iwasa, Y.; Yamashita, Y. Relationships of Variations in the Tongue Microbiota and Pneumonia Mortality in Nursing Home Residents. J. Gerontol. A Biol. Sci. Med. Sci. 2018, 73, 1097–1102. [Google Scholar] [CrossRef]
- Mammen, M.J.; Scannapieco, F.A.; Sethi, S. Oral-lung microbiome interactions in lung diseases. Periodontol 2000 2020, 83, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Segal, L.N.; Alekseyenko, A.V.; Clemente, J.C.; Kulkarni, R.; Wu, B.; Gao, Z.; Chen, H.; Berger, K.I.; Goldring, R.M.; Rom, W.N.; et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 2013, 1, 19, Erratum in Microbiome 2014, 2, 21. [Google Scholar] [CrossRef]
- Giron, L.B.; Peluso, M.J.; Ding, J.; Kenny, G.; Zilberstein, N.F.; Koshy, J.; Hong, K.Y.; Rasmussen, H.; Miller, G.E.; Bishehsari, F.; et al. Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-kappaB signaling. JCI Insight 2022, 7, e160989. [Google Scholar] [CrossRef]
- Candel, S.; Tyrkalska, S.D.; Alvarez-Santacruz, C.; Mulero, V. The nasopharyngeal microbiome in COVID-19. Emerg. Microbes Infect. 2023, 12, e2165970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Weng, S.; Xia, C.; Ren, Y.; Liu, Z.; Xu, Y.; Yang, X.; Wu, R.; Peng, L.; Sun, L.; et al. Gastrointestinal symptoms of long COVID-19 related to the ectopic colonization of specific bacteria that move between the upper and lower alimentary tract and alterations in serum metabolites. BMC Med. 2023, 21, 264. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; An, S.; Park, K.; Lee, S.; Han, Y.M.; Koh, S.J.; Lee, J.; Gim, H.; Kim, D.; Seo, H. Gut Microbial Signatures in Long COVID: Potential Biomarkers and Therapeutic Targets. Infect. Dis. Ther. 2025, 14, 1461–1475. [Google Scholar] [CrossRef]
- Cheng, X.; Zhang, Y.; Li, Y.; Wu, Q.; Wu, J.; Park, S.-K.; Guo, C.; Lu, J. Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases. BMC Microbiol. 2022, 22, 274. [Google Scholar] [CrossRef]
- Kopera, K.; Gromowski, T.; Wydmański, W.; Skonieczna-Żydecka, K.; Muszyńska, A.; Zielińska, K.; Wierzbicka-Woś, A.; Kaczmarczyk, M.; Kadaj-Lipka, R.; Cembrowska-Lech, D.; et al. Gut microbiome dynamics and predictive value in hospitalized COVID-19 patients: A comparative analysis of shallow and deep shotgun sequencing. Front. Microbiol. 2024, 15, 1342749. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef]
- Bars-Cortina, D.; Ramon, E.; Rius-Sansalvador, B.; Guinó, E.; Garcia-Serrano, A.; Mach, N.; Khannous-Lleiffe, O.; Saus, E.; Gabaldón, T.; Ibáñez-Sanz, G.; et al. Comparison between 16S rRNA and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota. BMC Genom. 2024, 25, 730. [Google Scholar] [CrossRef]
- Righi, E.; Dalla Vecchia, I.; Auerbach, N.; Morra, M.; Górska, A.; Sciammarella, C.; Lambertenghi, L.; Gentilotti, E.; Mirandola, M.; Tacconelli, E.; et al. Gut Microbiome Disruption Following SARS-CoV-2: A Review. Microorganisms 2024, 12, 131. [Google Scholar] [CrossRef]
- Li, J.; Ghosh, T.S.; McCann, R.; Mallon, P.; Hill, C.; Draper, L.; Schult, D.; Fanning, L.J.; Shannon, R.; Sadlier, C.; et al. Robust cross-cohort gut microbiome associations with COVID-19 severity. Gut Microbes 2023, 15, 2242615. [Google Scholar] [CrossRef] [PubMed]
- Donkers, A.; Seel, W.; Klümpen, L.; Simon, M.C. The Multiple Challenges of Nutritional Microbiome Research During COVID-19-A Perspective and Results of a Single-Case Study. Nutrients 2024, 16, 3693. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef] [PubMed]
- Anthony, W.E.; Wang, B.; Sukhum, K.V.; D’Souza, A.W.; Hink, T.; Cass, C.; Seiler, S.; Reske, K.A.; Coon, C.; Dubberke, E.R.; et al. Acute and persistent effects of commonly used antibiotics on the gut microbiome and resistome in healthy adults. Cell Rep. 2022, 39, 110649. [Google Scholar] [CrossRef]
- Straume, Z.; Krūmiņa, N.; Elbere, I.; Rozenberga, M.; Erts, R.; Rudzīte, D.; Proskurina, A.; Krumina, A. Impact of Vitamins, Antibiotics, Probiotics, and History of COVID-19 on the Gut Microbiome in Ulcerative Colitis Patients: A Cross-Sectional Study. Medicina 2025, 61, 284. [Google Scholar] [CrossRef]
- Kim, H.N.; Joo, E.J.; Lee, C.W.; Ahn, K.S.; Kim, H.L.; Park, D.I.; Park, S.K. Reversion of Gut Microbiota during the Recovery Phase in Patients with Asymptomatic or Mild COVID-19: Longitudinal Study. Microorganisms 2021, 9, 1237. [Google Scholar] [CrossRef]
- de Nies, L.; Galata, V.; Martin-Gallausiaux, C.; Despotovic, M.; Busi, S.B.; Snoeck, C.J.; Delacour, L.; Budagavi, D.P.; Laczny, C.C.; Habier, J.; et al. Altered infective competence of the human gut microbiome in COVID-19. Microbiome 2023, 11, 46. [Google Scholar] [CrossRef]
- Ng, S.C.; Peng, Y.; Zhang, L.; Mok, C.K.; Zhao, S.; Li, A.; Ching, J.Y.; Liu, Y.; Yan, S.; Chan, D.L.S.; et al. Gut microbiota composition is associated with SARS-CoV-2 vaccine immunogenicity and adverse events. Gut 2022, 71, 1106–1116. [Google Scholar] [CrossRef]
- Hong, M.; Lan, T.; Li, Q.; Li, B.; Yuan, Y.; Xu, F.; Wang, W. A comprehensive perspective on the interaction between gut microbiota and COVID-19 vaccines. Gut Microbes 2023, 15, 2233146. [Google Scholar] [CrossRef]
- Boston, R.H.; Guan, R.; Kalmar, L.; Beier, S.; Horner, E.C.; Beristain-Covarrubias, N.; Yam-Puc, J.C.; Pereyra Gerber, P.; Faria, L.; Kuroshchenkova, A.; et al. Stability of gut microbiome after COVID-19 vaccination in healthy and immuno-compromised individuals. Life Sci. Alliance 2024, 7, e202302529. [Google Scholar] [CrossRef]
- Chanda, D.; De, D. Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift. Gut Microbes 2024, 16, 2304900. [Google Scholar] [CrossRef]
- Safarchi, A.; Al-Qadami, G.; Tran, C.D.; Conlon, M. Understanding dysbiosis and resilience in the human gut microbiome: Biomarkers, interventions, and challenges. Front. Microbiol. 2025, 16, 1559521. [Google Scholar] [CrossRef]
- Shen, Y.; Fan, N.; Ma, S.X.; Cheng, X.; Yang, X.; Wang, G. Gut Microbiota Dysbiosis: Pathogenesis, Diseases, Prevention, and Therapy. MedComm 2020 2025, 6, e70168. [Google Scholar] [CrossRef] [PubMed]
