Predicting Antibiotic Resistance in Listeria monocytogenes from Food and Food-Processing Environments Using Next-Generation Sequencing: A Systematic Review
Abstract
1. Introduction
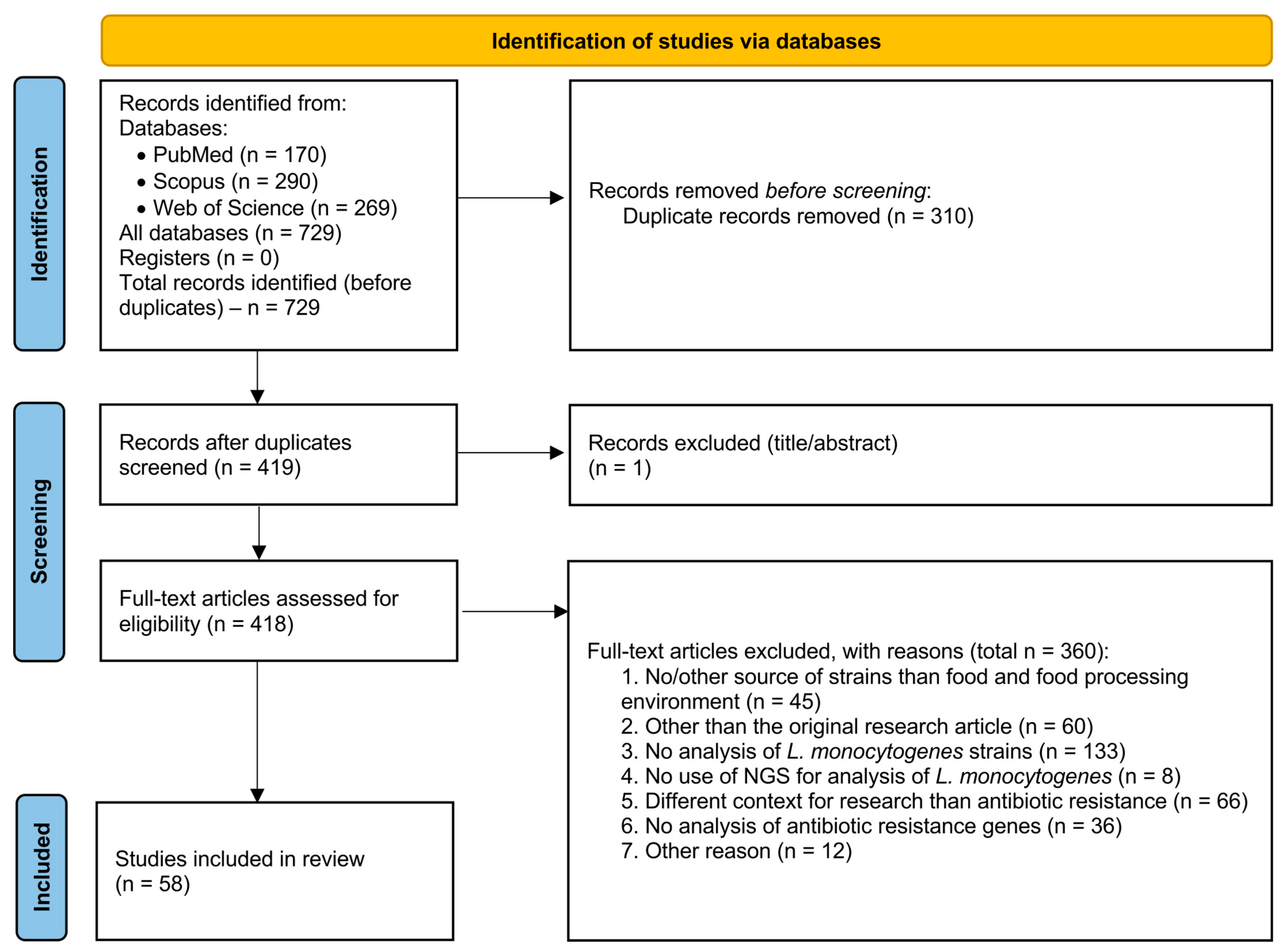
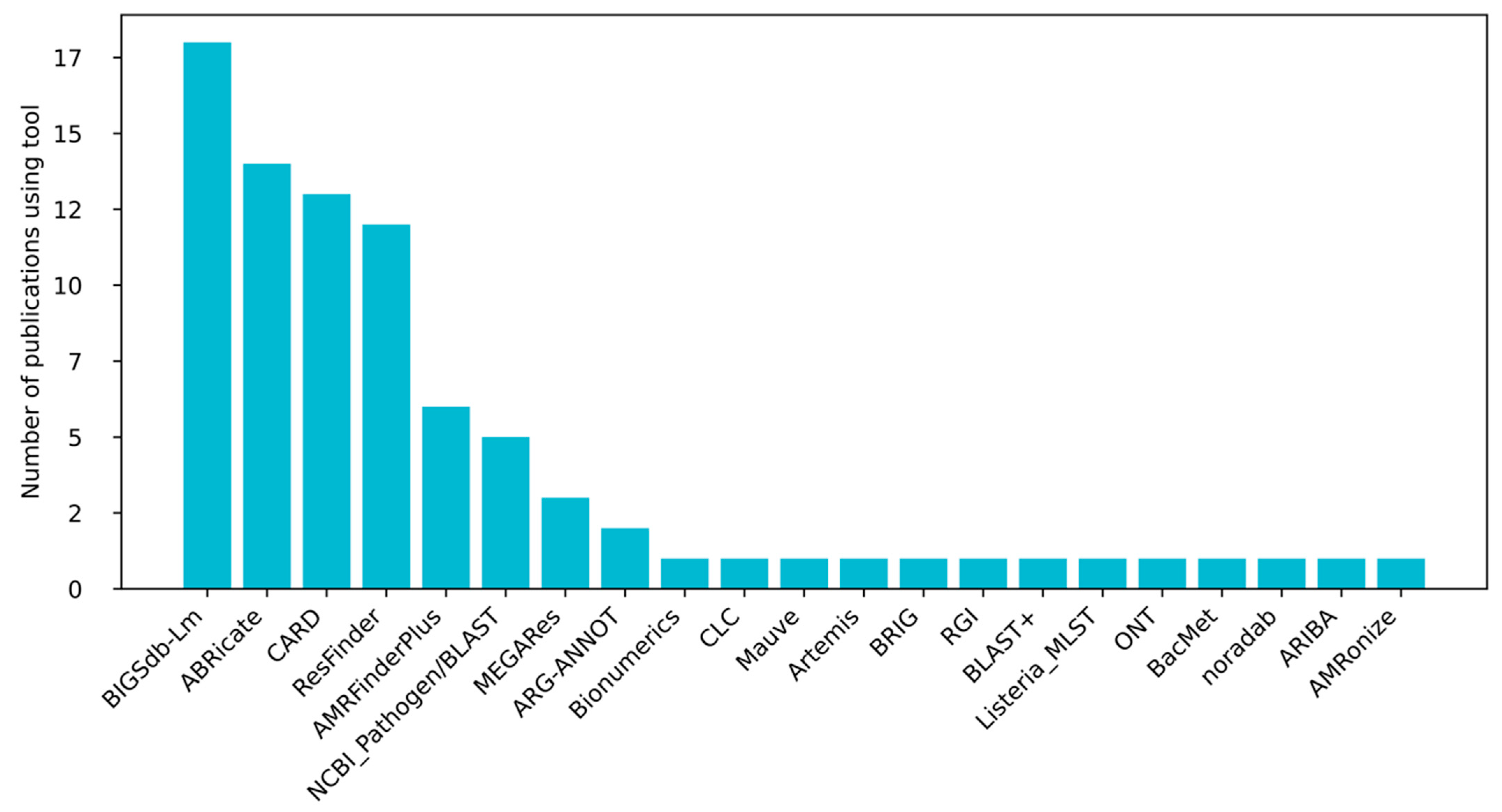
2. Methods
3. Results and Discussion
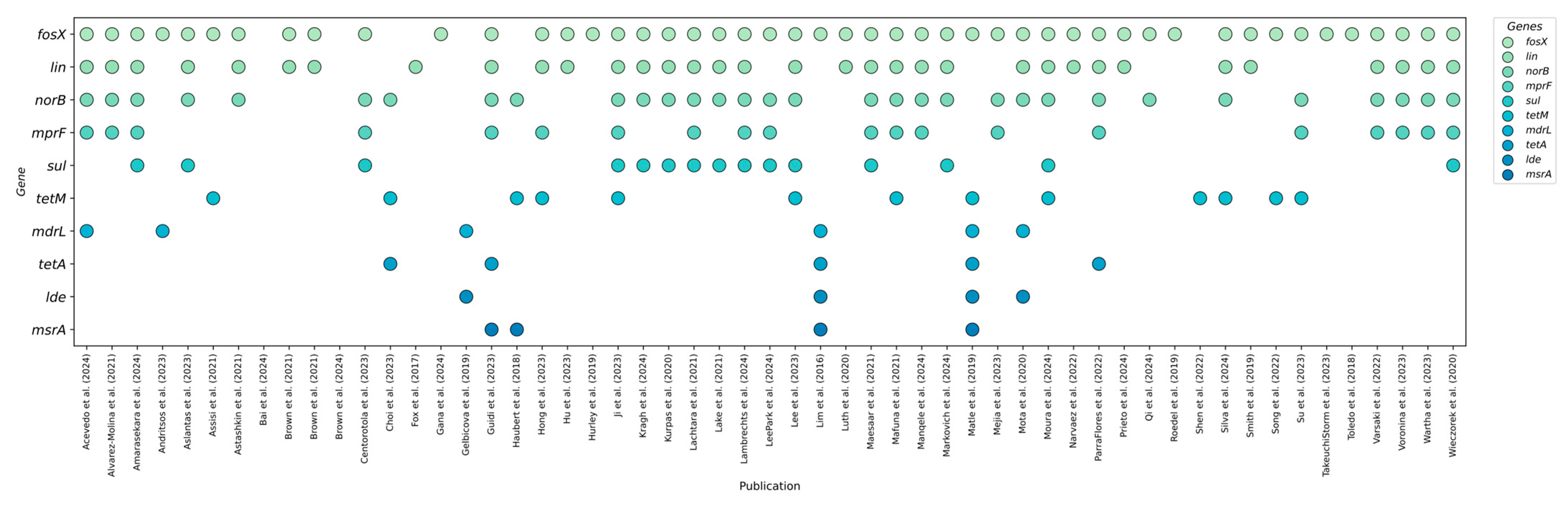
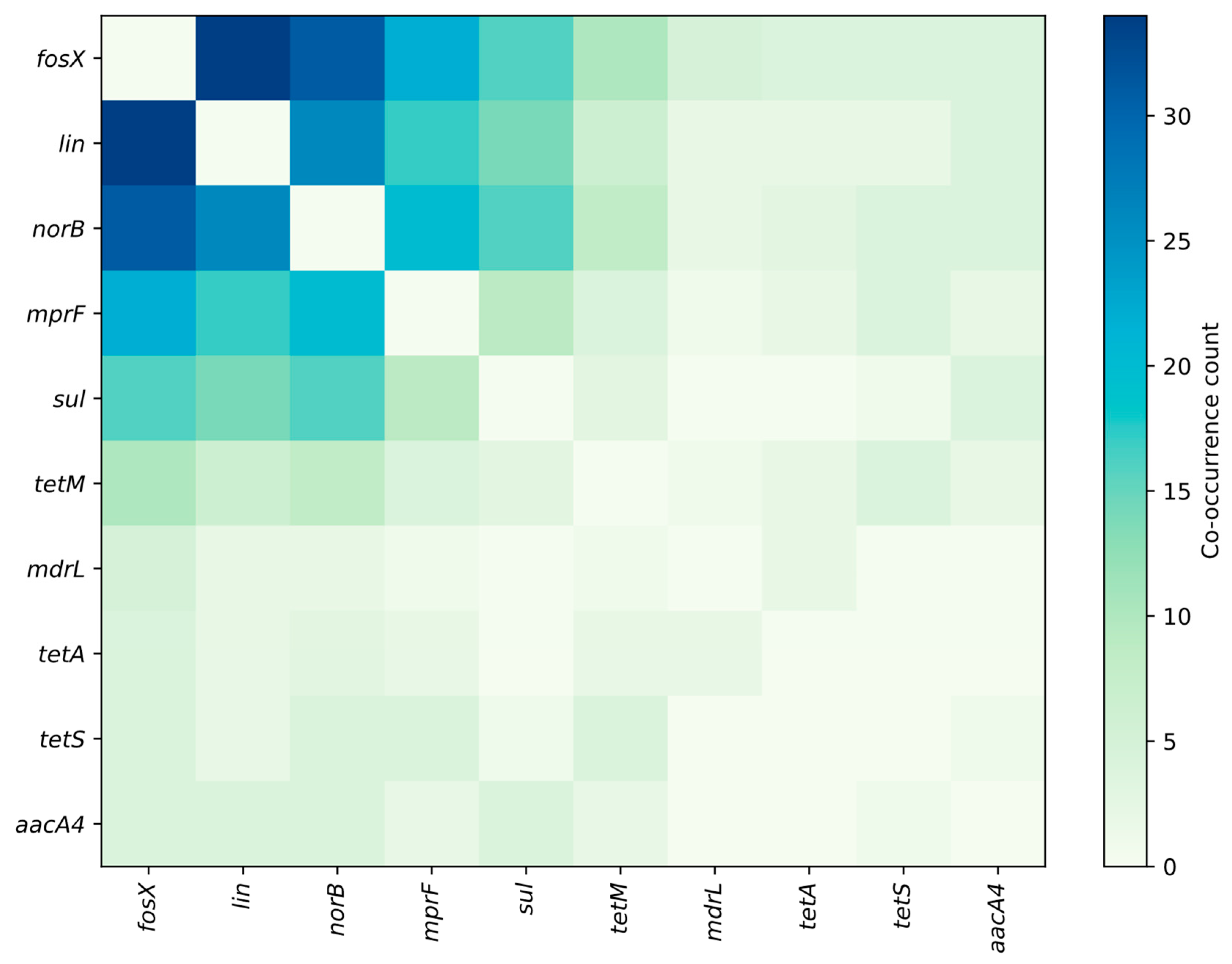
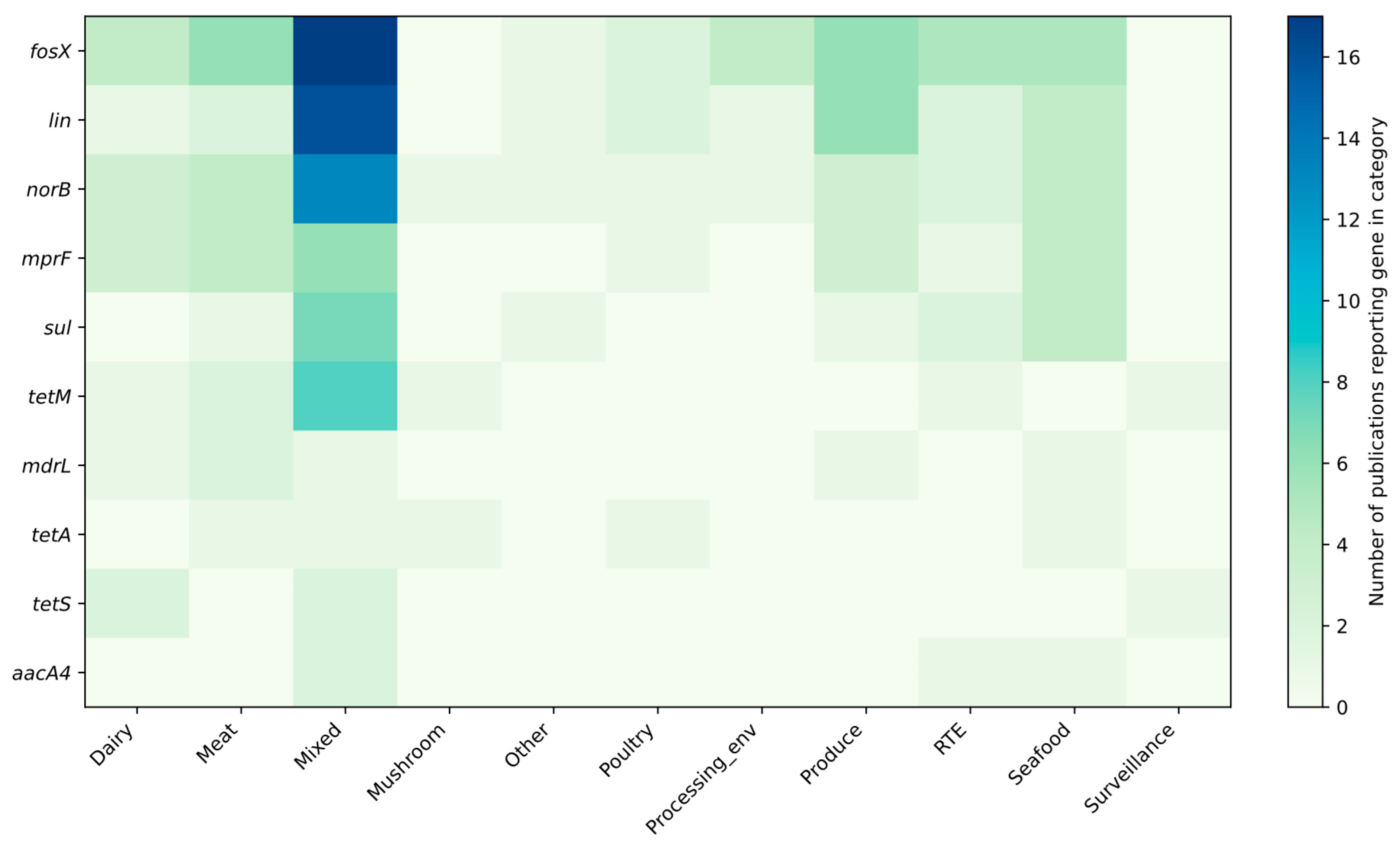
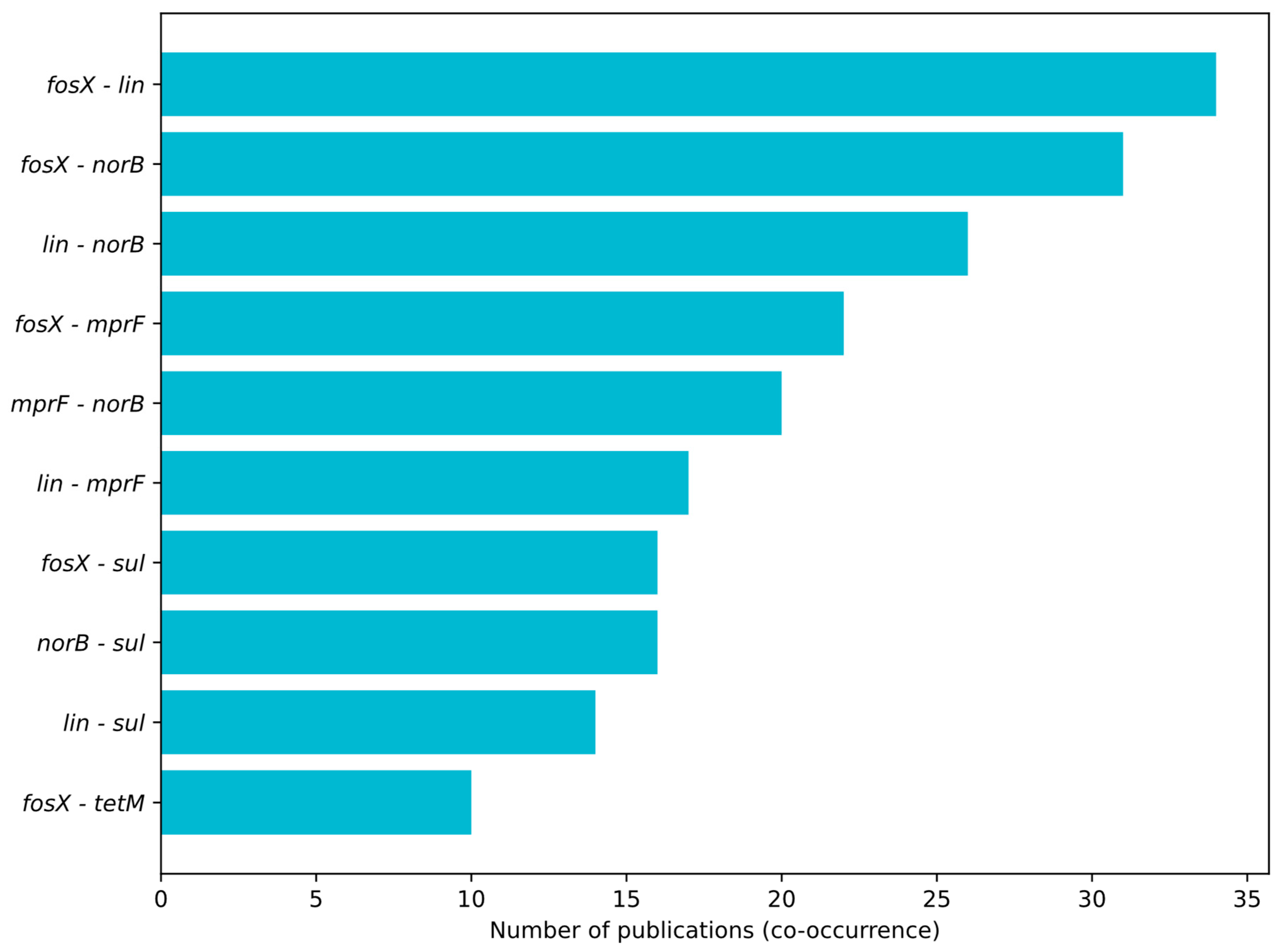
3.1. Profiling Resistance Genes as Phenotypic Predictors in L. monocytogenes Isolated from Food and Food-Production Environments
3.2. Prevalent Resistance Genes and Their Predictive Value
3.2.1. Fosfomycin Resistance
3.2.2. Lincosamides Resistance
3.2.3. Resistance to Fluoroquinolones
3.2.4. Antimicrobial Peptides Resistance
3.2.5. Sulfonamides Resistance
3.2.6. Tetracycline Resistance
3.2.7. Aminoglycosides Resistance
3.2.8. Efflux Proteins/Multidrug Resistance
3.2.9. Other Genes Detected in the Analyzed Studies
4. Epidemiological and Health Consequences of Detected Resistance Patterns
5. Conclusions and Limitations of the Research
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manyi-Loh, C.E.; Okoh, A.I.; Lues, R. Occurrence and Multidrug Resistance in Strains of Listeria monocytogenes Recovered from the Anaerobic Co-Digestion Sludge Contained in a Single Stage Steel Biodigester: Implications for Antimicrobial Stewardship. Microorganisms 2023, 11, 725. [Google Scholar] [CrossRef]
- Rippa, A.; Bilei, S.; Peruzy, M.F.; Marrocco, M.G.; Leggeri, P.; Bossù, T.; Murru, N. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated in Food and Food-Processing Environments in Italy. Antibiotics 2024, 13, 525. [Google Scholar] [CrossRef]
- Castello, A.; Alio, V.; Torresi, M.; Centorotola, G.; Chiaverini, A.; Pomilio, F.; Arrigo, I.; Giammanco, A.; Fasciana, T.; Ortoffi, M.F.; et al. Molecular Characterization and Antimicrobial Resistance Evaluation of Listeria monocytogenes Strains from Food and Human Samples. Pathogens 2025, 14, 294. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses Report. Eur. Food Saf. Auth. 2024, 22, 62–73. [Google Scholar] [CrossRef]
- Pagliano, P.; Arslan, F.; Ascione, T. Epidemiology and Treatment of the Commonest Form of Listeriosis: Meningitis and Bacteraemia. Infez. Med. 2017, 25, 210–216. [Google Scholar]
- Kayode, A.J.; Okoh, A.I. Antimicrobial-Resistant Listeria monocytogenes in Ready-to-Eat Foods: Implications for Food Safety and Risk Assessment. Foods 2023, 12, 1346. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Zakrzewski, A.J.; Zadernowska, A.; Chajęcka-Wierzchowska, W. Antimicrobial Resistance and Virulence Characterization of Listeria monocytogenes Strains Isolated from Food and Food Processing Environments. Pathogens 2022, 11, 1099. [Google Scholar] [CrossRef]
- Moura, A.; Leclercq, A.; Vales, G.; Tessaud-Rita, N.; Bracq-Dieye, H.; Thouvenot, P.; Madec, Y.; Charlier, C.; Lecuit, M. Phenotypic and Genotypic Antimicrobial Resistance of Listeria monocytogenes: An Observational Study in France. Lancet Reg. Health. Eur. 2024, 37, 100800. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing to Clinical Colleagues: On Recent Changes in Clinical Microbiology Susceptibility Reports—New Interpretation of Susceptibility Categories S, I and R. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Guidance_documents/To_clinical_colleagues_on_recent_changes_in_clinical_microbiology_susceptibility_reports_9_July2021.pdf (accessed on 9 August 2025).
- Ellington, M.J.; Ekelund, O.; Aarestrup, F.M.; Canton, R.; Doumith, M.; Giske, C.; Grundman, H.; Hasman, H.; Holden, M.T.G.; Hopkins, K.L.; et al. The Role of Whole Genome Sequencing in Antimicrobial Susceptibility Testing of Bacteria: Report from the EUCAST Subcommittee. Clin. Microbiol. Infect. 2017, 23, 2–22. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Xin, X.; Cao, X.; Nasifu, L.; Nie, Z.; He, B. Phenotypic and Genotypic Perspectives on Detection Methods for Bacterial Antimicrobial Resistance in a One Health Context: Research Progress and Prospects. Arch. Microbiol. 2024, 206, 409. [Google Scholar] [CrossRef]
- Manqele, A.; Adesiyun, A.; Mafuna, T.; Pierneef, R.; Moerane, R.; Gcebe, N. Virulence Potential and Antimicrobial Resistance of Listeria monocytogenes Isolates Obtained from Beef and Beef-Based Products Deciphered Using Whole-Genome Sequencing. Microorganisms 2024, 12, 1166. [Google Scholar] [CrossRef] [PubMed]
- Lakicevic, B.; Jankovic, V.; Pietzka, A.; Ruppitsch, W. Wholegenome Sequencing as the Gold Standard Approach for Control of Listeria monocytogenes in the Food Chain. J. Food Prot. 2023, 86, 100003. [Google Scholar] [CrossRef] [PubMed]
- Méndez Acevedo, M.; Rolon, M.L.; Johnson, B.B.; Burns, L.H.; Stacy, J.; Aurand-Cravens, A.; LaBorde, L.; Kovac, J. Sanitizer Resistance and Persistence of Listeria monocytogenes Isolates in Tree Fruit Packing Facilities. J. Food Prot. 2024, 87, 100354. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the Emergence and Population Diversity of Listeria monocytogenes in a Newly Built Meat Facility through Whole Genome Sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef]
- Amarasekara, N.R.; Swamy, A.S.; Paudel, S.K.; Jiang, W.; Li, K.; Shen, C.; Zhang, Y. Hypervirulent Clonal Complex (CC) of Listeria monocytogenes in Fresh Produce from Urban Communities. Front. Microbiol. 2024, 15, 1307610. [Google Scholar] [CrossRef]
- Andritsos, N.D.; Mataragas, M. Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods 2023, 12, 1200. [Google Scholar] [CrossRef]
- Aslantaş, Ö.; Büyükaltay, K.; Keskin, O.; Güllü Yücetepe, A.; Adigüzel, A. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes from Food and Animal Clinical Cases. Kafkas. Univ. Vet. Fak. Derg. 2023, 29, 221–230. [Google Scholar] [CrossRef]
- Assisi, C.; Forauer, E.; Oliver, H.F.; Etter, A.J. Genomic and Transcriptomic Analysis of Biofilm Formation in Persistent and Transient Listeria monocytogenes Isolates from the Retail Deli Environment Does Not Yield Insight into Persistence Mechanisms. Foodborne Pathog. Dis. 2021, 18, 179–188. [Google Scholar] [CrossRef]
