Molecular Docking and Simulation Analysis of Glioblastoma Cell Surface Receptors and Their Ligands: Identification of Inhibitory Drugs Targeting Fibronectin Ligand to Potentially Halt Glioblastoma Pathogenesis
Abstract
1. Introduction
2. Results
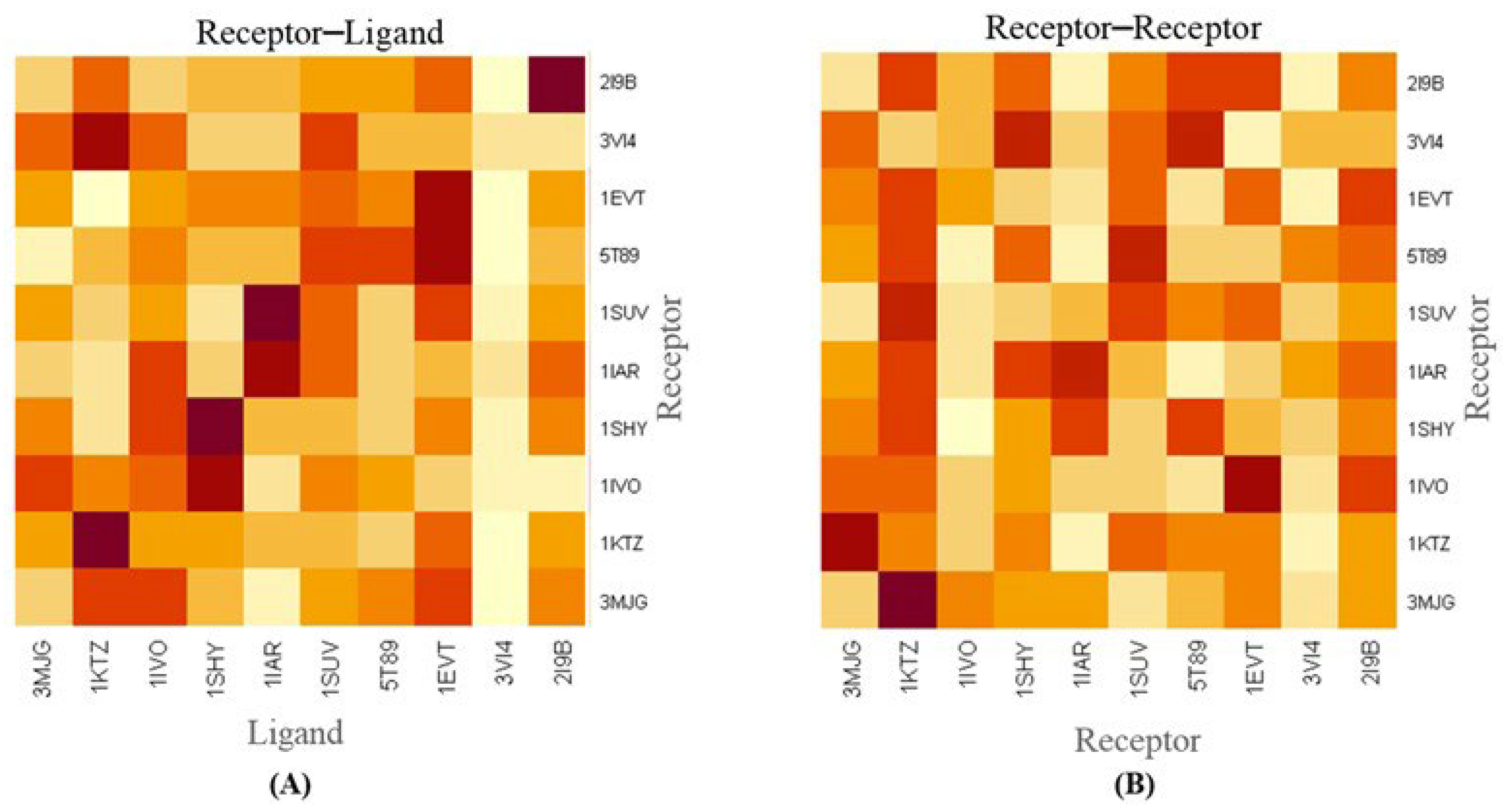
2.1. Docking Analysis of Targeted Proteins
2.2. Molecular Docking and Molecular Dynamic Simulation Analysis of Surface Receptors Binding vs. Extracellular Fibronectin Ligand
2.3. Molecular Docking Analysis (Fibronectin Ligand vs. Drugs)
3. Discussion
4. Materials and Methods
4.1. Retrieval of Protein Receptors
4.2. Retrieval of Approved Drugs for Glioblastoma
4.3. Molecular Docking Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, M.; Sherwani, S.; Khan, S.; Alouffi, S.; Alam, M.; Al-Motair, K.; Khan, S. Insights into multifunctional nanoparticle-based drug delivery systems for glioblastoma treatment. Molecules 2021, 26, 2262. [Google Scholar] [CrossRef] [PubMed]
- Wirsching, H.G.; Galanis, E.; Weller, M. Glioblastoma. Handb. Clin. Neurol. 2016, 134, 381–397. [Google Scholar]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3. [Google Scholar]
- Angom, R.S.; Nakka, N.M.; Bhattacharya, S. Advances in Glioblastoma Therapy: An Update on Current Approaches. Brain Sci. 2023, 13, 1536. [Google Scholar] [CrossRef]
- Ferrari, S.; Pellati, F.; Costi, M.P. Protein–Protein Interaction Inhibitors: Case Studies on Small Molecules and Natural Compounds. In Disruption of Protein–Protein Interfaces; Mangani, S., Ed.; Springer: Berlin, Germany, 2013; pp. 31–60. [Google Scholar]
