Diagnostic Performance of Universal Transport Medium for Viral Polymerase Chain Reaction in Aqueous Humor Samples of Suspected Viral Uveitis: A Pilot Methods Study
Abstract
1. Introduction
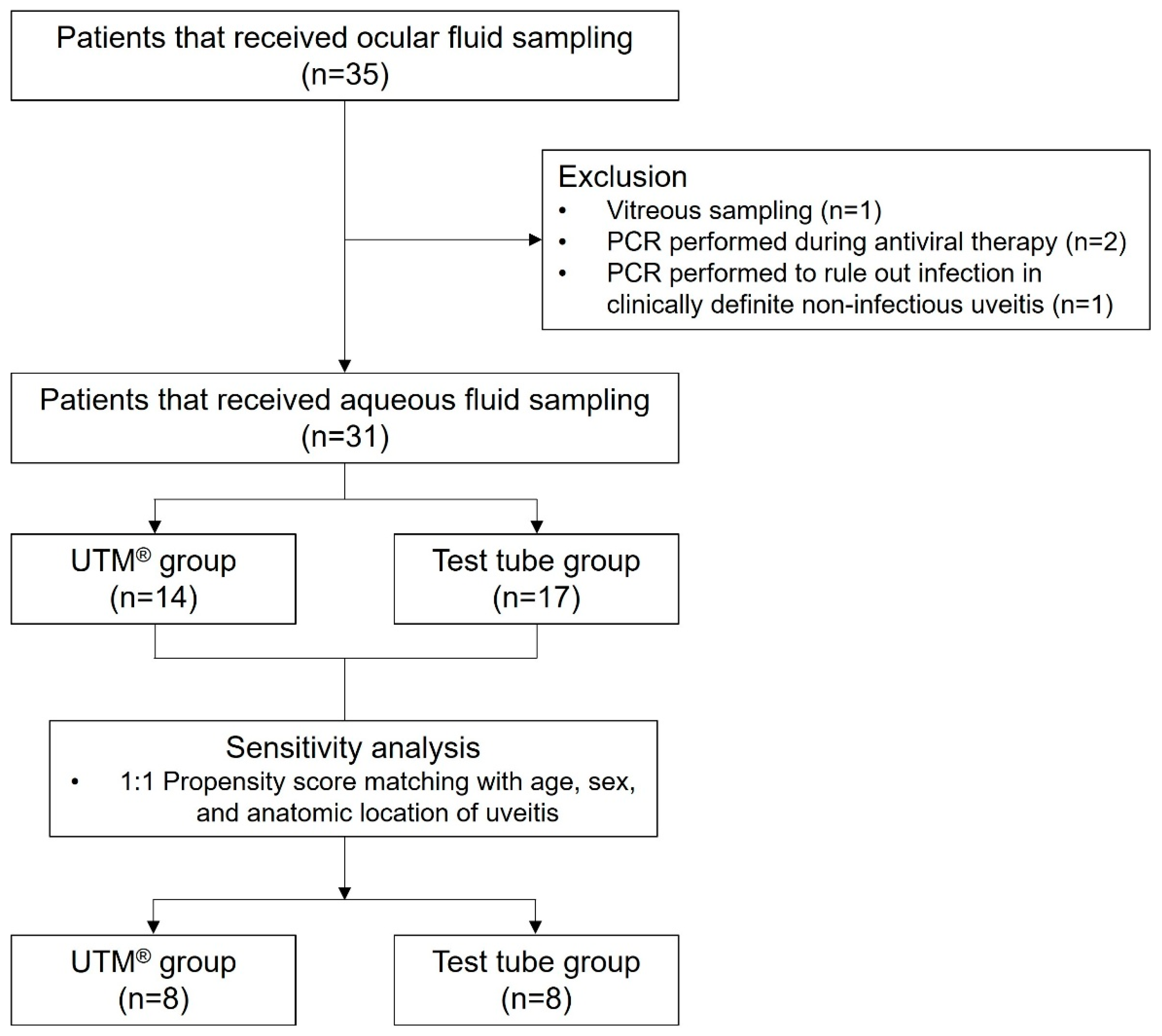
2. Results
2.1. Demographics and Ocular Characteristics
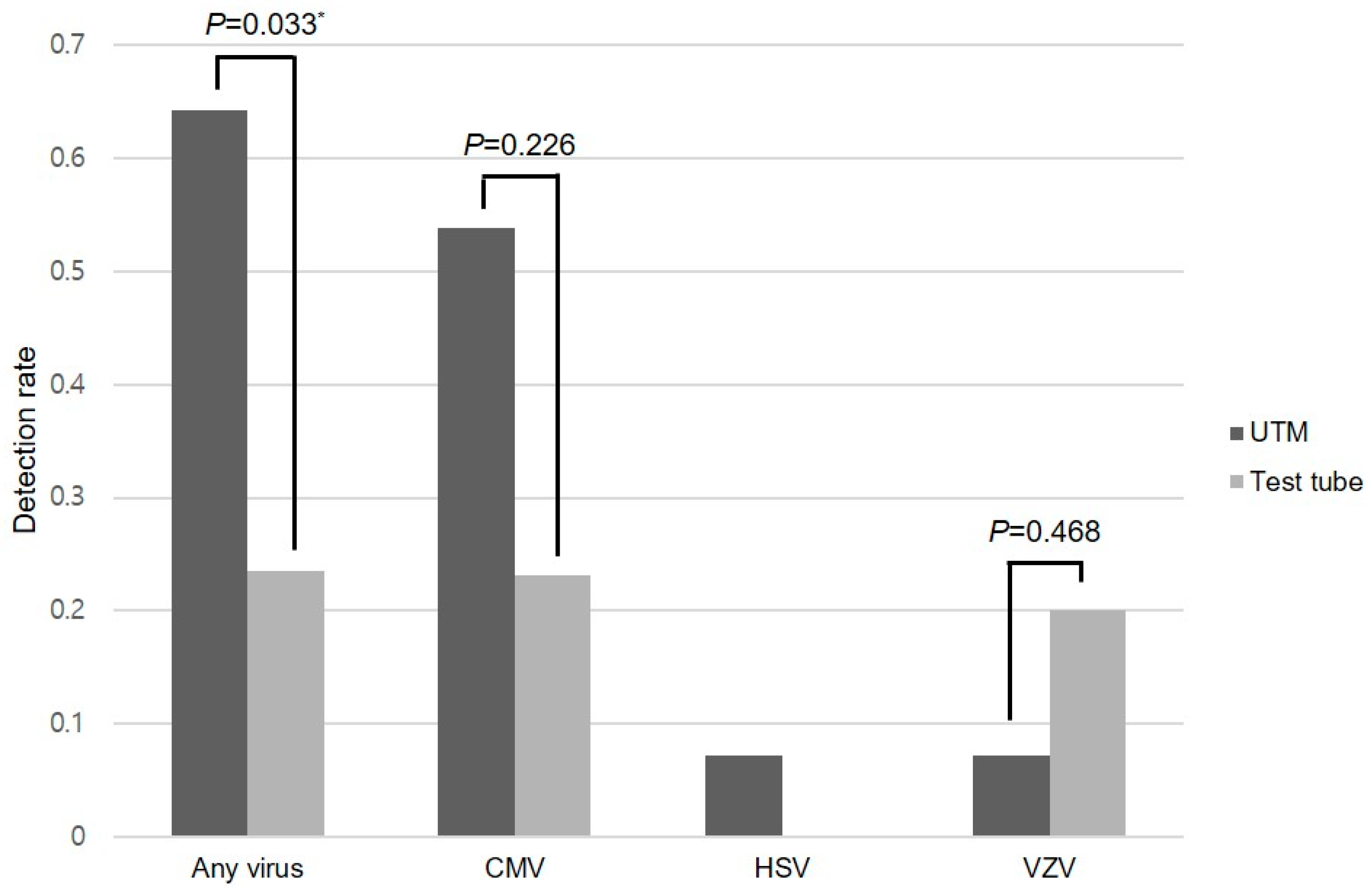
2.2. Viral Detection
2.3. Sensitivity Analyses
3. Discussion
4. Materials and Methods
4.1. Patient Enrollment and Study Design
4.2. Procedure
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CI | Confidence interval |
| CMV | Cytomegalovirus |
| COVID-19 | Coronavirus disease 2019 |
| Ct | Cycle threshold |
| DNA | Deoxyribonucleic acid |
| EBV | Epstein–Barr virus |
| HSV | Herpes simplex virus |
| OR | Odds ratio |
| PCR | Polymerase chain reaction |
| RNA | Ribonucleic acid |
| SUN | Standardization of Uveitis Nomenclature |
| UTM | Universal transport medium |
| VZV | Varicella-zoster virus |
Appendix A
Appendix A.1. Molecular Assay Characteristics for CMV PCR
- Extraction kits and platforms: All specimens were processed on fully automated cobas® 6800 Systems (Roche Diagnostics, Basel, Switzerland), which integrate nucleic acid extraction, purification, amplification, and detection. No external extraction kit was used.