- Astashkin, E.I.; Alekseeva, E.A.; Borzenkov, V.N.; Kislichkina, A.A.; Mukhina, T.N.; Platonov, M.E.; Svetoch, E.A.; Shepelin, A.P.; Fursova, N.K. Molecular-Genetic Characteristics of Polyresistant Listeria monocytogenes Strains and Identification of New Sequence Types. Mol. Genet. Microbiol. Virol. 2021, 36, 159–169. [Google Scholar] [CrossRef]
- Bai, Y.; Li, J.; Huang, M.; Yan, S.; Li, F.; Xu, J.; Peng, Z.; Wang, X.; Ma, J.; Sun, J.; et al. Prevalence and Characterization of Foodborne Pathogens Isolated from Fresh-Cut Fruits and Vegetables in Beijing, China. Int. J. Food Microbiol. 2024, 421, 110804. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.R.B.; Bland, R.; McIntyre, L.; Shyng, S.; Weisberg, A.J.; Riutta, E.R.; Chang, J.H.; Kovacevic, J. Genomic Characterization of Listeria monocytogenes Recovered from Dairy Facilities in British Columbia, Canada from 2007 to 2017. Front. Microbiol. 2024, 15, 1304734. [Google Scholar] [CrossRef]
- Centorotola, G.; Guidi, F.; D’aurizio, G.; Salini, R.; Di Domenico, M.; Ottaviani, D.; Petruzzelli, A.; Fisichella, S.; Duranti, A.; Tonucci, F.; et al. Intensive Environmental Surveillance Plan for Listeria monocytogenes in Food Producing Plants and Retail Stores of Central Italy: Prevalence and Genetic Diversity. Foods 2021, 10, 1944. [Google Scholar] [CrossRef]
- Choi, S.; Choi, Y.; Seo, Y.; Lee, S.; Yoon, Y. Identification of Genetic Variations Related to Pathogenicity by Whole Genome Sequencing of Listeria monocytogenes SMFM2019-FV16 Isolated from Enoki Mushroom. J. Food Saf. 2023, 43, 13076. [Google Scholar] [CrossRef]
- Fox, E.M.; Casey, A.; Jordan, K.; Coffey, A.; Gahan, C.G.M.; McAuliffe, O. Whole Genome Sequence Analysis; an Improved Technology That Identifies Underlying Genotypic Differences between Closely Related Listeria monocytogenes Strains. Innov. Food Sci. Emerg. Technol. 2017, 44, 89–96. [Google Scholar] [CrossRef]
- Gana, J.; Gcebe, N.; Pierneef, R.E.; Chen, Y.; Moerane, R.; Adesiyun, A.A. Whole Genome Sequence Analysis of Listeria monocytogenes Isolates Obtained from the Beef Production Chain in Gauteng Province, South Africa. Microorganisms 2024, 12, 1003. [Google Scholar] [CrossRef] [PubMed]
- Gelbicova, T.; Florianova, M.; Tomastikova, Z.; Pospisilova, L.; Kolackova, I.; Karpiskova, R. Prediction of Persistence of Listeria monocytogenes ST451 in a Rabbit Meat Processing Plant in the Czech Republic. J. Food Prot. 2019, 82, 1350–1356. [Google Scholar] [CrossRef]
- Guidi, F.; Centorotola, G.; Chiaverini, A.; Iannetti, L.; Schirone, M.; Visciano, P.; Cornacchia, A.; Scattolini, S.; Pomilio, F.; D’Alterio, N.; et al. The Slaughterhouse as Hotspot of CC1 and CC6 Listeria monocytogenes Strains with Hypervirulent Profiles in an Integrated Poultry Chain of Italy. Microorganisms 2023, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- Haubert, L.; Kremer, F.S.; da Silva, W.P. Whole-Genome Sequencing Identification of a Multidrug-Resistant Listeria monocytogenes Serotype 1/2a Isolated from Fresh Mixed Sausage in Southern Brazil. Infect. Genet. Evol. 2018, 65, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Yang, S.M.; Kim, E.; Kim, H.J.; Park, S.H. Comprehensive Metagenomic Analysis of Stress-Resistant and -Sensitive Listeria monocytogenes. Appl. Microbiol. Biotechnol. 2023, 107, 6047–6056. [Google Scholar] [CrossRef]
- Hu, L.; Brown, E.W.; Zhang, G. Diversity of Antimicrobial Resistance, Stress Resistance, and Virulence Factors of Salmonella, Shiga Toxin-Producing Escherichia coli, and Listeria monocytogenes from Produce, Spices, and Tree Nuts by Whole Genome Sequencing. Front. Sustain. Food Syst. 2023, 7, 360–368. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-Genome Sequencing-Based Characterization of 100 Listeria monocytogenes Isolates Collected from Food Processing Environments over a Four-Year Period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef]
- Ji, S.; Song, Z.; Luo, L.; Wang, Y.; Li, L.; Mao, P.; Ye, C.; Wang, Y. Whole-Genome Sequencing Reveals Genomic Characterization of Listeria monocytogenes from Food in China. Front. Microbiol. 2023, 13, 1049843. [Google Scholar] [CrossRef] [PubMed]
- Kragh, M.L.; Ivanova, M.; Dalgaard, P.; Leekitcharoenphon, P.; Hansen, L.T. Sensitivity of 240 Listeria monocytogenes Isolates to Common Industrial Biocides Is More Dependent on the Presence of Residual Organic Matter or Biofilm than on Genetic Determinants. Food Control 2024, 158, 110244. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated from Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Lachtara, B.; Osek, J.; Wieczorek, K. Molecular Typing of Listeria monocytogenes IVb Serogroup Isolated from Food and Food Production Environments in Poland. Pathogens 2021, 10, 482. [Google Scholar] [CrossRef]
- Lake, F.B.; van Overbeek, L.S.; Baars, J.J.P.; Koomen, J.; Abee, T.; den Besten, H.M.W. Genomic Characteristics of Listeria monocytogenes Isolated during Mushroom (Agaricus bisporus) Production and Processing. Int. J. Food Microbiol. 2021, 360, 109438. [Google Scholar] [CrossRef]
- Lambrechts, K.; Gouws, P.; Rip, D. Genetic Diversity of Listeria monocytogenes from Seafood Products, Its Processing Environment, and Clinical Origin in the Western Cape, South Africa Using Whole Genome Sequencing. AIMS Microb. 2024, 10, 608–643. [Google Scholar] [CrossRef]
- Lee, M.R.; Park, K.T. Whole-Genome Analysis of CC224 Listeria monocytogenes Strain IJPL9-1, Clonally Related to the Listeriosis Outbreak Strain in 2018, Isolated from Pork in Korea. Microbiol. Biotechnol. Lett. 2024, 52, 328–330. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.; Park, S.H.; Koo, O.K. Whole-Genome Sequencing of Listeria monocytogenes Isolated from the First Listeriosis Foodborne Outbreak in South Korea. Front. Microbiol. 2023, 14, 1182090. [Google Scholar] [CrossRef]