- Galeeva, J.S.; Fedorov, D.E.; Starikova, E.V.; Manolov, A.I.; Pavlenko, A.V.; Selezneva, O.V.; Klimina, K.M.; Veselovsky, V.A.; Morozov, M.D.; Yanushevich, O.O.; et al. Microbial Signatures in COVID-19: Distinguishing Mild and Severe Disease via Gut Microbiota. Biomedicines 2024, 12, 996. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Whalley, J.P.; Knight, J.C.; Wicker, L.S.; Todd, J.A.; Ferreira, R.C. SARS-CoV-2 infection induces a long-lived pro-inflammatory transcriptional profile. Genome Med. 2023, 15, 69. [Google Scholar] [CrossRef]
- Ren, Z.; Wang, H.; Cui, G.; Lu, H.; Wang, L.; Luo, H.; Chen, X.; Ren, H.; Sun, R.; Liu, W.; et al. Alterations in the human oral and gut microbiomes and lipidomics in COVID-19. Gut 2021, 70, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Delwiche, E.A.; Pestka, J.J.; Tortorello, M.L. The veillonellae: Gram-negative cocci with a unique physiology. Annu. Rev. Microbiol. 1985, 39, 175–193. [Google Scholar] [CrossRef] [PubMed]
- Georges, F.M.; Do, N.T.; Seleem, D. Oral dysbiosis and systemic diseases. Front. Dent. Med. 2022, 3–2022, 1–7. [Google Scholar] [CrossRef]
- Zollner, A.; Koch, R.; Jukic, A.; Pfister, A.; Meyer, M.; Rossler, A.; Kimpel, J.; Adolph, T.E.; Tilg, H. Postacute COVID-19 is Characterized by Gut Viral Antigen Persistence in Inflammatory Bowel Diseases. Gastroenterology 2022, 163, 495–506.e498. [Google Scholar] [CrossRef]
- Peluso, M.J.; Deeks, S.G. Mechanisms of long COVID and the path toward therapeutics. Cell 2024, 187, 5500–5529. [Google Scholar] [CrossRef]
- Doykov, I.; Hallqvist, J.; Gilmour, K.C.; Grandjean, L.; Mills, K.; Heywood, W.E. ‘The long tail of Covid-19’-The detection of a prolonged inflammatory response after a SARS-CoV-2 infection in asymptomatic and mildly affected patients. F1000Res 2020, 9, 1349. [Google Scholar] [CrossRef] [PubMed]
- Inciardi, R.M.; Solomon, S.D.; Ridker, P.M.; Metra, M. Coronavirus 2019 Disease (COVID-19), Systemic Inflammation, and Cardiovascular Disease. J. Am. Heart Assoc. 2020, 9, e017756. [Google Scholar] [CrossRef] [PubMed]
- Guermazi, D.; Guermazi, E. Post-COVID Gut Dysbiosis and Its Role in Persistent Skin Disorders: A Gut–Skin Axis Perspective. COVID 2025, 5, 48. [Google Scholar] [CrossRef]
- Barichello, T.; Kluwe-Schiavon, B.; Borba, L.A.; Pedro, L.C.; Niero, F.S.; Dos Santos, L.N.; Leonardo, L.M.; Ignacio, Z.M.; Morales, R.; Ceretta, L.B.; et al. Alterations in Gut Microbiome Composition and Increased Inflammatory Markers in Post-COVID-19 Individuals. Mol. Neurobiol. 2025, 62, 8038–8047. [Google Scholar] [CrossRef]
- Yang, X.; Xu, H.; Liang, X.; Yuan, G.; Gao, Q.; Tan, X.; Yang, Y.; Xiao, Y.; Huang, Z.; Dai, W.; et al. Exploring the casual association between gut microbiome, circulating inflammatory cytokines and chronic pancreatitis: A Mendelian randomization analysis. Medicine 2024, 103, e37959. [Google Scholar] [CrossRef]
- Deng, G.; Guo, M.; Fan, J.; Wang, W.; Jiang, M.L.; Zhang, C.J. Interleukin-17 family in health and immune diseases: From origin to clinical implications. Neural Regen. Res. 2025. [Google Scholar] [CrossRef]
- Rodrigues, V.F.; Elias-Oliveira, J.; Pereira, I.S.; Pereira, J.A.; Barbosa, S.C.; Machado, M.S.G.; Carlos, D. Akkermansia muciniphila and Gut Immune System: A Good Friendship That Attenuates Inflammatory Bowel Disease, Obesity, and Diabetes. Front. Immunol. 2022, 13, 934695. [Google Scholar] [CrossRef]
- Mussabay, K.; Kozhakhmetov, S.; Dusmagambetov, M.; Mynzhanova, A.; Nurgaziyev, M.; Jarmukhanov, Z.; Vinogradova, E.; Dusmagambetova, A.; Daulbaeva, A.; Chulenbayeva, L.; et al. Gut Microbiome and Cytokine Profiles in Post-COVID Syndrome. Viruses 2024, 16, 722. [Google Scholar] [CrossRef]
- Rohrhofer, J.; Wolflehner, V.; Schweighardt, J.; Koidl, L.; Stingl, M.; Zehetmayer, S.; Seneca, J.; Pjevac, P.; Untersmayr, E. Gastrointestinal Barrier Disruption in Post-COVID Syndrome Fatigue Patients. Allergy 2025, 80, 2610–2621. [Google Scholar] [CrossRef]
- Prucha, M.; Herold, I.; Zazula, R.; Dubska, L.; Dostal, M.; Hildebrand, T.; Hyanek, J. Significance of lipopolysaccharide-binding protein (an acute phase protein) in monitoring critically ill patients. Crit. Care 2003, 7, R154–R159. [Google Scholar] [CrossRef]
- Yin, J.X.; Agbana, Y.L.; Sun, Z.S.; Fei, S.W.; Zhao, H.Q.; Zhou, X.N.; Chen, J.H.; Kassegne, K. Increased interleukin-6 is associated with long COVID-19: A systematic review and meta-analysis. Infect. Dis. Poverty 2023, 12, 43. [Google Scholar] [CrossRef]
- Devaux, C.A.; Lagier, J.C.; Raoult, D. New Insights Into the Physiopathology of COVID-19: SARS-CoV-2-Associated Gastrointestinal Illness. Front. Med. 2021, 8, 640073. [Google Scholar] [CrossRef]
- Wang, B.; Zhang, L.; Wang, Y.; Dai, T.; Qin, Z.; Zhou, F.; Zhang, L. Alterations in microbiota of patients with COVID-19: Potential mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Ji, W.; Cao, X. Integrated analysis of gut microbiome and its metabolites in ACE2-knockout and ACE2-overexpressed mice. Front. Cell Infect. Microbiol. 2024, 14, 1404678. [Google Scholar] [CrossRef]
- De, R.; Dutta, S. Role of the Microbiome in the Pathogenesis of COVID-19. Front. Cell Infect. Microbiol. 2022, 12, 736397. [Google Scholar] [CrossRef] [PubMed]
- Naidu, A.S.; Wang, C.K.; Rao, P.; Mancini, F.; Clemens, R.A.; Wirakartakusumah, A.; Chiu, H.F.; Yen, C.H.; Porretta, S.; Mathai, I.; et al. Precision nutrition to reset virus-induced human metabolic reprogramming and dysregulation (HMRD) in long-COVID. NPJ Sci. Food 2024, 8, 19, Erratum in NPJ Sci. Food 2024, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.; Sarapultsev, A. Exploring the Pathophysiology of Long COVID: The Central Role of Low-Grade Inflammation and Multisystem Involvement. Int. J. Mol. Sci. 2024, 25, 6389. [Google Scholar] [CrossRef]
- Arneth, B.M. Gut-brain axis biochemical signalling from the gastrointestinal tract to the central nervous system: Gut dysbiosis and altered brain function. Postgrad. Med. J. 2018, 94, 446–452. [Google Scholar] [CrossRef]
- Bicknell, B.; Liebert, A.; Borody, T.; Herkes, G.; McLachlan, C.; Kiat, H. Neurodegenerative and Neurodevelopmental Diseases and the Gut-Brain Axis: The Potential of Therapeutic Targeting of the Microbiome. Int. J. Mol. Sci. 2023, 24, 9577. [Google Scholar] [CrossRef] [PubMed]