- Camps-Fajol, C.; Cavero, D.; Minguillon, J.; Surralles, J. Targeting protein-protein interactions in drug discovery: Modulators approved or in clinical trials for cancer treatment. Pharmacol. Res. 2025, 211, 107544. [Google Scholar] [CrossRef]
- Vidal, M.; Cusick, M.E.; Barabási, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Arkin, M.R.; Whitty, A. The road less traveled: Modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr. Opin. Chem. Biol. 2009, 13, 284–290. [Google Scholar] [CrossRef]
- Nero, T.L.; Morton, C.J.; Holien, J.; Wielens, J.; Parker, M.W. Oncogenic protein interfaces: Small molecules, big challenges. Nat. Rev. Cancer 2014, 14, 248–262. [Google Scholar] [CrossRef]
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 2016, 15, 533–550. [Google Scholar] [CrossRef] [PubMed]
- Nada, H.; Choi, Y.; Kim, S.; Jeong, K.S.; Meanwell, N.A.; Lee, K. New insights into protein–protein interaction modulators in drug discovery and therapeutic advance. Signal Transduct. Target. Ther. 2024, 9, 341. [Google Scholar] [CrossRef]
- Lorusso, G.; Rüegg, C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem. Cell Biol. 2008, 130, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Seebacher, N.A.; Krchniakova, M.; Stacy, A.E.; Skoda, J.; Jansson, P.J. Tumour microenvironment stress promotes the development of drug resistance. Antioxidants 2021, 10, 1801. [Google Scholar] [CrossRef]