- Lim, S.Y.; Yap, K.-P.; Thong, K.L. Comparative Genomics Analyses Revealed Two Virulent Listeria monocytogenes Strains Isolated from Ready-to-Eat Food. Gut Pathog. 2016, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Lüth, S.; Halbedel, S.; Rosner, B.; Wilking, H.; Holzer, A.; Roedel, A.; Dieckmann, R.; Vincze, S.; Prager, R.; Flieger, A.; et al. Backtracking and Forward Checking of Human Listeriosis Clusters Identified a Multiclonal Outbreak Linked to Listeria monocytogenes in Meat Products of a Single Producer. Emerg. Microbes Infect. 2020, 9, 1600–1608. [Google Scholar] [CrossRef]
- Mäesaar, M.; Mamede, R.; Elias, T.; Roasto, M. Retrospective Use of Whole-Genome Sequencing Expands the Multicountry Outbreak Cluster of Listeria monocytogenes St1247. Int. J. Genom. 2021, 2021, 6636138. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole Genome-Based Characterization of Listeria monocytogenes Isolates Recovered from the Food Chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef]
- Markovich, Y.; Palacios-Gorba, C.; Gomis, J.; Gómez-Martín, Á.; Ortolá, S.; Quereda, J.J. Phenotypic and Genotypic Antimicrobial Resistance of Listeria spp. in Spain. Vet. Microbiol. 2024, 293, 110086. [Google Scholar] [CrossRef]
- Matle, I.; Pierneef, R.; Mbatha, K.R.; Magwedere, K.; Madoroba, E. Genomic Diversity of Common Sequence Types of Listeria monocytogenes Isolated from Ready-to-Eat Products of Animal Origin in South Africa. Genes 2019, 10, 1007. [Google Scholar] [CrossRef]
- Mejía, L.; Espinosa-Mata, E.; Freire, A.L.; Zapata, S.; González-Candelas, F. Listeria monocytogenes, a Silent Foodborne Pathogen in Ecuador. Front. Microbiol. 2023, 14, 1278860. [Google Scholar] [CrossRef]
- Mota, M.I.; Vázquez, S.; Cornejo, C.; D’Alessandro, B.; Braga, V.; Caetano, A.; Betancor, L.; Varela, G. Does Shiga Toxin-Producing Escherichia coli and Listeria monocytogenes Contribute Significantly to the Burden of Antimicrobial Resistance in Uruguay? Front. Vet. Sci. 2020, 7, 583930. [Google Scholar] [CrossRef] [PubMed]
- Narvaez, S.A.; Shen, Z.; Yan, L.; Stenger, B.L.S.; Goodman, L.B.; Lim, A.; Nissly, R.H.; Nair, M.S.; Zhang, S.; Sanchez, S. Optimized Conditions for Listeria, Salmonella and Escherichia Whole Genome Sequencing Using the Illumina iSeq100 Platform with Point-and-Click Bioinformatic Analysis. PLoS ONE 2022, 17, e0277659. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Chen, Y.; Parsons, C.; Brown, E.; Loessner, M.J.; Shen, Y.; Kathariou, S. Whole Genome Sequence Analysis of Phage-Resistant Listeria monocytogenes Serotype 1/2a Strains from Turkey Processing Plants. Pathogens 2021, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Holý, O.; Bustamante, F.; Lepuschitz, S.; Pietzka, A.; Contreras-Fernández, A.; Castillo, C.; Ovalle, C.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; et al. Virulence and Antibiotic Resistance Genes in Listeria monocytogenes Strains Isolated from Ready-to-Eat Foods in Chile. Front. Microbiol. 2022, 12, 796040. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Chen, Y.; Siletzky, R.; Parsons, C.; Jaykus, L.-A.; Eifert, J.; Ryser, E.; Logue, C.M.; Stam, C.; Brown, E.; et al. Harnessing Whole Genome Sequence Data for Facility-Specific Signatures for Listeria monocytogenes: A Case Study with Turkey Processing Plants in the United States. Front. Sustain. Food Syst. 2021, 5, 742353. [Google Scholar] [CrossRef]
- Daza Prieto, B.; Pietzka, A.; Martinovic, A.; Ruppitsch, W.; Zuber Bogdanovic, I. Surveillance and Genetic Characterization of Listeria monocytogenes in the Food Chain in Montenegro during the Period 2014–2022. Front. Microbiol. 2024, 15, 1418333. [Google Scholar] [CrossRef]
- Qi, Y.; Cao, Q.; Zhao, X.; Tian, C.; Li, T.; Shi, W.; Wei, H.; Song, C.; Xue, H.; Gou, H. Comparative Genomic Analysis of Pathogenic Factors of Listeria spp. Using Whole-Genome Sequencing. BMC Genom. 2024, 25, 935. [Google Scholar] [CrossRef]
- Roedel, A.; Dieckmann, R.; Brendebach, H.; Hammerl, J.A.; Kleta, S.; Noll, M.; Al Dahouk, S.; Vinczea, S. Biocide-Tolerant Listeria monocytogenes Isolates from German Food Production Plants Do Not Show Crossresistance to Clinically Relevant Antibiotics. Appl. Environ. Microbiol. 2019, 85, e01253-19. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, G.; Yang, J.; Zhao, L.; Jiang, Y.; Guo, D.; Wang, X.; Zhi, S.; Xu, X.; Dong, Q.; et al. Prevalence, Antibiotic Resistance, and Molecular Epidemiology of Listeria monocytogenes Isolated from Imported Foods in China during 2018 to 2020. Int. J. Food Microbiol. 2022, 382, 109916. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Silva, V.; Gomes, J.P.; Coelho, A.; Batista, R.; Saraiva, C.; Esteves, A.; Martins, Â.; Contente, D.; Diaz-Formoso, L.; et al. Listeria monocytogenes from Food Products and Food Associated Environments: Antimicrobial Resistance, Genetic Clustering and Biofilm Insights. Antibiotics 2024, 13, 447. [Google Scholar] [CrossRef]
- Smith, A.; Hearn, J.; Taylor, C.; Wheelhouse, N.; Kaczmarek, M.; Moorhouse, E.; Singleton, I. Listeria monocytogenes Isolates from Ready to Eat Plant Produce Are Diverse and Have Virulence Potential. Int. J. Food Microbiol. 2019, 299, 23–32. [Google Scholar] [CrossRef]
- Song, Z.; Ji, S.; Wang, Y.; Luo, L.; Wang, Y.; Mao, P.; Li, L.; Jiang, H.; Ye, C. The Population Structure and Genetic Diversity of Listeria monocytogenes ST9 Strains Based on Genomic Analysis. Front. Microbiol. 2022, 13, 982220. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Wen, Y.; Prabakusuma, A.S.; Tang, X.; Huang, A.; Li, L. Prevalence, Antibiotic Resistance and Virulence Feature of Listeria monocytogenes Isolated from Bovine Milk in Yunnan, Southwest China. Int. Dairy J. 2023, 144, 105703. [Google Scholar] [CrossRef]
- Takeuchi-Storm, N.; Hansen, L.T.; Nielsen, N.L.; Andersen, J.K. Presence and Persistence of Listeria monocytogenes in the Danish Ready-to-Eat Food Production Environment. Hygiene 2023, 3, 18–32. [Google Scholar] [CrossRef]