- Rusch, J.A.; Layden, B.T.; Dugas, L.R. Signalling cognition: The gut microbiota and hypothalamic-pituitary-adrenal axis. Front. Endocrinol. 2023, 14, 1130689. [Google Scholar] [CrossRef] [PubMed]
- Smail, S.W.; Albarzinji, N.; Salih, R.H.; Taha, K.O.; Hirmiz, S.M.; Ismael, H.M.; Noori, M.F.; Azeez, S.S.; Janson, C. Microbiome dysbiosis in SARS-CoV-2 infection: Implication for pathophysiology and management strategies of COVID-19. Front. Cell Infect. Microbiol. 2025, 15, 1537456. [Google Scholar] [CrossRef]
- Nakhal, M.M.; Yassin, L.K.; Alyaqoubi, R.; Saeed, S.; Alderei, A.; Alhammadi, A.; Alshehhi, M.; Almehairbi, A.; Al Houqani, S.; BaniYas, S.; et al. The Microbiota-Gut-Brain Axis and Neurological Disorders: A Comprehensive Review. Life 2024, 14, 1234. [Google Scholar] [CrossRef]
- Lau, R.I.; Su, Q.; Ng, S.C. Long COVID and gut microbiome: Insights into pathogenesis and therapeutics. Gut Microbes 2025, 17, 2457495. [Google Scholar] [CrossRef]
- Baindara, P.; Chakraborty, R.; Holliday, Z.M.; Mandal, S.M.; Schrum, A.G. Oral probiotics in coronavirus disease 2019: Connecting the gut-lung axis to viral pathogenesis, inflammation, secondary infection and clinical trials. New Microbes New Infect. 2021, 40, 100837. [Google Scholar] [CrossRef]
- Ortega, M.A.; Álvarez-Mon, M.A.; García-Montero, C.; Fraile-Martínez, Ó.; Monserrat, J.; Martinez-Rozas, L.; Rodríguez-Jiménez, R.; Álvarez-Mon, M.; Lahera, G. Microbiota-gut-brain axis mechanisms in the complex network of bipolar disorders: Potential clinical implications and translational opportunities. Mol. Psychiatry 2023, 28, 2645–2673. [Google Scholar] [CrossRef]
- Vakili, K.; Fathi, M.; Yaghoobpoor, S.; Sayehmiri, F.; Nazerian, Y.; Nazerian, A.; Mohamadkhani, A.; Khodabakhsh, P.; Réus, G.Z.; Hajibeygi, R.; et al. The contribution of gut-brain axis to development of neurological symptoms in COVID-19 recovered patients: A hypothesis and review of literature. Front. Cell Infect. Microbiol. 2022, 12, 983089. [Google Scholar] [CrossRef] [PubMed]
- Plummer, A.M.; Matos, Y.L.; Lin, H.C.; Ryman, S.G.; Birg, A.; Quinn, D.K.; Parada, A.N.; Vakhtin, A.A. Gut-brain pathogenesis of post-acute COVID-19 neurocognitive symptoms. Front. Neurosci. 2023, 17, 1232480. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, J.; Chen, Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients 2021, 13, 2099. [Google Scholar] [CrossRef]
- Blackett, J.W.; Sun, Y.; Purpura, L.; Margolis, K.G.; Elkind, M.S.V.; O’Byrne, S.; Wainberg, M.; Abrams, J.A.; Wang, H.H.; Chang, L.; et al. Decreased Gut Microbiome Tryptophan Metabolism and Serotonergic Signaling in Patients With Persistent Mental Health and Gastrointestinal Symptoms After COVID-19. Clin. Transl. Gastroenterol. 2022, 13, e00524. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Devotta, H.; Li, J.; Lunjani, N.; Sadlier, C.; Lavelle, A.; Albrich, W.C.; Walter, J.; O’Toole, P.W.; O’Mahony, L. Dysrupted microbial tryptophan metabolism associates with SARS-CoV-2 acute inflammatory responses and long COVID. Gut Microbes 2024, 16, 2429754. [Google Scholar] [CrossRef]
- McMillan, P.; Turner, A.J.; Uhal, B.D. Mechanisms of Gut-Related Viral Persistence in Long COVID. Viruses 2024, 16, 1266. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, M.; Song, P.; Guan, W.; Shen, X. Mucosal-associated invariant T cells in digestive tract: Local guardians or destroyers? Immunology 2023, 170, 167–179. [Google Scholar] [CrossRef]
- Talwar, S.; Harker, J.A.; Openshaw, P.J.M.; Thwaites, R.S. Autoimmunity in long COVID. J. Allergy Clin. Immunol. 2025, 155, 1082–1094. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Saidara, E.; Maes, M. Persistent SARS-CoV-2 Infection, EBV, HHV-6 and Other Factors May Contribute to Inflammation and Autoimmunity in Long COVID. Viruses 2023, 15, 400. [Google Scholar] [CrossRef]
- Marino Gammazza, A.; Legare, S.; Lo Bosco, G.; Fucarino, A.; Angileri, F.; Conway de Macario, E.; Macario, A.J.; Cappello, F. Human molecular chaperones share with SARS-CoV-2 antigenic epitopes potentially capable of eliciting autoimmunity against endothelial cells: Possible role of molecular mimicry in COVID-19. Cell Stress. Chaperones 2020, 25, 737–741. [Google Scholar] [CrossRef]
- Bao, L.; Zhang, C.; Dong, J.; Zhao, L.; Li, Y.; Sun, J. Oral Microbiome and SARS-CoV-2: Beware of Lung Co-infection. Front. Microbiol. 2020, 11, 1840. [Google Scholar] [CrossRef]
- Visvabharathy, L.; Hanson, B.A.; Orban, Z.S.; Lim, P.H.; Palacio, N.M.; Jimenez, M.; Clark, J.R.; Graham, E.L.; Liotta, E.M.; Tachas, G.; et al. Neuro-PASC is characterized by enhanced CD4+ and diminished CD8+ T cell responses to SARS-CoV-2 Nucleocapsid protein. Front. Immunol. 2023, 14, 1155770, Erratum in Front. Immunol. 2023, 14, 1275925. [Google Scholar] [CrossRef]
- Wiertsema, S.P.; van Bergenhenegouwen, J.; Garssen, J.; Knippels, L.M.J. The Interplay between the Gut Microbiome and the Immune System in the Context of Infectious Diseases throughout Life and the Role of Nutrition in Optimizing Treatment Strategies. Nutrients 2021, 13, 886. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587, Erratum in Microorganisms 2020, 8, 2046. [Google Scholar] [CrossRef]
- Drosos, A.A.; Pelechas, E.; Voulgari, P.V. Long COVID from rheumatology perspective: A simple mimicker or promoter of autoimmunity? Clin. Rheumatol. 2022, 41, 957–958. [Google Scholar] [CrossRef]
- Hromic-Jahjefendic, A.; Mahmutovic, L.; Sezer, A.; Becirevic, T.; Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. The intersection of microbiome and autoimmunity in long COVID-19: Current insights and future directions. Cytokine Growth Factor Rev. 2025, 82, 43–54. [Google Scholar] [CrossRef]
- Paranga, T.G.; Mitu, I.; Pavel-Tanasa, M.; Rosu, M.F.; Miftode, I.L.; Constantinescu, D.; Obreja, M.; Plesca, C.E.; Miftode, E. Cytokine Storm in COVID-19: Exploring IL-6 Signaling and Cytokine-Microbiome Interactions as Emerging Therapeutic Approaches. Int. J. Mol. Sci. 2024, 25, 11411. [Google Scholar] [CrossRef]