- Neophytou, C.M.; Panagi, M.; Stylianopoulos, T.; Papageorgis, P. The role of tumor microenvironment in cancer metastasis: Molecular mechanisms and therapeutic opportunities. Cancers 2021, 13, 2053. [Google Scholar] [CrossRef] [PubMed]
- Brady-Kalnay, S.M. Molecular mechanisms of cancer cell-cell interactions: Cell-cell adhesion-dependent signaling in the tumor microenvironment. Cell Adhes. Migr. 2012, 6, 344–345. [Google Scholar] [CrossRef]
- Kiefel, H.; Bondong, S.; Hazin, J.; Ridinger, J.; Schirmer, U.; Riedle, S.; Altevogt, P. L1CAM: A major driver for tumor cell invasion and motility. Cell Adhes. Migr. 2012, 6, 374–386. [Google Scholar] [CrossRef]
- Lin, T.C.; Yang, C.H.; Cheng, L.H.; Chang, W.T.; Lin, Y.R.; Cheng, H.C. Fibronectin in Cancer: Friend or Foe. Cells 2019, 9, 27. [Google Scholar] [CrossRef]
- Chang, Y.H.; Lee, S.H.; Liao, I.C.; Huang, S.H.; Cheng, H.C.; Liao, P.C. Secretomic analysis identifies alpha-1 antitrypsin (A1AT) as a required protein in cancer cell migration, invasion, and pericellular fibronectin assembly for facilitating lung colonization of lung adenocarcinoma cells. Mol. Cell. Proteom. 2012, 11, 1320–1339. [Google Scholar] [CrossRef]
- Wang, Y.J.; Lin, J.F.; Cheng, L.H.; Chang, W.T.; Kao, Y.H.; Chang, M.M.; Wang, B.J.; Cheng, H.C. Pterostilbene prevents AKT-ERK axis-mediated polymerization of surface fibronectin on suspended lung cancer cells independently of apoptosis and suppresses metastasis. J. Hematol. Oncol. 