- Toledo, V.; den Bakker, H.C.; Hormazábal, J.C.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A.I. Genomic Diversity of Listeria monocytogenes Isolated from Clinical and Non-Clinical Samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Hernández, M.; Abad, D.; Rodríguez-Grande, J.; Ocampo-Sosa, A.A.; Martínez-Suárez, J.V. Prevalence and Population Diversity of Listeria monocytogenes Isolated from Dairy Cattle Farms in the Cantabria Region of Spain. Animals 2022, 12, 2477. [Google Scholar] [CrossRef] [PubMed]
- Voronina, O.L.; Kunda, M.S.; Ryzhova, N.N.; Aksenova, E.I.; Kustova, M.A.; Karpova, T.I.; Melkumyan, A.R.; Klimova, E.A.; Gruzdeva, O.A.; Tartakovsky, I.S. Listeria monocytogenes ST37 Distribution in the Moscow Region and Properties of Clinical and Foodborne Isolates. Life 2023, 13, 2167. [Google Scholar] [CrossRef]
- Wartha, S.; Bretschneider, N.; Dangel, A.; Hobmaier, B.; Hörmansdorfer, S.; Huber, I.; Murr, L.; Pavlovic, M.; Sprenger, A.; Wenning, M.; et al. Genetic Characterization of Listeria from Food of Non-Animal Origin Products and from Producing and Processing Companies in Bavaria, Germany. Foods 2023, 12, 1120. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes from Fish and Fish Production Environments in Poland. Int. J. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Yan, S.; Li, M.; Luque-Sastre, L.; Wang, W.; Hu, Y.; Peng, Z.; Dong, Y.; Gan, X.; Nguyen, S.; Anes, J.; et al. Susceptibility (Re)-Testing of a Large Collection of Listeria monocytogenes from Foods in China from 2012 to 2015 and WGS Characterization of Resistant Isolates. J. Antimicrob. Chemother. 2019, 74, 1786–1794. [Google Scholar] [CrossRef]
- Zakrzewski, A.J.; Gajewska, J.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Insights into the Genetic Diversity of Listeria monocytogenes from Bivalves. Sci. Total Environ. 2024, 908, 168481. [Google Scholar] [CrossRef]
- Zuber, I.; Lakicevic, B.; Pietzka, A.; Milanov, D.; Djordjevic, V.; Karabasil, N.; Teodorovic, V.; Ruppitsch, W.; Dimitrijevic, M. Molecular Characterization of Listeria monocytogenes Isolates from a Small-Scale Meat Processor in Montenegro, 2011–2014. Food Microbiol. 2019, 79, 116–122. [Google Scholar] [CrossRef]
- Schirmer, M.; D’Amore, R.; Ijaz, U.Z.; Hall, N.; Quince, C. Illumina Error Profiles: Resolving Fine-Scale Variation in Metagenomic Sequencing Data. BMC Bioinform. 2016, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Commichaux, S.; Javkar, K.; Ramachandran, P.; Nagarajan, N.; Bertrand, D.; Chen, Y.; Reed, E.; Gonzalez-Escalona, N.; Strain, E.; Rand, H.; et al. Evaluating the Accuracy of Listeria monocytogenes Assemblies from Quasimetagenomic Samples Using Long and Short Reads. BMC Genom. 2021, 22, 389. [Google Scholar] [CrossRef]
- Lee, Y.; Son, B.; Cha, Y.; Ryu, S. Characterization and Genomic Analysis of PALS2, a Novel Staphylococcus Jumbo Bacteriophage. Front. Microbiol. 2021, 12, 622755. [Google Scholar] [CrossRef]
- Rigsby, R.E.; Fillgrove, K.L.; Beihoffer, L.A.; Armstrong, R.N. Fosfomycin Resistance Proteins: A Nexus of Glutathione Transferases and Epoxide Hydrolases in a Metalloenzyme Superfamily. Methods Enzymol. 2005, 401, 367–379. [Google Scholar]
- Scortti, M.; Han, L.; Alvarez, S.; Leclercq, A.; Moura, A.; Lecuit, M.; Vazquez-Boland, J. Epistatic Control of Intrinsic Resistance by Virulence Genes in Listeria. PLoS Genet. 2018, 14, e1007525, Erratum in: PLoS Genet. 2018, 14, e1007727. [Google Scholar] [CrossRef]
- Wiśniewski, P.; Chajęcka-Wierzchowska, W.; Zadernowska, A. High-Pressure Processing—Impacts on the Virulence and Antibiotic Resistance of Listeria monocytogenes Isolated from Food and Food Processing Environments. Foods 2023, 12, 3899. [Google Scholar] [CrossRef]
- Kinde, M.Z.; Kerisew, B.; Eshetu, T.; Gessese, A.T. Genomic Analysis of Listeria monocytogenes Strains from Dairy Products in Ethiopia. Front. Bioinform. 2025, 5, 1572241. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Petinaki, E.; Guérin-Faublée, V.; Pichereau, V.; Villers, C.; Achard, A.; Malbruny, B.; Leclercq, R. Lincomycin Resistance Gene Lnu (D) in Streptococcus uberis. Antimicrob. Agents Chemother. 2008, 52, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Morar, M.; Bhullar, K.; Hughes, D.W.; Junop, M.; Wright, G.D. Structure and Mechanism of the Lincosamide Antibiotic Adenylyltransferase LinB. Structure 2009, 17, 1649–1659. [Google Scholar] [CrossRef]
- Vester, B.; Douthwaite, S. Macrolide Resistance Conferred by Base Substitutions in 23S rRNA. Antimicrob. Agents Chemother. 2001, 45, 1–12. [Google Scholar] [CrossRef]
- Roberts, M.C.; Facinelli, B.; Giovanetti, E.; Varaldo, P.E. Transferable Erythromycin Resistance in Listeria spp. Isolated from Food. Appl. Environ. Microbiol. 1996, 62, 269–270. [Google Scholar] [CrossRef]
- Wilson, A.; Gray, J.; Chandry, P.S.; Fox, E.M. Phenotypic and Genotypic Analysis of Antimicrobial Resistance among Listeria monocytogenes Isolated from Australian Food Production Chains. Genes 2018, 9, 80. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes Isolates from Food and Food Environment Harbouring tetM and ermB Resistance Genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef]
- Novotna, G.; Janata, J. A New Evolutionary Variant of the Streptogramin a Resistance Protein, Vga(A)LC, from Staphylococcus haemolyticus with Shifted Substrate Specificity towards Lincosamides. Antimicrob. Agents Chemother. 2006, 50, 4070–4076. [Google Scholar] [CrossRef]
- Zhu, X.-Q.; Wang, X.-M.; Li, H.; Shang, Y.-H.; Pan, Y.-S.; Wu, C.-M.; Wang, Y.; Du, X.-D.; Shen, J.-Z. Novel Lnu (G) Gene Conferring Resistance to Lincomycin by Nucleotidylation, Located on Tn 6260 from Enterococcus faecalis E531. J. Antimicrob. Chemother. 2016, 72, 993–997. [Google Scholar] [CrossRef]
- Ding, Y.; Onodera, Y.; Lee, J.C.; Hooper, D.C. NorB, an Efflux Pump in Staphylococcus aureus Strain MW2, Contributes to Bacterial Fitness in Abscesses. J. Bacteriol. 2008, 190, 7123–7129. [Google Scholar] [CrossRef] [PubMed]
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The Major Facilitator Superfamily and Antimicrobial Resistance Efflux Pumps of the ESKAPEE Pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343. [Google Scholar] [CrossRef] [PubMed]