- van den Bogert, B.; Meijerink, M.; Zoetendal, E.G.; Wells, J.M.; Kleerebezem, M. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS ONE 2014, 9, e114277. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Xia, Y.; Sun, J. Microbiome and intestinal pathophysiology in post-acute sequelae of COVID-19. Genes Dis. 2023, 11, 100978. [Google Scholar] [CrossRef]
- Catalan, E.A.; Seguel-Fuentes, E.; Fuentes, B.; Aranguiz-Varela, F.; Castillo-Godoy, D.P.; Rivera-Asin, E.; Bocaz, E.; Fuentes, J.A.; Bravo, D.; Schinnerling, K.; et al. Oral Pathobiont-Derived Outer Membrane Vesicles in the Oral-Gut Axis. Int. J. Mol. Sci. 2024, 25, 11141. [Google Scholar] [CrossRef]
- Schwartz, J.; Capistrano, K.; Hussein, H.; Hafedi, A.; Shukla, D.; Naqvi, A. Oral SARS-CoV-2 Infection and Risk for Long Covid. Rev. Med. Virol. 2025, 35, e70029. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.; Capistrano, K.J.; Gluck, J.; Hezarkhani, A.; Naqvi, A.R. SARS-CoV-2, periodontal pathogens, and host factors: The trinity of oral post-acute sequelae of COVID-19. Rev. Med. Virol. 2024, 34, e2543. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Schwartz, J.; Brandini, D.A.; Schaller, S.; Hussein, H.; Valverde, A.; Naqvi, R.A.; Shukla, D. COVID-19 and oral diseases: Assessing manifestations of a new pathogen in oral infections. Int. Rev. Immunol. 2022, 41, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Mendes de Almeida, V.; Engel, D.F.; Ricci, M.F.; Cruz, C.S.; Lopes, Í.S.; Alves, D.A.; d’ Auriol, M.; Magalhães, J.; Machado, E.C.; Rocha, V.M.; et al. Gut microbiota from patients with COVID-19 cause alterations in mice that resemble post-COVID symptoms. Gut Microbes 2023, 15, 2249146. [Google Scholar] [CrossRef]
- Clerbaux, L.A.; Fillipovska, J.; Muñoz, A.; Petrillo, M.; Coecke, S.; Amorim, M.J.; Grenga, L. Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways. J. Clin. Med. 2022, 11, 5400. [Google Scholar] [CrossRef]
- Pathak, A.; Agrawal, D.K. Role of Gut Microbiota in Long COVID: Impact on Immune Function and Organ System Health. Arch. Microbiol. Immunol. 2025, 9, 38–53. [Google Scholar] [CrossRef]
- Soranno, D.E.; Coopersmith, C.M.; Brinkworth, J.F.; Factora, F.N.F.; Muntean, J.H.; Mythen, M.G.; Raphael, J.; Shaw, A.D.; Vachharajani, V.; Messer, J.S. A review of gut failure as a cause and consequence of critical illness. Crit. Care 2025, 29, 91. [Google Scholar] [CrossRef]
- Basting, C.M.; Langat, R.; Broedlow, C.A.; Guerrero, C.R.; Bold, T.D.; Bailey, M.; Velez, A.; Schroeder, T.; Short-Miller, J.; Cromarty, R.; et al. SARS-CoV-2 infection is associated with intestinal permeability, systemic inflammation, and microbial dysbiosis in hospitalized patients. Microbiol. Spectr. 2024, 12, e0068024. [Google Scholar] [CrossRef]
- Manor, O.; Dai, C.L.; Kornilov, S.A.; Smith, B.; Price, N.D.; Lovejoy, J.C.; Gibbons, S.M.; Magis, A.T. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 2020, 11, 5206. [Google Scholar] [CrossRef]
- Kandasamy, S.; Letchumanan, V.; Hong, K.W.; Chua, K.-O.; Mutalib, N.; Ng, A.L.O.; Ming, L.C.; Lim, H.; Thurairajasingam, S.; Law, J.; et al. The Role of Human Gut Microbe Ruminococcus gnavus in Inflammatory Diseases. Prog. Progress. Microbes Mol. Biol. 2023, 6, a0000396. [Google Scholar] [CrossRef]
- Crost, E.H.; Coletto, E.; Bell, A.; Juge, N. Ruminococcus gnavus: Friend or foe for human health. FEMS Microbiol. Rev. 2023, 47, fuad014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jian, Y.P.; Zhang, Y.N.; Li, Y.; Gu, L.T.; Sun, H.H.; Liu, M.D.; Zhou, H.L.; Wang, Y.S.; Xu, Z.X. Short-chain fatty acids in diseases. Cell Commun. Signal 2023, 21, 212. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Weiss, S.T.; Liu, Y.Y. Dissecting the role of the human microbiome in COVID-19 via metagenome-assembled genomes. Nat. Commun. 2022, 13, 5235. [Google Scholar] [CrossRef] [PubMed]
- Cyprian, F.; Sohail, M.U.; Abdelhafez, I.; Salman, S.; Attique, Z.; Kamareddine, L.; Al-Asmakh, M. SARS-CoV-2 and immune-microbiome interactions: Lessons from respiratory viral infections. Int. J. Infect. Dis. 2021, 105, 540–550. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Shankar, E.M. SARS-CoV-2-Indigenous Microbiota Nexus: Does Gut Microbiota Contribute to Inflammation and Disease Severity in COVID-19? Front. Cell Infect. Microbiol. 2021, 11, 590874. [Google Scholar] [CrossRef]
- Rossini, V.; Tolosa-Enguis, V.; Frances-Cuesta, C.; Sanz, Y. Gut microbiome and anti-viral immunity in COVID-19. Crit. Rev. Food Sci. Nutr. 2024, 64, 4587–4602. [Google Scholar] [CrossRef]
- Brīvība, M.; Silamiķele, L.; Birzniece, L.; Ansone, L.; Megnis, K.; Silamiķelis, I.; Pelcmane, L.; Borisova, D.; Rozenberga, M.; Jagare, L.; et al. Gut Microbiome Composition and Dynamics in Hospitalized COVID-19 Patients and Patients with Post-Acute COVID-19 Syndrome. Int. J. Mol. Sci. 2024, 25, 567. [Google Scholar] [CrossRef] [PubMed]
- Meringer, H.; Mehandru, S. Gastrointestinal post-acute COVID-19 syndrome. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 345–346. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.X.; Khalid, K.; Abdullah, A.D.I.; Lee, L.H.; Raja Ali, R.A. Subphenotypes of Long COVID and the clinical applications of probiotics. Biomed. Pharmacother. 2025, 183, 117855. [Google Scholar] [CrossRef]
- Raman, B.; Ramasamy, M.N. Synbiotics in post-acute COVID-19 syndrome-a potential new treatment framework? Lancet Infect. Dis. 2024, 24, 219–221. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, L.; Lin, W.; Tang, W.; Chan, F.K.L.; Ng, S.C. Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci. Technol. 2021, 108, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Moreno, N.; Gomez-Sanchez, L.; Navarro-Caceres, A.; Arroyo-Romero, S.; Dominguez-Martin, A.; Lugones-Sanchez, C.; Tamayo-Morales, O.; Gonzalez-Sanchez, S.; Castro-Rivero, A.B.; Rodriguez-Sanchez, E.; et al. Association of Mediterranean Diet with Cardiovascular Risk Factors and with Metabolic Syndrome in Subjects with Long COVID: BioICOPER Study. Nutrients 2025, 17, 656. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; de la, O.V.; Higuera-Gomez, A.; Chero-Sandoval, L.; de Cuevillas, B.; Martinez-Urbistondo, M.; Moreno-Torres, V.; Pintos-Pascual, I.; Castejon, R.; Martinez, J.A. Mediterranean Diet and Olive Oil Redox Interactions on Lactate Dehydrogenase Mediated by Gut Oscillibacter in Patients with Long-COVID-19 Syndrome. Antioxidants 2024, 13, 1358. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Rodriguez-Quintero, C.M.; Hromic-Jahjefendic, A.; Uversky, V.N.; Redwan, E.M.; Brogna, C. The essential role of prebiotics in restoring gut health in long COVID. Prog. Mol. Biol. Transl. Sci. 2025, 213, 385–411. [Google Scholar] [CrossRef] [PubMed]