2017, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Moschetta, M.; Pretto, F.; Berndt, A.; Galler, K.; Richter, P.; Bassi, A.; Oliva, P.; Micotti, E.; Valbusa, G.; Schwager, K.; et al. Paclitaxel enhances therapeutic efficacy of the F8-IL2 immunocytokine to EDA-fibronectin-positive metastatic human melanoma xenografts. Cancer Res. 2012, 72, 1814–1824. [Google Scholar] [CrossRef]
- Han, H.-J.; Russo, J.; Kohwi, Y.; Kohwi-Shigematsu, T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008, 452, 187–193. [Google Scholar] [CrossRef]
- Wong, F.H.; Huang, C.Y.; Su, L.J.; Wu, Y.C.; Lin, Y.S.; Hsia, J.Y.; Tsai, H.T.; Lee, S.A.; Lin, C.H.; Tzeng, C.H.; et al. Combination of microarray profiling and protein-protein interaction databases delineates the minimal discriminators as a metastasis network for esophageal squamous cell carcinoma. Int. J. Oncol. 2009, 34, 117–128. [Google Scholar]
- Reuter, J.A.; Ortiz-Urda, S.; Kretz, M.; Garcia, J.; Scholl, F.A.; Pasmooij, A.M.; Cassarino, D.; Chang, H.Y.; Khavari, P.A. Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell 2009, 15, 477–488. [Google Scholar] [CrossRef]
- Stevens, S.; Schrader, A.J.; Vetter, G.; Eggers, H.; Blasig, H.; Becker, J.; Kuczyk, M.A.; Serth, J. Fibronectin 1 protein expression in clear cell renal cell carcinoma. Oncol. Lett. 2012, 3, 787–790. [Google Scholar] [CrossRef]
- Ma, L.J.; Lee, S.W.; Lin, L.C.; Chen, T.J.; Chang, I.W.; Hsu, H.P.; Chang, K.Y.; Huang, H.Y.; Li, C.F. Fibronectin overexpression is associated with latent membrane protein 1 expression and has independent prognostic value for nasopharyngeal carcinoma. Tumor Biol. 2014, 35, 1703–1712. [Google Scholar] [CrossRef]