- Centorotola, G.; Ziba, M.W.; Cornacchia, A.; Chiaverini, A.; Torresi, M.; Guidi, F.; Cammà, C.; Bowa, B.; Mtonga, S.; Magambwa, P.; et al. Listeria monocytogenes in Ready to Eat Meat Products from Zambia: Phenotypical and Genomic Characterization of Isolates. Front. Microbiol. 2023, 14, 1228726. [Google Scholar] [CrossRef]
- Guérin, F.; Galimand, M.; Tuambilangana, F.; Courvalin, P.; Cattoir, V. Overexpression of the Novel MATE Fluoroquinolone Efflux Pump FepA in Listeria monocytogenes Is Driven by Inactivation of Its Local Repressor FepR. PLoS ONE 2014, 9, e106340. [Google Scholar] [CrossRef]
- Nowak, J.; Visnovsky, S.B.; Cruz, C.D.; Fletcher, G.C.; van Vliet, A.H.M.; Hedderley, D.; Butler, R.; Flint, S.; Palmer, J.; Pitman, A.R. Inactivation of the Gene Encoding the Cationic Antimicrobial Peptide Resistance Factor MprF Increases Biofilm Formation but Reduces Invasiveness of Listeria monocytogenes. J. Appl. Microbiol. 2021, 130, 464–477. [Google Scholar] [CrossRef]
- Bao, Y.; Sakinc, T.; Laverde, D.; Wobser, D.; Benachour, A.; Theilacker, C.; Hartke, A.; Huebner, J. Role of mprF1 and mprF2 in the Pathogenicity of Enterococcus faecalis. PLoS ONE 2012, 7, e38458. [Google Scholar] [CrossRef]
- Guidi, F.; Lorenzetti, C.; Centorotola, G.; Torresi, M.; Cammà, C.; Chiaverini, A.; Pomilio, F.; Blasi, G. Atypical Serogroup IVb-v1 of Listeria monocytogenes Assigned to New ST2801, Widely Spread and Persistent in the Environment of a Pork-Meat Producing Plant of Central Italy. Front. Microbiol. 2022, 13, 930895. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Moon, J.-S.; Lee, Y.J.; Kim, H.-Y. Whole-Genome Sequencing-Based Characterization of Listeria monocytogenes Isolated from Cattle and Pig Slaughterhouses. Infect. Genet. Evol. 2025, 130, 105737. [Google Scholar] [CrossRef]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.H.; Guan, Z. Discovery of a Cardiolipin Synthase Utilizing Phosphatidylethanolamine and Phosphatidylglycerol as Substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef]
- Lee, H.; Hsu, F.-F.; Turk, J.; Groisman, E.A. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J. Bacteriol. 2004, 186, 4124–4133. [Google Scholar] [CrossRef]
- Razavi, M.; Marathe, N.P.; Gillings, M.R.; Flach, C.-F.; Kristiansson, E.; Joakim Larsson, D.G. Discovery of the Fourth Mobile Sulfonamide Resistance Gene. Microbiome 2017, 5, 160. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide Resistance: Mechanisms and Trends. Drug Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Adamski, P.; Zakrzewski, A.; Wiśniewski, P.; Chajęcka-Wierzchowska, W.; Zadernowska, A. High-Pressure Processing (HPP) Alters Tetracycline Resistance in Listeria monocytogenes: A Phenotypic and Genotypic Study. NFS J. 2025, 38, 100223. [Google Scholar] [CrossRef]
- Bertsch, D.; Uruty, A.; Anderegg, J.; Lacroix, C.; Perreten, V.; Meile, L. Tn6198, a Novel Transposon Containing the Trimethoprim Resistance Gene dfrG Embedded into a Tn916 Element in Listeria monocytogenes. J. Antimicrob. Chemother. 2013, 68, 986–991. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Rather, P.N.; Munayyer, H.; Mann, P.A.; Hare, R.S.; Miller, G.H.; Shaw, K.J. Genetic Analysis of Bacterial Acetyltransferases: Identification of Amino Acids Determining the Specificities of the Aminoglycoside 6′-N-Acetyltransferase Lb and Ila Proteins. J. Bacteriol. 1992, 174, 3196–3203. [Google Scholar] [CrossRef]
- Dzyubak, E.; Yap, M.-N.F. The Expression of Antibiotic Resistance Methyltransferase Correlates with mRNA Stability Independently of Ribosome Stalling. Antimicrob. Agents Chemother. 2016, 60, 7178–7188. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Wang, W.; Koteva, K.; Barton, H.A.; McArthur, A.G.; Wright, G.D. A Diverse Intrinsic Antibiotic Resistome from a Cave Bacterium. Nat. Commun. 2016, 7, 13803. [Google Scholar] [CrossRef]
- Li, Q.; Pellegrino, J.; Lee, D.J.; Tran, A.A.; Chaires, H.A.; Wang, R.; Park, J.E.; Ji, K.; Chow, D.; Zhang, N.; et al. Synthetic Group A Streptogramin Antibiotics That Overcome Vat Resistance. Nature 2020, 586, 145–150. [Google Scholar] [CrossRef]
- Srinivasan, V.B.; Rajamohan, G.; Gebreyes, W.A. Role of AbeS, a Novel Efflux Pump of the SMR Family of Transporters, in Resistance to Antimicrobial Agents in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 5312–5316. [Google Scholar] [CrossRef]
- Idrees, M.M.; Saeed, K.; Shahid, M.A.; Akhtar, M.; Qammar, K.; Hassan, J.; Khaliq, T.; Saeed, A. Prevalence of mecA- and mecC-Associated Methicillin-Resistant Staphylococcus aureus in Clinical Specimens, Punjab, Pakistan. Biomedicines 2023, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Lv, M.; Yang, K.; Zhao, G.; Fu, Y. A Case Report of Diagnosis and Dynamic Monitoring of Listeria monocytogenes Meningitis with NGS. Open Life Sci. 2023, 18, 20220738. [Google Scholar] [CrossRef]
- Shi, Q.; Zhang, X.; Wang, D.; Zhang, W.; Jin, X.; Sun, Y.; Huang, A. Genetic Diversity, Antimicrobial Resistance, and Virulence Profiles of Listeria monocytogenes Isolates from Nantong, China (2020–2023). Foodborne Pathog. Dis. 2025. [Google Scholar] [CrossRef] [PubMed]
- Andriyanov, P.A.; Zhurilov, P.A.; Liskova, E.A.; Karpova, T.I.; Sokolova, E.V.; Yushina, Y.K.; Zaiko, E.V.; Bataeva, D.S.; Voronina, O.L.; Psareva, E.K.; et al. Antimicrobial Resistance of Listeria monocytogenes Strains Isolated from Humans, Animals, and Food Products in Russia in 1950–1980, 2000–2005, and 2018–2021. Antibiotics 2021, 10, 1206. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, E.; Hosseini, H.; Abdi Moghadam, Z.; Halberg Larsen, M.; Haslberger, A.; Alebouyeh, M. Importance of Listeria monocytogenes in Food Safety: A Review of Its Prevalence, Detection, and Antibiotic Resistance. Iran. J. Vet. Res. 2019, 20, 241–254. [Google Scholar]
- Sołtysiuk, M.; Przyborowska, P.; Wiszniewska-Łaszczych, A.; Tobolski, D. Prevalence and Antimicrobial Resistance Profile of Listeria spp. Isolated from Raw Fish. BMC Vet. Res. 2025, 21, 333. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, P.; Trymers, M.; Chajęcka-Wierzchowska, W.; Tkacz, K.; Zadernowska, A.; Modzelewska-Kapituła, M. Antimicrobial Resistance in the Context of Animal Production and Meat Products in Poland—A Critical Review and Future Perspective. Pathogens 2024, 13, 1123. [Google Scholar] [CrossRef] [PubMed]