- Target gene: Conserved region of the UL54 DNA polymerase gene of human cytomegalovirus (CMV).
- Positivity thresholds and quantification range: Results were reported in IU/mL, traceable to the WHO International Standard for CMV. Results were categorized as: not detected, <LLoQ, within quantifiable range, or >ULoQ. The lower limit of detection (LOD) was 20.6 IU/mL (95% CI, 17.9–24.3); the lower limit of quantification (LLoQ) was 34.5 IU/mL; and the upper limit of quantification (ULoQ) was 1.0 × 107 IU/mL. The validated linear range extended from 2.45 × 101 to 1.34 × 107 IU/mL. According to the manufacturer, detection of CMV DNA above the LOD (20.6 IU/mL) is reported as positive, whereas results ≥ LLoQ (34.5 IU/mL) are considered within the validated quantifiable range.
- Internal controls: Each run included an exogenous DNA quantitation standard (DNA-QS) spiked into all samples prior to extraction, along with manufacturer-provided controls (CMV Low Positive Control, CMV High Positive Control, and Normal Human Plasma Negative Control).
- Sample input volumes: A minimum of 525 µL of specimen was required, of which 350 µL was aspirated into the cobas secondary tube for analysis. The same input requirements were applied to aqueous humor samples, and identical input volumes were maintained across all media and laboratories. Although the cobas CMV assay is formally validated for EDTA plasma, aqueous humor was processed under the same protocol; this off-label application was consistently implemented and is acknowledged in the main text as a study limitation.
Appendix A.2. Molecular Assay Characteristics for Externally Performed PCR Assays (HSV, VZV, EBV, and Rubella Virus)
- Extraction kits and platforms: HSV and VZV assays were performed at Green Cross Laboratories (Yongin, Republic of Korea).Nucleic acids were extracted using the MagNA Pure 96 system (Roche Diagnostics) with the MagNA Pure 96 DNA and Viral NA Small Volume Kit.Subsequent amplification was performed on different PCR platforms: HSV testing was conducted using the CFX96 real-time PCR system with the HSV 1/2 Real-Time PCR Kit (Biocore Co., Seoul, Republic of Korea), while VZV testing was performed on the ProFlex conventional PCR platform using the VZV PCR Real-Time PCR Kit (Biocore Co., Ltd., Seoul, Republic of Korea).EBV and rubella virus assays were performed at Seoul Clinical Laboratories (Seoul, Republic of Korea).For EBV, extraction and amplification were conducted using the Alinity m Sample Preparation Kit (Abbott, Abbott Park, IL, USA) on the Alinity m platform, which integrates extraction, amplification, and detection.For rubella virus, extraction was carried out using the MagNA Pure 96 system with the MagNA Pure 96 DNA and Viral NA Small Volume Kit, followed by amplification on the SLAN 96P real-time PCR platform with the Rubella Real-Time PCR Kit (Biocore Co., Ltd., Seoul, Republic of Korea).
- Target gene: The BioCore HSV 1/2 Real-Time PCR Kit (Biocore Co., Seoul, Republic of Korea) amplifies the UL30 DNA polymerase gene conserved across HSV-1 and HSV-2.The BioCore VZV PCR Kit (Biocore Co., Ltd., Seoul, Republic of Korea) targets the ORF62 gene, a well-conserved immediate-early regulatory region of the VZV genome.The Alinity m EBV AMP Kit (Abbott, Abbott Park, IL, USA) amplifies the BamHI W region of the EBV genome.The BioCore Rubella Real-Time PCR Kit (Biocore Co., Ltd., Seoul, Republic of Korea) targets the E1 envelope glycoprotein gene of rubella virus.
- Positivity thresholds and quantification range: For HSV, the assay had a limit of detection (LOD) of 155 copies/mL for HSV-1 and 163 copies/mL for HSV-2. For VZV, the limit of detection was 197 copies/mL. For EBV, the limit of detection was 50 IU/mL, traceable to the WHO International Standard, and results ≥ 50 IU/mL were reported as positive. For rubella virus, this qualitative assay had a manufacturer-validated positivity cut-off of Ct ≤ 45. Additional assay performance characteristics such as the linear quantification range or LOQ were unavailable.
- Internal controls: Each assay included manufacturer-provided internal controls to monitor extraction and amplification efficiency. However, detailed specifications of these internal control materials were not disclosed by the laboratories.
- Sample input volumes: For HSV, VZV, and rubella virus assays, 200 µL of sample was used for extraction, while 800 µL was used for EBV testing. Identical input volumes were maintained across transport media (UTM® vs. test tube) within each virus-specific assay. Although these assays were originally validated for use with plasma, serum, or cerebrospinal fluid specimens, the same protocols were applied to aqueous humor samples to ensure standardized processing across all media. This represents an off-label application, which was implemented consistently and is acknowledged in the main text as a study limitation.