- Lee, J.J.; Ng, K.Y.; Bakhtiar, A. Extracellular matrix: Unlocking new avenues in cancer treatment. Biomark. Res. 2025, 13, 78. [Google Scholar] [CrossRef]
- Serres, E.; Debarbieux, F.; Stanchi, F.; Maggiorella, L.; Grall, D.; Turchi, L.; Burel-Vandenbos, F.; Figarella-Branger, D.; Virolle, T.; Rougon, G.; et al. Fibronectin expression in glioblastomas promotes cell cohesion, collective invasion of basement membrane in vitro and orthotopic tumor growth in mice. Oncogene 2013, 33, 3451–3462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, T.; Pan, Y.; Liu, J. Exploring the mechanism of fibronectin extra domain B in the tumor microenvironment and implications for targeted immunotherapy and diagnostics. Mol. Med. Rep. 2025, 31, 160. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Nandi, S.; Hindi, E.S.; Wainwright, D.A.; Han, Y.; Lesniak, M.S. Short hairpin RNA-mediated fibronectin knockdown delays tumor growth in a mouse glioma model. Neoplasia 2010, 12, 837–847. [Google Scholar] [CrossRef]
- Adamson, C.; Kanu, O.O.; Mehta, A.I.; Di, C.; Lin, N.; Mattox, A.K.; Bigner, D.D. Glioblastoma multiforme: A review of where we have been and where we are going. Expert Opin. Investig. Drugs. 2009, 18, 1061–1083. [Google Scholar] [CrossRef]
- Rose, M.; Cardon, T.; Aboulouard, S.; Hajjaji, N.; Kobeissy, F.; Duhamel, M.; Fournier, I.; Salzet, M. Surfaceome Proteomic of Glioblastoma Revealed Potential Targets for Immunotherapy. Front. Immunol. 2021, 12, 746168. [Google Scholar] [CrossRef] [PubMed]