- Bradford, D.; Lodhi, K.; Yuan, J.; Graham, D.; Graham, J.; Maldani, M.; White, E.; Arhin, A.; Kassem, M. Comparative Genomics of Foodborne Pathogens: Diversity, Virulence, and Epidemiological Relevance. Am. J. Bot. 2025, 846–855. [Google Scholar] [CrossRef]
| No | Number of Sequenced Strains | Serotype/Serogroup | Source of Samples | Sequencing Method | Genotypic Antibiotic Resistance | Phenotypic Antibiotic Resistance | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Method (Program) | Antibiotic Resistance Genes | Method | Phenotype of Resistance | ||||||
| 1. | 45 | NA | Three tree fruit packing facilities | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | ABRicate (v1.0.1) in GalaxyTrakr, using the MEGARes (v3.0) | fosX, lin, mdrL, mprF, norB | NA | NA | [14] |
| 2. | 18 | 1/2a, 1/2c | Meat-processing facility | Illumina HiSeq 1500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ABRicate (v.0.9.9), CARD, AMRFinderPlus, ResFinder) | norB, mprF (CARD), lin (ARG-ANNOT and CARD), fosX (ResFinder and NCBI AMRFinderPlus) | NA | NA | [15] |
| 3. | 12 | 1/2a, 4b | Leafy greens, root vegetables, and seeded vegetables | Illumina X10 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm database | fosX, lin, mprF, norB, sul | NA | NA | [16] |
| 4. | 54 | 1/2a, 3a,1/2b, 3b, 7 | Cheese and cheese-processing surfaces | Novogene Genomics Service (Novogene Co., Ltd., Cambridge, UK) | Bionumerics version 8.1.1 (bioMérieux, Marcy-l’Étoile, France) | fosX, mdrL, mdrM | Disk diffusion method, E-test | Ciprofloxacin (intermediately resistant) | [17] |
| 5. | 21 | IIa, IIb, IIc, IVb, L | Food | Illumina NovaSeq 6000 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm | fosX, lin, norB, sul | NA | NA | [18] |
| 6. | 21 | 4bV, 4b/4d/4e | Retail deli environments | Illumina MiSeq (San Diego, CA, USA) | ResFinder (v.3.2) | tetM, fosX | NA | NA | [19] |
| 7. | 9 | 1/2a–3a, 1/2b–3b-7, 1/2c–3c | Beef, fish, and poultry | Illumina MiSeq (San Diego, CA, USA) | BIGSdb-Lm database | fosX, lmo0441, lin, norB, lmo0224 | Microdilution method | Tetracyclines, macrolides, sulfonamides, beta-lactams. | [20] |
| 8. | 3 | NA | Resh-cut fruits and vegetables | Illumina HiSeq (San Diego, CA, USA) | ResFinder | No resistance genes detected | VITEK 2 AST- GN05/GP67 Test Kit (bioM ’erieux Corporate) | No resistant strains | [21] |
| 9. | 54 | NA | Dairy processing plants | Illumina HiSeq 1500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | NCBI BLASTN | tetR, emrE, emrC | NA | NA | [22] |
| 10. | 18 | NA | Rady-to-eat meat products | Illumina NextSeq 500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm database | fosX, mprF, norB, sul | Microdilution method with the Sensititre OptiRead Automated Fluorometric Plate Reading System (Thermo Scientific, Monza, Italy) | Cephalosporins, penicillin, tetracycline, trimethoprim/sulfamethoxazole | [23] |
| 11. | 7 | 1/2a-3a, 1/2b-3b | Enoki mushrooms | PacBio RS II platform (Pacific Biosciences, Menlo Park, CA, USA) | CLC Genomics Workbench version 12.0 (Qiagen) | pbp, pbpX, pbpH, ponA, pbpB, pbpF, tetA, tet(M), emrY, emrB, emrB, norB, norm, mepA, mdtL | Disk diffusion method | Penicillin G, ampicillin, tetracycline, clindamycin. | [24] |
| 12. | 4 | 1/2c, 4b | Food | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | Mauve, Artemis, and BRIG (BLAST Ring Image Generator) | A number of non-specific multidrug efflux genes, as well as genes with putative functions in increasing resistance of the bacterium to lincomycin, quinolone, β-lactams, and tetracycline | NA | NA | [25] |
| 13. | 60 | IVb, IVb, IIa | Cattle farms, cattle abattoirs, and retail outlets | Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | ABRicate | fosX, vga(G) | NA | NA | [26] |
| 14. | 4 | 1/2a | Rabbit meat | Illumina HiSeq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | BIGSdb-Lm database at Institute Pasteur | mdrL, lde | NA | NA | [27] |
| 15. | 122 | NA | Broiler chickens | Illumina NextSeq 500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm | norB, mprF, lin, fosX, parC, msrA, tetA | NA | NA | [28] |
| 16. | 1 | 1/2a | Fresh mixed sausage | Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | MEGARes database | tetM, mepA, msrA, norB, efTu, gyrA | Disk diffusion method | Streptomycin, erythromycin, clindamycin, rifampicin, meropenem, trimethoprim–sulfamethoxazole, tetracycline | [29] |
| 17. | 19 | NA | Foods | Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | Resistance gene identifier (RGI) software (v.5.1.1), CARD (v.3.1.1) | fosX, lin, mprF, tet(M); (tet(M) is not known to be among the isolates from food) | NA | NA | [30] |
| 18. | 20 | NA | Produce, nuts, and spices | Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | NCBI AMRFinder process in the NCBI Pathogen detection system, 4 based on the Bacterial Antimicrobial Resistance Reference Gene Database. | fosX, lin, abc-f | NA | NA | [31] |
| 19. | 100 | IIa, IIb, IIc, IVb, IVb-v1 | Meat and vegetable processing facilities | Illumina HiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | BLAST+ (v.2.9.0), ResFinder (v.3.1.0) | fosX | NA | NA | [32] |
| 20. | 322 | Ia, IIb, IIc, IVb and L | Raw meat, raw poultry, ready-to-eat food, aquatic products, and unknown food sources | Illumina HiSeq X PE150 (San Diego, CA, USA) (coverage rate of more than 100-fold) | BIGSdb-Lm database | fosX, lmo0441, lin, norB, mprF, sul, aacA4, tetS, tetM | NA | NA | [33] |
| 21. | 240 | 1/2a, 1/2b, 4b, 1/2c | Foods, food-processing environments | Illumina NextSeq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm database at Institute Pasteur | fosX, lin, norB, lmo0441, sul | NA | NA | [34] |
| 22. | 48 | IIb, IVb | Ready-to-eat food of animal origin (e.g., ham, sausages, or meat), and food-processing environments | Illumina MiSeq (San Diego, CA, USA), Illumina NextSeq500 (San Diego, CA, USA) | BIGSdb-Lm database | fosX, lin, norB, sul, aacA4 | NA | NA | [35] |