References
- Feng, Y.; Garcia, R.; Rojas-Carabali, W.; Cifuentes-González, C.; Putera, I.; Li, J.; La Distia Nora, R.; Mahendradas, P.; Gupta, V.; de-la-Torre, A.; et al. Viral anterior uveitis: A practical and comprehensive review of diagnosis and treatment. Ocul. Immunol. Inflamm. 2024, 32, 1804–1818. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, C.; Peng, X.; Lum, F.; McLeod, S.D.; Acharya, N.R. Use of anterior chamber paracentesis for diagnosis in viral anterior uveitis. Ophthalmology 2024, 131, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Standardization of Uveitis Nomenclature (SUN) Working Group. Classification criteria for varicella zoster virus anterior uveitis. Am. J. Ophthalmol. 2021, 228, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Standardization of Uveitis Nomenclature (SUN) Working Group. Classification criteria for herpes simplex virus anterior uveitis. Am. J. Ophthalmol. 2021, 228, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Standardization of Uveitis Nomenclature (SUN) Working Group. Classification criteria for cytomegalovirus anterior uveitis. Am. J. Ophthalmol. 2021, 228, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Standardization of Uveitis Nomenclature (SUN) Working Group. Classification criteria for juvenile idiopathic arthritis-associated chronic anterior uveitis. Am. J. Ophthalmol. 2021, 228, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Standardization of Uveitis Nomenclature (SUN) Working Group. Classification criteria for Spondyloarthritis/HLA-B27-Associated anterior uveitis. Am. J. Ophthalmol. 2021, 228, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Anwar, Z.; Galor, A.; Albini, T.A.; Miller, D.; Perez, V.; Davis, J.L. The diagnostic utility of anterior chamber paracentesis with polymerase chain reaction in anterior uveitis. Am. J. Ophthalmol. 2013, 155, 781–786. [Google Scholar] [CrossRef]
- Druce, J.; Garcia, K.; Tran, T.; Papadakis, G.; Birch, C. Evaluation of swabs, transport media, and specimen transport conditions for optimal detection of viruses by PCR. J. Clin. Microbiol. 2012, 50, 1064–1065. [Google Scholar] [CrossRef]
- Jo, S.J.; Kim, J.; Kim, S.; Kim, J.; Kwon, H.; Kwon, Y.; Kim, H.; Kim, H.H.; Lee, H.; Kim, S.W.; et al. Simple saliva sample collection for the detection of SARS-CoV-2 variants compared with nasopharyngeal swab sample. Arch. Pathol. Lab. Med. 2022, 146, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Chronopoulos, A.; Roquelaure, D.; Souteyrand, G.; Seebach, J.D.; Schutz, J.S.; Thumann, G. Aqueous humor polymerase chain reaction in uveitis—Utility and safety. BMC Ophthalmol. 2016, 16, 189. [Google Scholar] [CrossRef] [PubMed]
- Yoo, W.S.; Kim, G.N.; Chung, I.; Cho, M.C.; Han, Y.S.; Kang, S.S.; Yun, S.P.; Seo, S.W.; Kim, S.J. Clinical characteristics and prognostic factors in hypertensive anterior uveitis diagnosed with polymerase chain reaction. Sci. Rep. 2021, 11, 8814. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Kang, H.G.; Choi, E.Y.; Kim, S.S.; Kim, C.Y.; Koh, H.J.; Lee, S.C.; Kim, M. Clinical utility of aqueous humor polymerase chain reaction and serologic testing for suspected infectious uveitis: A single-center retrospective study in South Korea. BMC Ophthalmol. 2020, 20, 242. [Google Scholar] [CrossRef] [PubMed]