- Song, I.; Yang, J.; Saito, M.; Hartanto, T.; Nakayama, Y.; Ichinohe, T.; Fukuda, S. Prebiotic inulin ameliorates SARS-CoV-2 infection in hamsters by modulating the gut microbiome. NPJ Sci. Food 2024, 8, 18. [Google Scholar] [CrossRef]
- Thomas, R.; Williams, M.; Aldous, J.; Yanagisawa, Y.; Kumar, R.; Forsyth, R.; Chater, A. A Randomised, Double-Blind, Placebo-Controlled Trial Evaluating Concentrated Phytochemical-Rich Nutritional Capsule in Addition to a Probiotic Capsule on Clinical Outcomes among Individuals with COVID-19—The UK Phyto-V Study. COVID 2022, 2, 433–449. [Google Scholar]
- Suzuki, T.; Takahashi, J.; Yamamoto, M. Molecular Basis of the KEAP1-NRF2 Signaling Pathway. Mol. Cells 2023, 46, 133–141. [Google Scholar] [CrossRef]
- Marín-Palma, D.; Tabares-Guevara, J.H.; Zapata-Cardona, M.I.; Flórez-Álvarez, L.; Yepes, L.M.; Rugeles, M.T.; Zapata-Builes, W.; Hernandez, J.C.; Taborda, N.A. Curcumin Inhibits In Vitro SARS-CoV-2 Infection In Vero E6 Cells through Multiple Antiviral Mechanisms. Molecules 2021, 26, 6900. [Google Scholar] [CrossRef]
- Nittayananta, W.; Lerdsamran, H.; Chutiwitoonchai, N.; Promsong, A.; Srichana, T.; Netsomboon, K.; Prasertsopon, J.; Kerdto, J. A novel film spray containing curcumin inhibits SARS-CoV-2 and influenza virus infection and enhances mucosal immunity. Virol. J. 2024, 21, 26. [Google Scholar] [CrossRef]
- Nicoliche, T.; Bartolomeo, C.S.; Lemes, R.M.R.; Pereira, G.C.; Nunes, T.A.; Oliveira, R.B.; Nicastro, A.L.M.; Soares, É.N.; da Cunha Lima, B.F.; Rodrigues, B.M.; et al. Antiviral, anti-inflammatory and antioxidant effects of curcumin and curcuminoids in SH-SY5Y cells infected by SARS-CoV-2. Sci. Rep. 2024, 14, 10696. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Do, V.M.; Phan, T.T.; Nguyen Huynh, D.T. The Potential of Ameliorating COVID-19 and Sequelae From Andrographis paniculata via Bioinformatics. Bioinform. Biol. Insights 2023, 17, 11779322221149622. [Google Scholar] [CrossRef] [PubMed]
- Periferakis, A.; Periferakis, A.T.; Troumpata, L.; Periferakis, K.; Scheau, A.E.; Savulescu-Fiedler, I.; Caruntu, A.; Badarau, I.A.; Caruntu, C.; Scheau, C. Kaempferol: A Review of Current Evidence of Its Antiviral Potential. Int. J. Mol. Sci. 2023, 24, 16299. [Google Scholar] [CrossRef] [PubMed]
- Periferakis, A.; Periferakis, A.T.; Troumpata, L.; Periferakis, K.; Georgatos-Garcia, S.; Touriki, G.; Dragosloveanu, C.D.M.; Caruntu, A.; Savulescu-Fiedler, I.; Dragosloveanu, S.; et al. Pinosylvin: A Multifunctional Stilbenoid with Antimicrobial, Antioxidant, and Anti-Inflammatory Potential. Curr. Issues Mol. Biol. 2025, 47, 204. [Google Scholar] [CrossRef]
- Bodke, H.; Jogdand, S. Role of Probiotics in Human Health. Cureus 2022, 14, e31313. [Google Scholar] [CrossRef]
- Ranisavljev, M.; Stajer, V.; Todorovic, N.; Ostojic, J.; Cvejic, J.H.; Steinert, R.E.; Ostojic, S.M. The effects of 3-month supplementation with synbiotic on patient-reported outcomes, exercise tolerance, and brain and muscle metabolism in adult patients with post-COVID-19 chronic fatigue syndrome (STOP-FATIGUE): A randomized Placebo-controlled clinical trial. Eur. J. Nutr. 2024, 64, 28, Erratum in Eur. J. Nutr. 2025, 64, 80. [Google Scholar] [CrossRef]
- Santinelli, L.; Laghi, L.; Innocenti, G.P.; Pinacchio, C.; Vassalini, P.; Celani, L.; Lazzaro, A.; Borrazzo, C.; Marazzato, M.; Tarsitani, L.; et al. Oral Bacteriotherapy Reduces the Occurrence of Chronic Fatigue in COVID-19 Patients. Front. Nutr. 2021, 8, 756177. [Google Scholar] [CrossRef]
- Nilsen, M.S.; Jersin, R.A.; Ulvik, A.; Madsen, A.; McCann, A.; Svensson, P.A.; Svensson, M.K.; Nedrebo, B.G.; Gudbrandsen, O.A.; Tell, G.S.; et al. 3-Hydroxyisobutyrate, A Strong Marker of Insulin Resistance in Type 2 Diabetes and Obesity That Modulates White and Brown Adipocyte Metabolism. Diabetes 2020, 69, 1903–1916. [Google Scholar] [CrossRef] [PubMed]
- Hamamah, S.; Gheorghita, R.; Lobiuc, A.; Sirbu, I.O.; Covasa, M. Fecal microbiota transplantation in non-communicable diseases: Recent advances and protocols. Front. Med. 2022, 9, 1060581. [Google Scholar] [CrossRef]
- Cha, R.R.; Sonu, I. Fecal microbiota transplantation: Present and future. Clin. Endosc. 2025, 58, 352–359. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; He, L.; Xu, X.; Piao, M.; Wang, B.; Liu, T.; Cao, H. Gut microbiota in post-acute COVID-19 syndrome: Not the end of the story. Front. Microbiol. 2024, 15, 1500890. [Google Scholar] [CrossRef] [PubMed]
- Zollner, A.; Meyer, M.; Jukic, A.; Adolph, T.; Tilg, H. The Intestine in Acute and Long COVID: Pathophysiological Insights and Key Lessons. Yale J. Biol. Med. 2024, 97, 447–462. [Google Scholar] [CrossRef]
- Jiang, X.; Gao, X.; Ding, J.; Pang, B.; Pei, Y.; Zhao, Z.; Zhao, N.; Wang, Z.; Chen, C.; Gao, D.; et al. Fecal microbiota transplantation alleviates mild-moderate COVID-19 associated diarrhoea and depression symptoms: A prospective study of a randomized, double-blind clinical trial. J. Med. Virol. 2024, 96, e29812. [Google Scholar] [CrossRef]
- Fallah, A.; Sedighian, H.; Kachuei, R.; Fooladi, A.A.I. Human microbiome in post-acute COVID-19 syndrome (PACS). Curr. Res. Microb. Sci. 2025, 8, 100324. [Google Scholar] [CrossRef]
- Scalzo, P.L.; Marshall, A.G.; Soriano, S.; Curry, K.; Dulay, M.; Hodics, T.; Quigley, E.M.M.; Treangen, T.J.; Piskorz, M.M.; Villapol, S. Gut Microbiome dysbiosis and immune activation correlate with somatic and neuropsychiatric symptoms in COVID-19 patients. J. Transl. Med. 2025, 23, 327. [Google Scholar] [CrossRef]
- Vidal, A.; Barrows, B.; Bao, G.; Goudzwaard, A.; Papoutsis, A.; Hazan, S. S234 COVID-19 Long COVID and Gut Dysbiosis. Am. J. Gastroenterol. 2024, 119, S170. [Google Scholar] [CrossRef]