- Tilak, M.; Holborn, J.; New, L.A.; Lalonde, J.; Jones, N. Receptor Tyrosine Kinase Signaling and Targeting in Glioblastoma Multiforme. Int. J. Mol. Sci. 2021, 22, 1831. [Google Scholar] [CrossRef]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Jovčevska, I.; Kočevar, N.; Komel, R. Glioma and glioblastoma—How much do we (not) know? Mol. Clin. Oncol. 2013, 1, 935–941. [Google Scholar] [CrossRef]
- Ito, H.; Nakashima, H.; Chiocca, E.A. Molecular Responses to Immune Checkpoint Blockade in Glioblastoma. Nat. Med. 2019, 25, 359–361. [Google Scholar] [CrossRef]
- Roth, P.; Mason, W.P.; Richardson, P.G.; Weller, M. Proteasome Inhibition for the Treatment of Glioblastoma. Expert Opin. Investig. Drugs 2020, 29, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Agoni, C.; Fernández-Díaz, R.; Timmons, P.B.; Adelfio, A.; Gómez, H.; Shields, D.C. Molecular Modelling in Bioactive Peptide Discovery and Characterisation. Biomolecules 2025, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.H.M.; Sodero, A.C.R.; Jofily, P.; Silva-Jr, F.P. Key Topics in Molecular Docking for Drug Design. Int. J. Mol. Sci. 2019, 20, 4574. [Google Scholar] [CrossRef] [PubMed]
- Bornot, A.; Etchebest, C.; de Brevern, A.G. Predicting protein flexibility through the prediction of local structures. Proteins 2011, 79, 839–852. [Google Scholar] [CrossRef]