| 23. | 91 | NA | Raw meat, ready-to-eat food and food-production environments | Illumina MiSeq (San Diego, CA, USA) (50× average coverage) | BIGSdb-Lm | fosX, lin, norB, sul, mprF | NA | NA | [36] |
| 24. | 44 | 1/2a-3a, 1/2b-3b-7, 4b-4d-4e | Casing soil, mushrooms, equipment, and frozen mushrooms | Illumina Miseq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm database | fosX, sul, norB, lin | Disk diffusion method | No resistant strains | [37] |
| 25. | 38 | 1/2a, 1/2b, 4b | Seafood products and other food | CosmosID (Germantown, MD, USA), Inqaba Biotechnical Industries (Pretoria, South Africa) | ResFinder (v.4.1), BIGSdb-Lm | fosX, lin, mprF, norB, sul | Disk diffusion method | Tetracycline, sulphamethoxazole/trimethoprim, erythromycin, chloramphenicol | [38] |
| 26. | 1 | IIb | Pork | Illumina NextSeq P1 600 cycles (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | AMRFinderPlus (v.3.11.26), BIGSdb_LM | fosX, vga(G), mprF, norB, sul | NA | NA | [39] |
| 27. | 25 | 1/2b, 1/2a, 1/2c, 4b | Foods | Illumina MiSeq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads), Oxford Nanopore sequencing technologies (Oxford Nanopore Technologies, Oxford, UK) | Listeria MLST database | fosX, norB, sul, lin, tetM | Disk diffusion method | Lincomycin, penicillin G | [40] |
| 28. | 2 | 4b | Fried fish and salad | Illumina HiSeq 2000 (San Diego, CA, USA) | Resistance Gene Identifier (RID) of the CARD | tetA, lmrB, fosX, msrA, lde, mdrL | NA | NA | [41] |
| 29. | 48 | NA | Ready-to-eat meat products | Illumina HiSeq 1500 (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | ABRicate version (v.0.8) | lin, fosX, | NA | NA | [42] |
| 30. | 1 | IIa | Vacuum-packaged sliced salted salmon products | Illumina MiSeq (San Diego, CA, USA) (600 cycles) | BIGSdb database | fosX, lin, mprF, norB, sul | NA | NA | [43] |
| 31. | 152 | IIa, IIb, IIc, IVb, | Raw meat, processed meat, ready-to-eat meat products and environmental samples collected from a commercial pig farm environment | Illumina HiSeq and MiSeq (Illumina, San Diego, CA, USA) | ABRicate and NCBI | fosX, lin, norB, mprF, tetM, tetS | NA | NA | [44] |
| 32. | 24 | IIa, IVb, IIb, IIc | Beef and beef-based products | Illumina MiSeq (Illumina, San Diego, CA, USA) | ResFinder, CARD, and NCBI | lin, norB, fosX, mprF | NA | NA | [12] |
| 33. | 35 | IIa, IVb, IIc, IIb, | Corn, meat, vegetables, pepper, fish, pineapple, salad, easter sausage, smoked salmon, salami, barbecue chorizo, cooked peeled prawn, smoked sardine, smoked fish product, surface sponge, sweet chorizo, matured cheese, surface sponge, blood sausage, Majorcan sausage, and chorizo | Illumina NextSeq 500 (San Diego, CA, USA) | BIGSdb database | fosX, lin, norB, sul | Disk diffusion method, Microdilution method | No resistant strains | [45] |
| 34. | 6 | 2a, 4b | Meat and meat products | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | BacMet, MEGARes and nonredundant antibiotic resistance database (noradab) | fosX, tetA, tetM, mecC, mrB, msrA, lde, mdrL. | NA | NA | [46] |
| 35. | 45 | NA | Cheese | Illumina NextSeq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ARIBA software (v.2.11.1) (CARD, MEGAres, and ResFinder databases) | fosX, mprF, norB (CARD), Gyra_23, TUFAB-7 (MEGAres), tetS (Resfinder) | Microdilution technique, using panels of lyophilized antibiotics for Gram-positive bacteria (Sensititre 36GPALL1F, ThermoScientific®, Waltham, MA, USA) | No resistant strains | [47] |
| 36. | 21 | 1/2a, 1/2b, 4b | Frozen food, ready-to-eat food, deli meat, and cheese. | Illumina MiSeq (San Diego, CA, USA) | ABRicate software (ResFinder, CARD, NCBI, AMRFinderPlus, MEGARes BLAST tool) | fosX, lin, norB, lde, mdrL, fepA | Disk diffusion method | Ciprofloxacin | [48] |
| 37. | 2431 | NA | Food | Illumina NextSeq 500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | BIGSdb-Lm | fosX, norB, lin, sul, pbp, tetM, ermB, mphB, fexA, dfrD, InuG, aacA4, aphA, dfrD | NA | NA | [8] |
| 38. | 2 | NA | Mung bean sprouts | Illumina iSeq100 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ResFinder, AMRonize, AMRFinder, ABRicate, Staramr | fosX, lin | NA | NA | [49] |
| 39. | 54 | 1/2a | Processing plants | Illumina NextSeq 500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads), Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | NCBI Pathogen Detection Pipeline | fosX, lin | NA | NA | [50] |
| 40. | 14 | 1/2a, 1/2b | Cheeses, cooked meats (artisanal ham, pâté, sausages, and blood sausage), pre-processed fruits and vegetables (chopped fruit, fruit salads with strawberries, melon, peaches, and leafy vegetable salads), and meals and mixed dishes with raw and/or cooked ingredients | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | CARD, AMRFinderPlus (v.3.2.3) database | fosX, lin, norB, mprF, tetA, tetC | Disk diffusion method | Ampicillin | [51] |
| 41. | 54 | 4b, 1/2b, 4b, 1,2a, 1/2c, 4b (4b-v1) | Turkey processing plants | Illumina HiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | NCBI Pathogen Detection Pipeline | fosX, lin, abc-F | NA | NA | [52] |
| 42. | 160 | IIc, IIb, IIa, IVb | Raw milk and fresh meat | Illumina NextSeq2000 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | NCBI AMRFinder+ (v.3.11.2) | fosX, lin | NA | NA | [53] |
| 43. | 1 | 4b | Slaughterhouse | Illumina Novaseq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | CARD | pgs, arlR, bcrA, rphA, rpoB, vanR, vanG, mdtG, lmrC, lmrD, norB, adeC, FosX | Disk diffusion method | Polymyxin, ceftazidime | [54] |
| 44. | 93 | NA | Food-production plants | Illumina MiSeq (San Diego, CA, USA) (602 cycles, 301 bp paired-end reads) | ResFinder (v.3.0) | fosX | Commercial test system Micronaut S Listeria MHK-2 (Merlin Gesellschaft für Mikrobiologische Diagnostika mbH, Bornheim, Germany), | Daptomycin, tigecycline, meropenem, ciprofloxacin (susceptible, increased exposure [I]), and rifampin (I). | [55] |