- Bakhsh, M.A.; Khawandanah, J.; Naaman, R.K.; Alashmali, S. The impact of COVID-19 quarantine on dietary habits and physical activity in Saudi Arabia: A cross-sectional study. BMC Public Health 2021, 21, 1487. [Google Scholar] [CrossRef] [PubMed]
- Mohammed Salah Gabal, H.-A.; Manzour, A.F. Lifestyle and eating habits changes among adults during COVID-19 era in Egypt: A population-based study. BMC Nutr. 2024, 10, 52. [Google Scholar] [CrossRef]
- Ahsan, K.; Anwar, M.A.; Munawar, N. Gut microbiome therapeutic modulation to alleviate drug-induced hepatic damage in COVID-19 patients. World J. Gastroenterol. 2023, 29, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Narayanan, A.; Giske, C.G.; Neogi, U.; Sonnerborg, A.; Nowak, P. Altered Gut Microbiome under Antiretroviral Therapy: Impact of Efavirenz and Zidovudine. ACS Infect. Dis. 2021, 7, 1104–1115. [Google Scholar] [CrossRef]
- Biazzo, M.; Deidda, G. Fecal Microbiota Transplantation as New Therapeutic Avenue for Human Diseases. J. Clin. Med. 2022, 11, 4119. [Google Scholar] [CrossRef]
- Qiu, Y.; Mo, C.; Chen, L.; Ye, W.; Chen, G.; Zhu, T. Alterations in microbiota of patients with COVID-19: Implications for therapeutic interventions. MedComm 2020 2024, 5, e513. [Google Scholar] [CrossRef]
- Ceban, F.; Ling, S.; Lui, L.M.W.; Lee, Y.; Gill, H.; Teopiz, K.M.; Rodrigues, N.B.; Subramaniapillai, M.; Di Vincenzo, J.D.; Cao, B.; et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain Behav. Immun. 2022, 101, 93–135. [Google Scholar] [CrossRef] [PubMed]
- Bull-Otterson, L.; Baca, S.; Saydah, S.; Boehmer, T.; Adjei, S.; Gray, S.; Harris, A. Post–COVID Conditions Among Adult COVID-19 Survivors Aged 18–64 and ≥65 Years—United States, March 2020–November 2021. Morb. Mortal. Wkly. Rep. 2022, 71, 713–717. [Google Scholar] [CrossRef]
- Leviner, S. Recognizing the Clinical Sequelae of COVID-19 in Adults: COVID-19 Long-Haulers. J. Nurse Pr. Pract. 2021, 17, 946–949. [Google Scholar] [CrossRef] [PubMed]
| Year (Ref.) | Sample (Design) | N | Study Country | Time Since Infection (Months) | Sequencing Method | Microbiome Focus | Key Taxa Alterations | Clinical Outcomes | Main Findings | Limitation |
|---|---|---|---|---|---|---|---|---|---|---|
| 2025 Blankestijn et al. [29] | Post-COVID-19 clinic cohort 3–6 months post-infection; fecal metagenomic sequencing with clustering analysis | 79 | Netherlands | 3–6 | Shotgun metagenomics | Gut | ↓ F. prausnitzii, Eubacterium spp.; ↑ R. gnavus, E. coli, Veillonella, Streptococcus | Pulmonary function (FEV1, FVC, DLCO); severity correlation | Patients with dysbiotic microbiota had poorer lung recovery; dysbiosis linked to severe acute COVID-19 history. | Small single-country study; observational; confounders (diet, meds) not fully controlled. |
| 2024 Su et al. [30] | Multi-cohort machine learning study predicting PACS phenotypes from microbiome data | Several hundred | China (Hong Kong) | 3–6 | Shotgun metagenomics | Gut | Enterotypes: one depleted in butyrate-producers, enriched in R. gnavus, E. coli | PACS symptom clusters (respiratory, neuro, GI) | Gut enterotypes predicted symptom type with ~89% accuracy; microbiome heterogeneity underpins symptom diversity. | Complex model; regional limitation; predictive, not causal. |
| 2024 Lau et al. [31] | Non-randomized open-label trial: 30 received multi-donor FMT (capsules), 30 received control | 60 | China (Hong Kong) | 9 | Shotgun metagenomics | Gut | ↑ Gemmiger formicilis, ↑ donor-like microbiota composition | Insomnia, fatigue, anxiety, cortisol levels | FMT improved sleep quality and reduced fatigue/anxiety; safe with no serious AEs. | Open-label, moderate N, short follow-up; limited generalizability. |
| 2023 Lau et al. [32] | RCT: Long COVID patients randomized 1:1 to synbiotic (SIM01) vs. placebo for 6 months | 463 | China (Hong Kong) | 4–10 | Shotgun metagenomics | Gut | ↑ Bifidobacterium adolescentis, SCFA-producers; ↑ diversity | Symptom improvement (fatigue, memory loss, GI issues, well-being) | Synbiotic improved symptoms and microbiome diversity; microbiome correlated with recovery. | Subjective outcomes; single-center; no objective function gains. |
| 2023 Zhang et al. [13] | Recovered patients 1 year post-hospitalization (84 long COVID, 103 recovered, 32 controls) | 219 | China (Wuhan) | 12 | 16S rRNA sequencing | Gut | ↓ Eubacterium hallii, Subdoligranulum, Ruminococcus, Agathobacter; ↑ Veillonella | Presence of long COVID symptoms at 12 months | Long COVID associated with reduced diversity and SCFA-producing genera; dysbiosis persisted ≥1 year. | Cross-sectional; single region; no diet control; association only. |
| 2022 Liu et al. [17] | Prospective cohort followed from diagnosis to 6 months; pre-pandemic controls included | 174 | China (Hong Kong) | 6 | Shotgun metagenomics | Gut | ↓ Faecalibacterium, Bifidobacterium; ↑ R. gnavus, B. vulgatus | Persistence of ≥1 symptom at 6 months (fatigue, respiratory, GI, neuro) | Baseline microbiota predicted long COVID; persistent dysbiosis at 6 months in long COVID. | Single-region cohort; no causal inference; did not adjust for acute severity. |
| 2021 Haran et al. [33] | Adult COVID-19 outpatients; tongue swabs collected during acute illness; followed until symptom resolution (~37% developed long COVID) | 27 | USA | 0–3 | Shotgun metagenomics | Oral | ↑ Prevotella, Veillonella | Symptom duration; presence of long COVID | Oral dysbiosis linked to prolonged inflammation; microbiota resembled chronic fatigue syndrome. | Small sample; no uninfected controls; observational study. |
| 2020 Zuo et al. [9] | 15 COVID-19 in-patients with varying severity; 15 uninfected controls; 6 pneumonia controls; longitudinal stool sampling during hospitalization | 36 | China (Hong Kong) | 0–1 | Shotgun metagenomics | Gut | ↑ Coprobacillus, C. ramosum, C. hathewayi; ↓ F. prausnitzii | Acute COVID-19 severity; microbiome persistence post-clearance | Gut dysbiosis persisted from admission to discharge; opportunistic pathogens correlated with severity. | Small hospitalized cohort; no follow-up in mild/asymptomatic cases. |