- De Brevern, A.G. Impact of protein dynamics on secondary structure prediction. Biochimie 2020, 179, 14–22. [Google Scholar] [CrossRef]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N. Protein-protein interaction detection: Methods and analysis. Int. J. Proteom. 2014, 2014, 147648. [Google Scholar] [CrossRef]
- Planas-Iglesias, J.; Bonet, J.; García-García, J.; Marín-López, M.A.; Feliu, E.; Oliva, B. Understanding protein-protein interactions using local structural features. J. Mol. Biol. 2013, 425, 1210–1224. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal Component Analysis: A Method for Determining the Essential Dynamics of Proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar]
- Bahar, I.; Rader, A.J. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct Biol. 2005, 15, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Im, M.; Roh, J.; Jang, W.; Kim, W. FN1 and VEGFA are potential therapeutic targets in glioblastoma as determined by bioinformatics analysis. Cancer Genomics Proteom. 2025, 22, 70–80. [Google Scholar] [CrossRef]
- Kumra, H.; Reinhardt, D.P. Fibronectin-targeted drug delivery in cancer. Adv. Drug Deliv. Rev. 2016, 97, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, B.; Huang, S.; Qu, M.; Lin, Q.; Gong, T.; Huang, Y.; Sun, X.; He, Q.; Zhang, Z.; et al. Novel fibronectin-targeted nanodisk drug delivery system displayed superior efficacy against prostate cancer compared with nanospheres. Nano Res. 2019, 12, 2451–2459, Erratum in Nano Res. 2020, 13, 597. [Google Scholar] [CrossRef]