- Bang, L.L.; Tornby, D.R.; Pham, S.T.D.; Assing, K.; Möller, S.; Palarasah, Y.; Madsen, L.W.; Thomsen, K.G.; Johansen, I.S.; Pedersen, R.M.; et al. Culturing of SARS-CoV-2 from patient samples: Protocol for optimal virus recovery and assessment of infectious viral load. J. Virol. Methods 2024, 326, 114912. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.A.; Baumann, R.E.; Borillo, G.A.; Kagan, R.M.; Batterman, H.J.; Galdzicka, M.M.; Marlowe, E.M. Evaluation of Transport Media and Specimen Transport Conditions for the Detection of SARS-CoV-2 by Use of Real-Time Reverse Transcription-PCR. J. Clin. Microbiol. 2020, 58, e00708-20. [Google Scholar] [CrossRef] [PubMed]
- Chan, N.S.W.; Chee, S.P.; Caspers, L.; Bodaghi, B. Clinical features of CMV-associated anterior uveitis. Ocul. Immunol. Inflamm. 2018, 26, 107–115. [Google Scholar] [CrossRef] [PubMed]

| Variable | n (%) or Mean ± SD |
|---|---|
| No. of patients | 31 |
| Age (years) | 52.9 ± 15.2 |
| Sex, female | 17 (55%) |
| Ocular findings | |
| Intraocular pressure (mmHg) | 17.5 ± 8.8 |
| Keratic precipitates | 21 (68%) |
| Anterior chamber cells | 27 (87%) |
| Ocular diagnosis | |
| Keratitis/Endotheliitis | 6 (19%) |
| Anterior uveitis | 13 (42%) |
| Retinitis/Retinal vasculitis | 9 (29%) |
| Acute retinal necrosis | 3 (10%) |
| Use of UTM | 14 (45%) |
| Use of conventional test tube | 17 (55%) |
| PCR tests performed | |
| Cytomegalovirus | 26 (84%) |
| Herpes simplex virus | 21 (68%) |
| Varicella-zoster virus | 19 (61%) |
| Epstein–Barr virus | 4 (13%) |
| Rubella virus | 2 (6%) |
| Virus | UTM® | Test Tube | p-Value † |
|---|---|---|---|
| Cytomegalovirus | 7/13 (53.8%) | 3/13 (23.1%) | 0.226 |
| Herpes simplex virus | 1/14 (7.1%) | 0/7 (0%) | N/A ‡ |
| Varicella-zoster virus | 1/14 (7.1%) | 1/5 (20.0%) | 0.468 * |
| Test Group | n | Mean ± SD | Min–Max | Median (IQR) |
|---|---|---|---|---|
| All patients | 10 | 962,572 ± 2,875,170 | 258–9,140,000 | 3735 (147,614) |
| UTM® | 7 | 20,548 ± 33,456 | 258–85,500 | 2090 (47,825) |
| Test tube | 3 | 3,160,630 ± 5,181,045 | 1890–9,140,000 | 340,000 (9,138,110) |
| Variable | OR (95% CI) | p-Value |
|---|---|---|
| Age | 1.069 (1.000–1.143) | 0.051 |
| Sex (female) | 0.625 (0.147–2.664) | 0.525 |
| Use of UTM® | 5.850 (1.222–27.994) | 0.027 * |
| Anatomic location of uveitis (anterior segment) | 1.018 (0.235–4.407) | 0.981 |
| Intraocular pressure | 0.944 (0.858–1.040) | 0.242 |
| Presence of keratic precipitates | 4.400 (0.749–25.842) | 0.101 |
| Presence of anterior chamber cells | 2.400 (0.221–26.116) | 0.472 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.; Chung, Y.-R.; Song, J.H.; Choi, Y.J.; Kim, H.R. Diagnostic Performance of Universal Transport Medium for Viral Polymerase Chain Reaction in Aqueous Humor Samples of Suspected Viral Uveitis: A Pilot Methods Study. Int. J. Mol. Sci. 2025, 26, 10091. https://doi.org/10.3390/ijms262010091
Kim C, Chung Y-R, Song JH, Choi YJ, Kim HR. Diagnostic Performance of Universal Transport Medium for Viral Polymerase Chain Reaction in Aqueous Humor Samples of Suspected Viral Uveitis: A Pilot Methods Study. International Journal of Molecular Sciences. 2025; 26(20):10091. https://doi.org/10.3390/ijms262010091
Chicago/Turabian StyleKim, Chungwoon, Yoo-Ri Chung, Ji Hun Song, Young Joon Choi, and Hae Rang Kim. 2025. "Diagnostic Performance of Universal Transport Medium for Viral Polymerase Chain Reaction in Aqueous Humor Samples of Suspected Viral Uveitis: A Pilot Methods Study" International Journal of Molecular Sciences 26, no. 20: 10091. https://doi.org/10.3390/ijms262010091
APA StyleKim, C., Chung, Y.-R., Song, J. H., Choi, Y. J., & Kim, H. R. (2025). Diagnostic Performance of Universal Transport Medium for Viral Polymerase Chain Reaction in Aqueous Humor Samples of Suspected Viral Uveitis: A Pilot Methods Study. International Journal of Molecular Sciences, 26(20), 10091. https://doi.org/10.3390/ijms262010091