| Year (Ref.) | Sample (Design) | Study Subjects | Pathophysiologic Mechanism | Main Findings | Limitations |
|---|---|---|---|---|---|
| 2025 Barichello et al. [76] | Cross-Sectional Study | Humans | Inflammation | Significant increases in MPIF-1, IL-1 and triglycerides in long-COVID19 patients. β-diversity was reduced, including decreased abundance of Akkermansia spp. No differences were observed in α-diversity data | Could not identify species in 16S rRNA analysis Cognitive assessment 3–4 weeks post-COVID-19 may be suboptimal Findings may be specific to study population or methodology Akkermansia depletion not consistently reported in other studies Psychiatric symptom assessment influenced by multifactorial factors |
| 2025 Rohrhofer et al. [81] | Prospective Observational Study | Humans | Intestinal Barrier Disruption | Significant associations were present between gastrointestinal and neuropsychiatric symptoms in long COVID. In the post-acute phase, patients showed higher LBP/sCD14, lower IL-33 and higher IL-6 levels, indicating a proinflammatory state and intestinal barrier disruption | Low sample size Self-reported data |
| 2024 Mussabay et al. [80] | Prospective Cohort Study | Humans | Inflammation | Severe COVID19, complicated by pneumonia, increased presence of proinflammatory bacterial species. In their post-acute phase various cytokines and chemokines such as MDC, IL-1b, TNF-α, FGF-2, EGF, IL-1RA, IFN-α, IL-10, sCD40L, IL-8, IL-12p40 and MIP-1b displayed a proinflammatory profile | Relatively small sample size (n = 60) Included patients that had severe COVID19 disease only |
| 2024 Song et al. [86] | Experimental Study | Murine | ACE2 | ACE2 knockout mice has increase inflammatory bacterial genera including Deferribacteres, Parasutterella, Catenibacterium, Anaerotruncus, with concomitant decreases in SCFA-producing bacteria. Contrarily, ACE2-overexpression enhanced concentrations of SCFA-producing bacteria such as Lactobacillus, Bifidobacterium, Alisipes, etc. | Study was performed in murine models and not in humans |
| 2024 Yao et al. [102] | Experimental (Shotgun Metagenomic) with a Meta-Analysis | Human | Neurotransmitters Inflammation | Serum levels of tryptophan and IPA were negatively correlated with inflammatory markers such as circulating cytokines, while C-Trp, ILA and IAA were positively correlated with proinflammatory markers in long COVID patients. Metagenomics of microbiota showed reduction in enzymes involved in tryptophan metabolism in hospitalized patients. Microbiota-derived tryptophan metabolites modified TH1 and TH17 associated cytokine responses and reduced innate cell proinflammatory responses to TLR3 and TLR4 | Only hospitalized patients included Confounding factors such as diet, medications, comorbidities and lifestyle were not adjusted for |
| 2023 Visvabharathy et al. [109] | Observational, Cross-Sectional Study | Humans | Adaptive Immune Response | Patients with neurological symptoms in the post-acute COVID19 phase have elevated CD4 T cell response and reduced CD8 activation. CD8 T cell production of IL-6 heightened severity of neurologic symptoms | Small sample size Unable to control for time of sample collection with respect to date of COVID19 symptom onset |
| 2022 Blackett et al. [101] | Randomized Control Trial | Human | Neurotransmitter Regulation | Gut microbiome L-tryptophan synthesis was decreased in patients with more severe gastrointestinal symptoms in acute COVID19. Similar biosynthesis pathways of tryptophan biosynthesis were also decreased in those with severe mental health symptoms in the post-acute phase. | Low sample size 5-HT concentrations were obtained postprandially 2 different cohorts were designed for separate studies, with differences in methods for self-reported mental health symptoms |
| 2021 Haran et al. [33] | Prospective Cohort Study | Humans | Oral Microbiota Inflammation | Patients with prolonged COVID19 symptoms and progression to long COVID had higher abundance of proinflammatory microbiota including Prevotella and Veillonella. The oral microbiome in long COVID patients were similar to those with chronic fatigue syndrome | Sample size |
| 2020 Gammazza et al. [107] | Bioinformatics Study | Computational | Molecular Mimicry | HSPs, which are considered human molecular chaperones, participate in molecular mimicry following COVID19 infection. Post-translational modifications can cause autoimmune endothelial damage in the post-acute phase | Hypothesis generated study Scanned for exact peptides No tissue confirmation |
| Category/Biomarker Type | Representative Studies (Year, Ref.) | Key Microbial Findings/Taxa or Functions | Potential Clinical or Diagnostic Relevance | Mechanistic or Functional Basis | Limitations/Validation Needs |
|---|---|---|---|---|---|
| Reduced SCFA Producers | Zhang et al., 2023 [13]; Liu et al., 2022 [17]; Zuo et al., 2020 [9]; Yeoh et al., 2021 [8] | ↓ Faecalibacterium, Eubacterium, Subdoligranulum, Anaerostipes, Bifidobacterium spp.; reduced fecal butyrate levels and SCFA synthesis genes. | Low SCFA-producer abundance may signal increased risk of long COVID or delayed recovery; stool SCFA quantification could aid monitoring. | Loss of butyrate-producing bacteria → reduced anti-inflammatory and mucosal healing capacity → chronic inflammation and barrier dysfunction. | Observed in multiple cohorts but not specific to COVID; further validation in prospective studies needed. |
| Enriched Pathobionts | Haran et al., 2021 [33]; Liu et al., 2022 [17]; Su et al., 2024 [30] | ↑ Ruminococcus gnavus, Bacteroides vulgatus (gut); ↑ Prevotella, Veillonella (oral); enrichment of proinflammatory Gram-negative species. | High abundance of these taxa correlates with persistent fatigue, GI, and neurological symptoms; may indicate ongoing mucosal inflammation. | Pathobiont overgrowth → LPS release → immune activation via TLR pathways; cytokine production (IL-6, IL-1β, TNF-α). | Requires cross-validation; ratios like R. gnavus, F. prausnitzii may improve specificity. |