| 45. | 89 | 1/2a, 3a, 1/2c, 3c | Frozen beef, frozen pork, fresh fish, fresh aquatic products except fish, frozen chicken, frozen sheep casing, and dairy food products | Illumina Hiseq×10 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ABRicate software | aph(4)Ia, ermC, fexA, tetK, tetM | Microdilution method | Oxacillin, daptomycin, chloramphenicol, tetracycline, ciprofloxacin, erythromycin, imipenem, clindamycin, ciprofloxacin (Intermediate levels of resistance), chloramphenicol (Intermediate levels of resistance) | [56] |
| 46. | 13 | NA | Food products or food-processing environments | Illumina NextSeq 2000 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ResFinder (v.2.0), CARD | fosX lin, norB, tetM | Disk diffusion method | Trimethoprim–sulfamethoxazole, erythromycin, penicillin | [57] |
| 47. | 15 | NA | Spinach, environmental swab (drain), red leaf lettuce, beetroot, pea shoots, and baby salad kale | Illumina HiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | ABRicate (v.0.8) (BLAST+ & EMBOSS, Resfinder) | fosX, lin | NA | NA | [58] |
| 48. | 142 | NA | Food | Illumina Hiseq PE150 by Novogene (Beijing, China) | ABRicate (v.1.0.0) pipeline by the Resfinder database | fosX, tetM | NA | NA | [59] |
| 49. | 8 | NA | Bovine milk | Illumina NovaSeq 6000 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | CARD | ANT(9)-Ia, ANT(6)-Ia, APH(3′)-IIIa, SRT-1, ACC-3, mecA, PBP2x, PBP1a, dfrG, FosX, fusA, fusB, fusE, vanHF, vanHD, vanRM, vanTE, vanYM, vanRG, vanSM, vanRE, vanTG, vanRF, vanRI, norB, patB, mdtG, mepA, lmrB, lmrC, lmrD, lsaA, lsaE, salA, macB, LpeA, oleC, abeS, efrA, msrC, bcrA, cmrA, fexA, vgaE, vgaALC, optrA, ErmB, ErmH, Erm(34), cfrA, lnuB, vatI, vatB, sul4, folP, tetA(60), tetA(48), tetA(46), tetT, tetC, tetD, tetM, tetS, tet(42), tetB(60), tetB(P), tcr3, PmrF, mprF, liaR, liaS, cls | Microdilution method | Penicillin, tetracycline, trimethoprim-sulfamethoxazole, erythromycin | [60] |
| 50. | 85 | NA | Ready-to-eat fish and meat | Illumina MiSeq (San Diego, CA, USA) (500 cycles, 250 bp paired-end reads) | AMRFinderPlus (v.3.10.14) | fosX, lin, Tn6188, tetR | NA | NA | [61] |
| 51. | 16 | 1/2a, 1/2b, 3a, 4a, 4b, 4c, 4e | Food and food-related environments. | Illumina NextSeq500 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ResFinder | fosX | NA | NA | [62] |
| 52. | 45 | IVb, IIa, IIb | Dairy cattle farms forage, water, raw tank milk, the tank milk filters, fresh feces, stored manure, and soil | Illumina MiSeq (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | Resfinder, CARD, ARG-ANNOT databases by using ABRicate | fosX, lin, norB, mprF | Microdilution method | Ampicillin, ciprofloxacin, erythromycin, tetracycline, vancomycin, meropenem | [63] |
| 53. | 16 | NA | Food | Illumina MiSeq and NextSeq 500/550 (San Diego, CA, USA) | BIGSdb-Lm database, CARD | fosX, lin, mprF, norB | NA | NA | [64] |
| 54. | 8 | IVb, IIa | Primary production and processing companies’ fresh fruit and frozen berries | Illumina NextSeq 550 (San Diego, CA, USA) (300 cycles, 150 bp paired-end reads) | ABRicate (v.1.0.1), CARD | fosX, mprF, lin, norB | BD Phoenix System (Becton Dickinson, Franklin Lakes, NJ, USA) | Penicillin, fosfomycin, ciprofloxacin, moxifloxacin | [65] |
| 55. | 28 | IIa, IVb | Raw fish material, i.e., fresh and frozen Atlantic salmon (Salmo salar), swabs from fish-production environments (n = 200) as well as samples from ready-to-eat (RTE) fish products, i.e., cold-smoked Atlantic salmon, marinated (gravlax) Atlantic salmon, and cold-smoked rainbow trout (Oncorhynchus mykiss) | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | BIGSdb-Lm, ABRicate software | fosX, lin, mprF, norB, sul, aacA4 | NA | NA | [66] |
| 56. | 28 | IIa, IIb, IIc, IVb | National food surveillance | Novogene (Beijing, China) on an Illumina HiSeq (San Diego, CA, USA) | ABRicate software package (ResFinder v.2.1.37, ARG-ANNOT v.438, CARD v.2.0.339 databases) | aadE, ant9, aph3, cat, erm(B), lsaE, lnuB, drfG, tet(S), tet(M) | Microdilution method | tetracycline, erythromycin, chloramphenicol, trimethoprim/sulfamethoxazole | [67] |
| 57. | 30 | NA | Bivalves | Illumina MiSeq (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | CARD | norB, lin, mprF, fosX, sul | Microdilution method | Fosfomycin, lincomycin, tetracycline | [68] |
| 58. | 20 | IIa, IVb | Traditional pork products, including drycured ham (Njeguški pršut), pork tenderloin, pancetta (thin, dry bacon), sausages, and processing environment (swabs from surfaces and drains) | Illumina MiSeq NexteraXT (San Diego, CA, USA) (600 cycles, 300 bp paired-end reads) | CARD | mprF, fosX | NA | NA | [69] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiśniewski, P.; Adamski, P.; Trymers, M.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Predicting Antibiotic Resistance in Listeria monocytogenes from Food and Food-Processing Environments Using Next-Generation Sequencing: A Systematic Review. Int. J. Mol. Sci. 2025, 26, 10112. https://doi.org/10.3390/ijms262010112
Wiśniewski P, Adamski P, Trymers M, Chajęcka-Wierzchowska W, Zadernowska A. Predicting Antibiotic Resistance in Listeria monocytogenes from Food and Food-Processing Environments Using Next-Generation Sequencing: A Systematic Review. International Journal of Molecular Sciences. 2025; 26(20):10112. https://doi.org/10.3390/ijms262010112
Chicago/Turabian StyleWiśniewski, Patryk, Patryk Adamski, Miłosz Trymers, Wioleta Chajęcka-Wierzchowska, and Anna Zadernowska. 2025. "Predicting Antibiotic Resistance in Listeria monocytogenes from Food and Food-Processing Environments Using Next-Generation Sequencing: A Systematic Review" International Journal of Molecular Sciences 26, no. 20: 10112. https://doi.org/10.3390/ijms262010112
APA StyleWiśniewski, P., Adamski, P., Trymers, M., Chajęcka-Wierzchowska, W., & Zadernowska, A. (2025). Predicting Antibiotic Resistance in Listeria monocytogenes from Food and Food-Processing Environments Using Next-Generation Sequencing: A Systematic Review. International Journal of Molecular Sciences, 26(20), 10112. https://doi.org/10.3390/ijms262010112