| Microbiome Diversity Index | Zhang et al., 2023 [13]; Liu et al., 2022 [17]; Yeoh et al., 2021 [8]; Haran et al., 2021 [33] | Reduced alpha diversity (Chao1, Shannon, Simpson indices) in long COVID vs. recovered or control groups. | Alpha diversity metrics may serve as global indicators of gut health and recovery trajectory after COVID-19. | Lower diversity reflects ecosystem instability and loss of protective taxa; correlates with immune dysregulation and persistent symptoms. | Requires standardization; diversity thresholds vary by population and sequencing platform. |
| Specific Species or Functional Genes | Liu et al., 2022 [17]; Lau et al., 2023 [32]; Su et al., 2024 [30] | ↑ Clostridium innocuum, Actinomyces naeslundii, R. gnavus; ↓ SCFA-related genes; ↑ genes for LPS and antibiotic resistance pathways. | Specific species and gene profiles may stratify long COVID subtypes (fatigue-dominant vs. neurocognitive). | Functional shift toward proinflammatory and oxidative pathways; depletion of metabolic and barrier-supportive functions. | Functional gene panels are promising but require longitudinal validation and standard bioinformatics pipelines. |
| Combined Microbiome Signatures | Haran et al., 2021 [33]; Su et al., 2024 [30]; Liu et al., 2022 [17] | Integrated microbial signatures combining taxa (e.g., Prevotella: Bifidobacterium ratio) and metabolic pathways via machine learning classifiers. | Predictive models could identify high-risk patients early; basis for stool- or saliva-based diagnostic assays. | Multi-omics integration (metagenomics + metabolomics + transcriptomics) enhances specificity for long COVID risk prediction. | Still experimental; reproducibility across populations and sequencing platforms remains unproven. |
| Year (Ref.) | Sample (Design) | N | Microbiome-Targeted Therapy | Main Findings | Limitations |
|---|---|---|---|---|---|
| 2025 Suarez-Morena et al. [139] | Cross-Sectional Study | 305 | Mediterranean Diet | Introduction of a Mediterranean diet in patients with Long COVID improved waist circumference and improved metabolic parameters | Sample size was predominately women and may not be generalizable to men Adherence was gathered through questionnaires |
| 2024 Cuevas-Sierra et al. [140] | Prospective Cohort Study | 188 | Mediterranean Diet | High adherence to the Mediterranean diet improved inflammatory and oxidative markers. Adherence was also correlated with reduction in LDL-cholesterol and glucose levels. These changes were accompanied by lower Oscillobacter concentrations in high adherence groups which was related to oxidative markers. | Relatively small sample size Only some components of the Mediterranean Diet were used (i.e., olive oil) |
| 2024 Song et al. [142] | Experimental | 19 | Prebiotics (Inulin) | Inulin supplementation improved concentrations of fecal SCFAs and secondary bile acids in the setting of COVID19 | Experiment was performed in murine models Small sample size |
| 2024 Lau et al. [32] | Double-Blind Placebo-Controlled Trial | 463 | Synbiotics (SIM01) | Six-month supplementation with a Bifidobacterium-based synbiotic significantly improved fatigue, mental fog, and gastrointestinal symptoms in long COVID patients compared to placebo | Difficult to assess based on severity of initial COVID-19 infection |
| 2024 Ranisavljev et al. [152] | Double-Blind Placebo-Controlled Trial | 26 | Synbiotic | Three-month supplementation of synbiotics and placebo improved fatigue in Long COVID. Synbiotics attenuated post-exercise malaise. Synbiotics increased choline levels within the thalamus | Sample limited to young-to-middle-aged adults Vaccination status, time since infection, and dietary factors not considered Multi-strain synbiotic and placebo may have confounded effects Small sample size; short 3-month follow-up Limited mechanistic biomarker assessment |
| 2024 Lau et al. [31] | Pilot Study | 30 | Fecal Microbiota Transplant | After 12 weeks, following FMT, the treatment group exhibited decreased symptoms associated with insomnia and anxiety. Serum cortisol levels were also lower. Depletion of microbial species that produce harmful derivatives were observed and microbiota was similar to donors at the study endpoint. | Low sample size |
| 2024 Jiang et al. [159] | Double-Blind Placebo-Controlled Trial | 40 | Fecal Microbiota Transplant | FMT showed improvements in diarrhea, depression and neuropsychiatric symptoms in COVID-19. Serum AST/ALT ratio was reduced following FMT | Low sample size |
| 2022 Santinelli et al. [153] | Retrospective Observational Study | 58 | Probiotics | Probiotic supplementation significantly lowered proportion of COVID-19 patients that reported fatigue. Concentrations of serum amino acids were increased and harmful microbiota byproducts were reduced | Lack of randomization |
| 2022 Thomas et al. [143] | Double-Blind Placebo-Controlled Trial | 147 | Phytochemical and Probiotics | Addition of phytochemical two-fold reduction in fatigue, three-fold reduction in cough, and two-fold improvement in quality-of-life scores, compared to probiotics alone | Non-randomization of the probiotic element within the study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caliman-Sturdza, O.A.; Hamamah, S.; Iatcu, O.C.; Lobiuc, A.; Bosancu, A.; Covasa, M. Microbiome and Long COVID-19: Current Evidence and Insights. Int. J. Mol. Sci. 2025, 26, 10120. https://doi.org/10.3390/ijms262010120
Caliman-Sturdza OA, Hamamah S, Iatcu OC, Lobiuc A, Bosancu A, Covasa M. Microbiome and Long COVID-19: Current Evidence and Insights. International Journal of Molecular Sciences. 2025; 26(20):10120. https://doi.org/10.3390/ijms262010120
Chicago/Turabian StyleCaliman-Sturdza, Olga A., Sevag Hamamah, Oana C. Iatcu, Andrei Lobiuc, Anca Bosancu, and Mihai Covasa. 2025. "Microbiome and Long COVID-19: Current Evidence and Insights" International Journal of Molecular Sciences 26, no. 20: 10120. https://doi.org/10.3390/ijms262010120
APA StyleCaliman-Sturdza, O. A., Hamamah, S., Iatcu, O. C., Lobiuc, A., Bosancu, A., & Covasa, M. (2025). Microbiome and Long COVID-19: Current Evidence and Insights. International Journal of Molecular Sciences, 26(20), 10120. https://doi.org/10.3390/ijms262010120