- Yuan, L.; Siegel, M.; Choi, K.; Khosla, C.; Miller, C.R.; Jackson, E.N.; Piwnica-Worms, D.; Rich, K.M. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene 2007, 26, 2563–2573. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Hinz, U.; UniProt Consortium. From protein sequences to 3D-structures and beyond: The example of the UniProt knowledgebase. Cell Mol. Life. Sci. 2010, 67, 1049–1064. [Google Scholar] [CrossRef]
- Berman, H.M.; Kleywegt, G.J.; Nakamura, H.; Markley, J.L. The Protein Data Bank archive as an open data resource. J. Comput. Aided Mol. Des. 2014, 28, 1009–1014. [Google Scholar] [CrossRef]
- BIOVIA Discovery Studio. Discovery Studio Modeling Environment; Release 4.5; Dassault Systèmes: San Diego, CA, USA, 2015. [Google Scholar]
- Kuriata, A.; Gierut, A.M.; Oleniecki, T.; Ciemny, M.P.; Kolinski, A.; Kurcinski, M.; Kmiecik, S. CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Res. 2018, 46, W338–W343. [Google Scholar] [CrossRef] [PubMed]
- Jamroz, M.; Kolinski, A.; Kmiecik, S. CABS-flex: Server for fast simulation of protein structure fluctuations. Nucleic Acids Res. 2013, 41, W427–W431. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-García, B.; Elez, K.; Koukos, P.I.; Bonvin, A.M.; Vangone, A. PRODIGY-crystal: A web-tool for classification of biological interfaces in protein complexes. Bioinformatics 2019, 35, 4821–4823. [Google Scholar] [CrossRef] [PubMed]


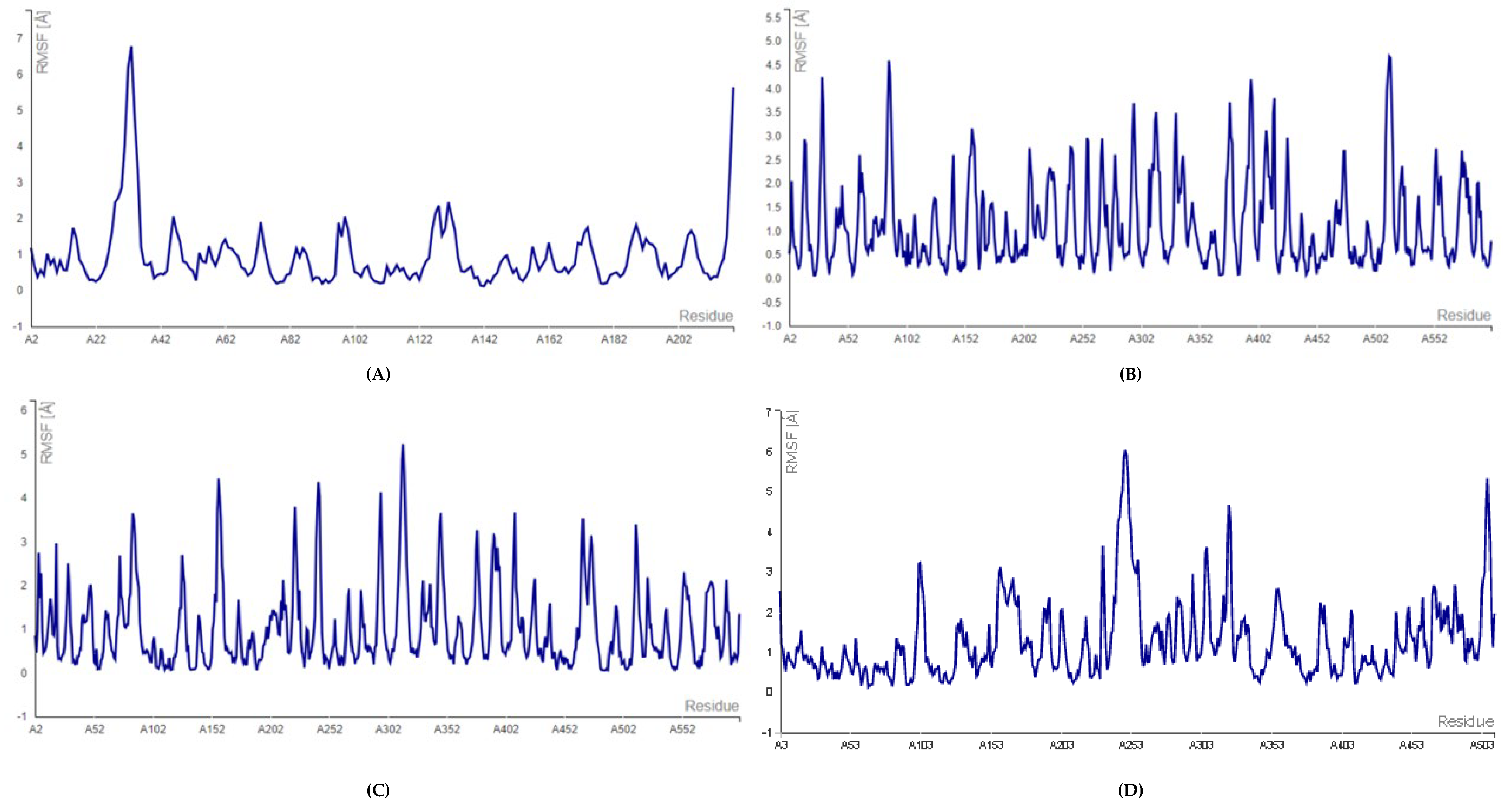
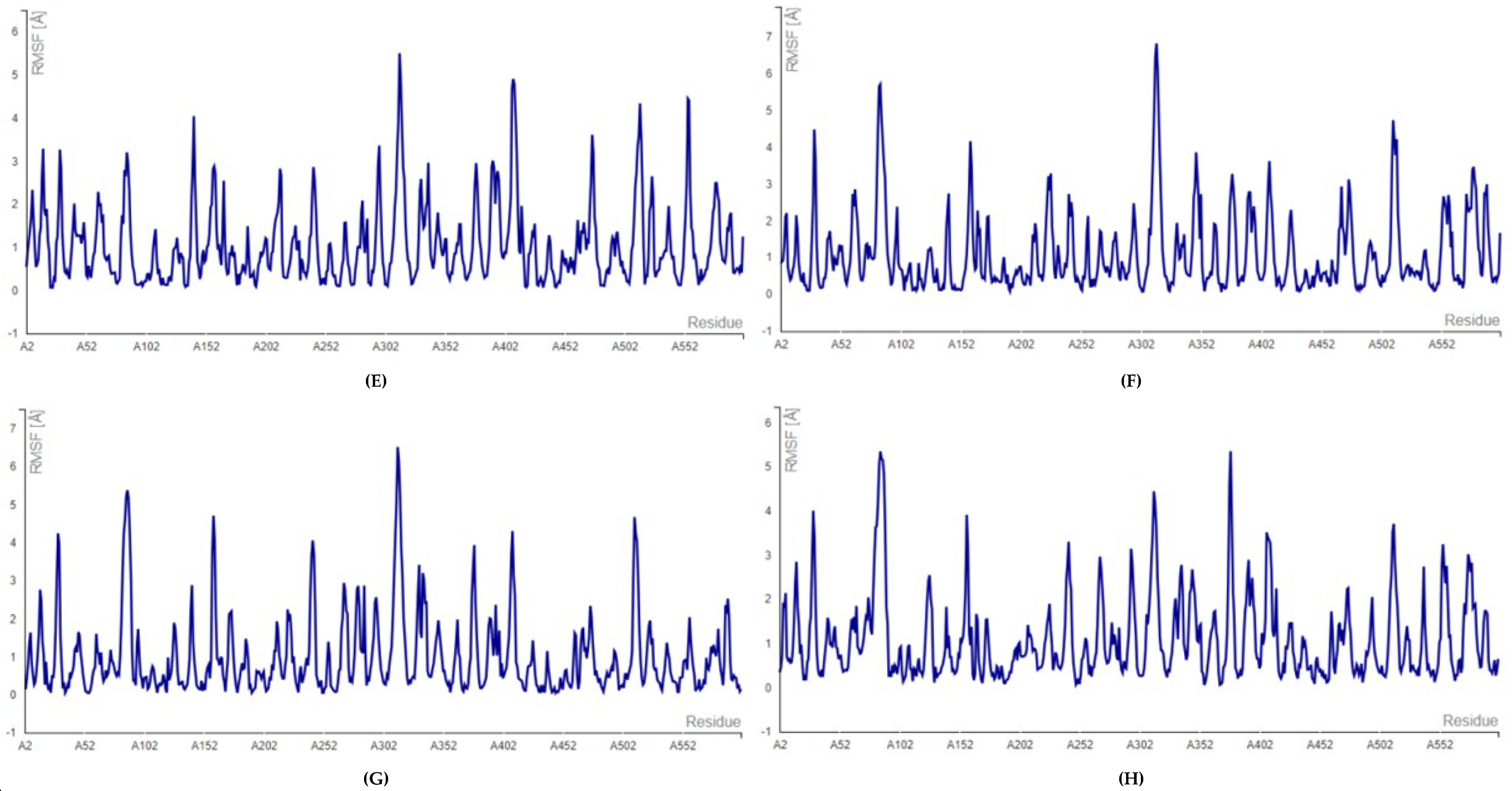
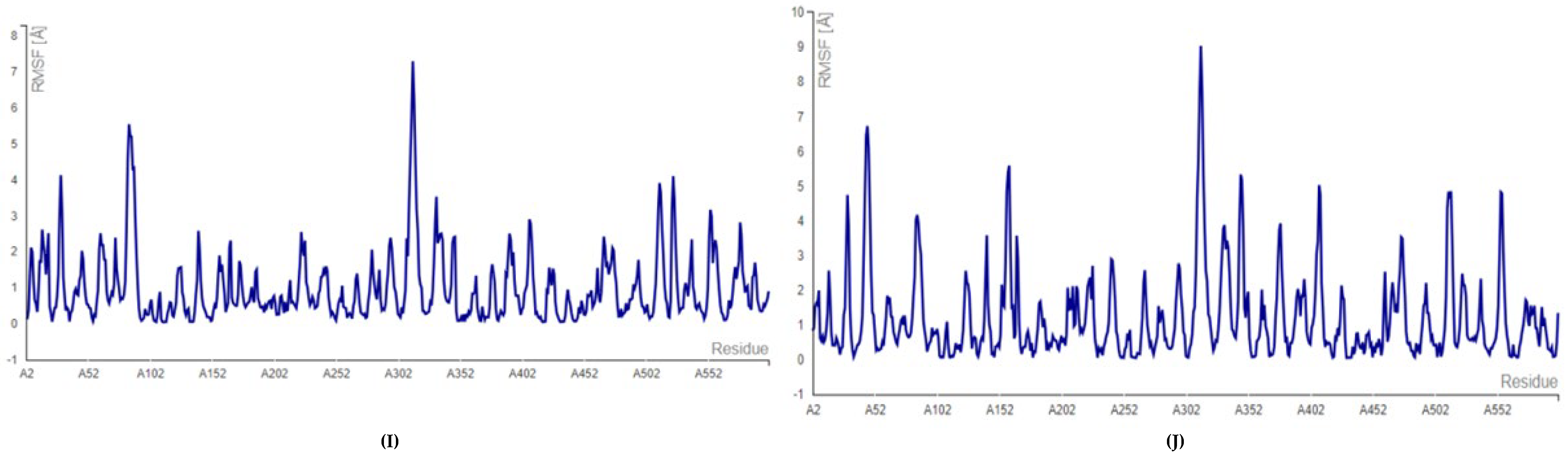
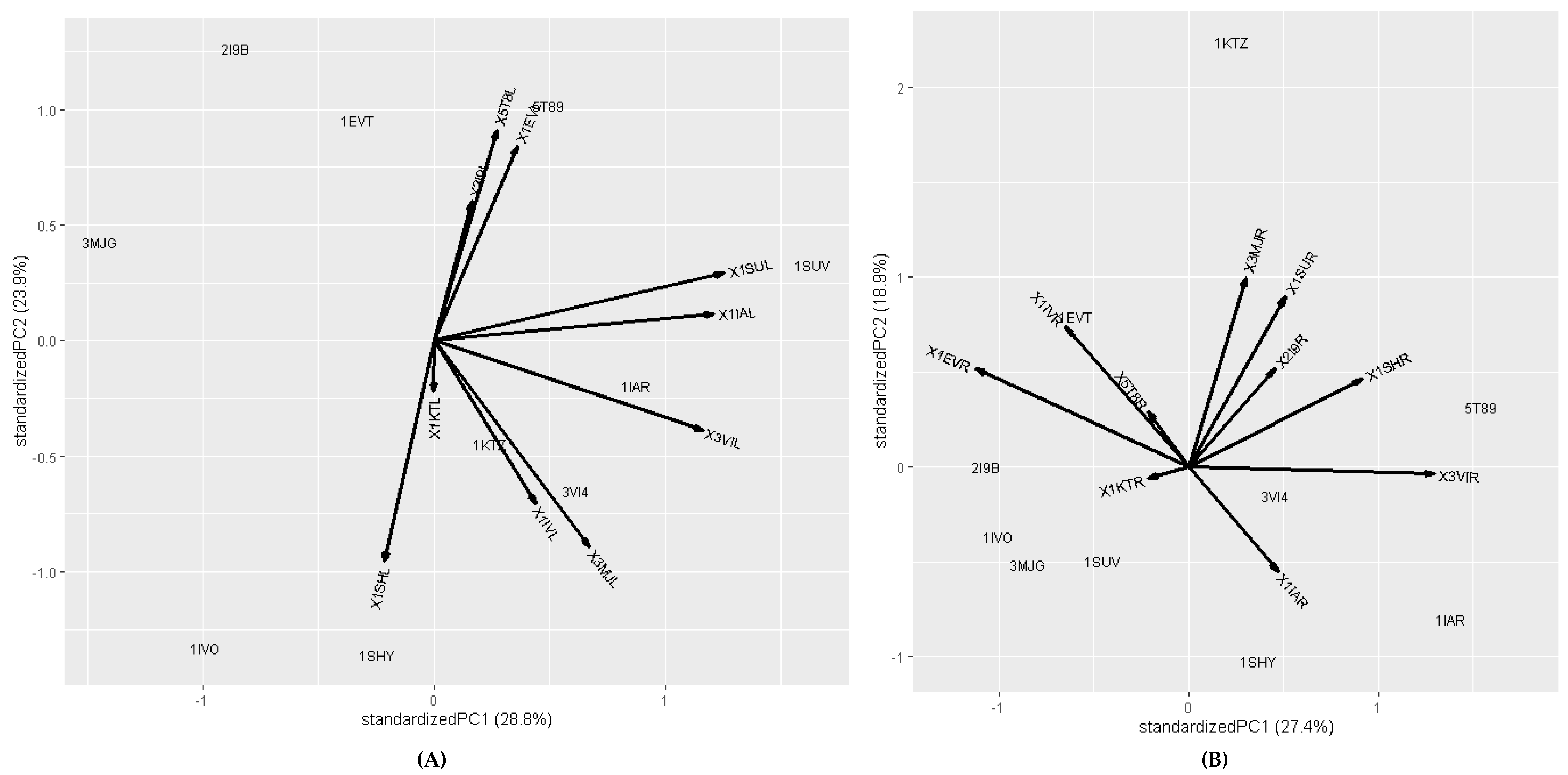
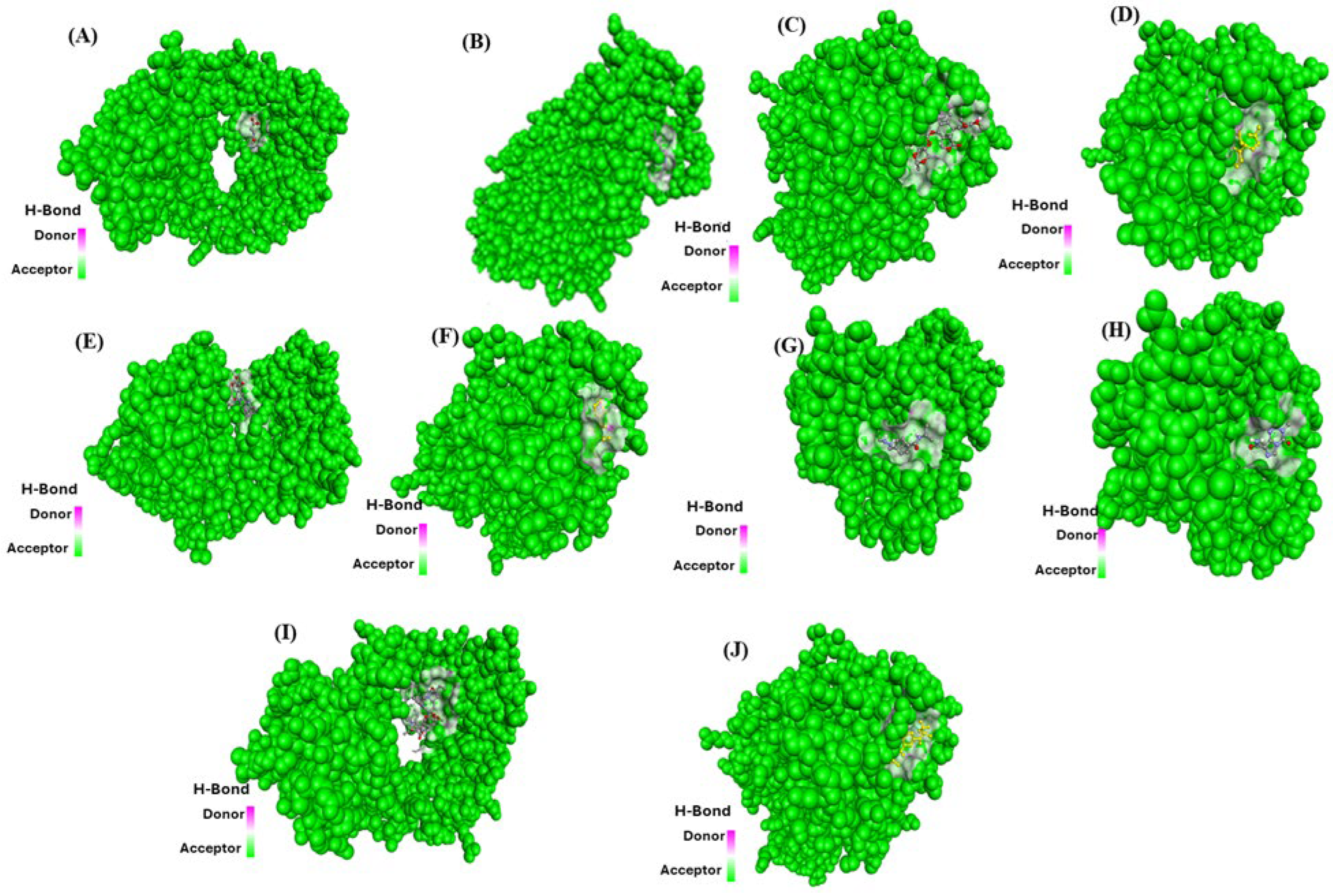
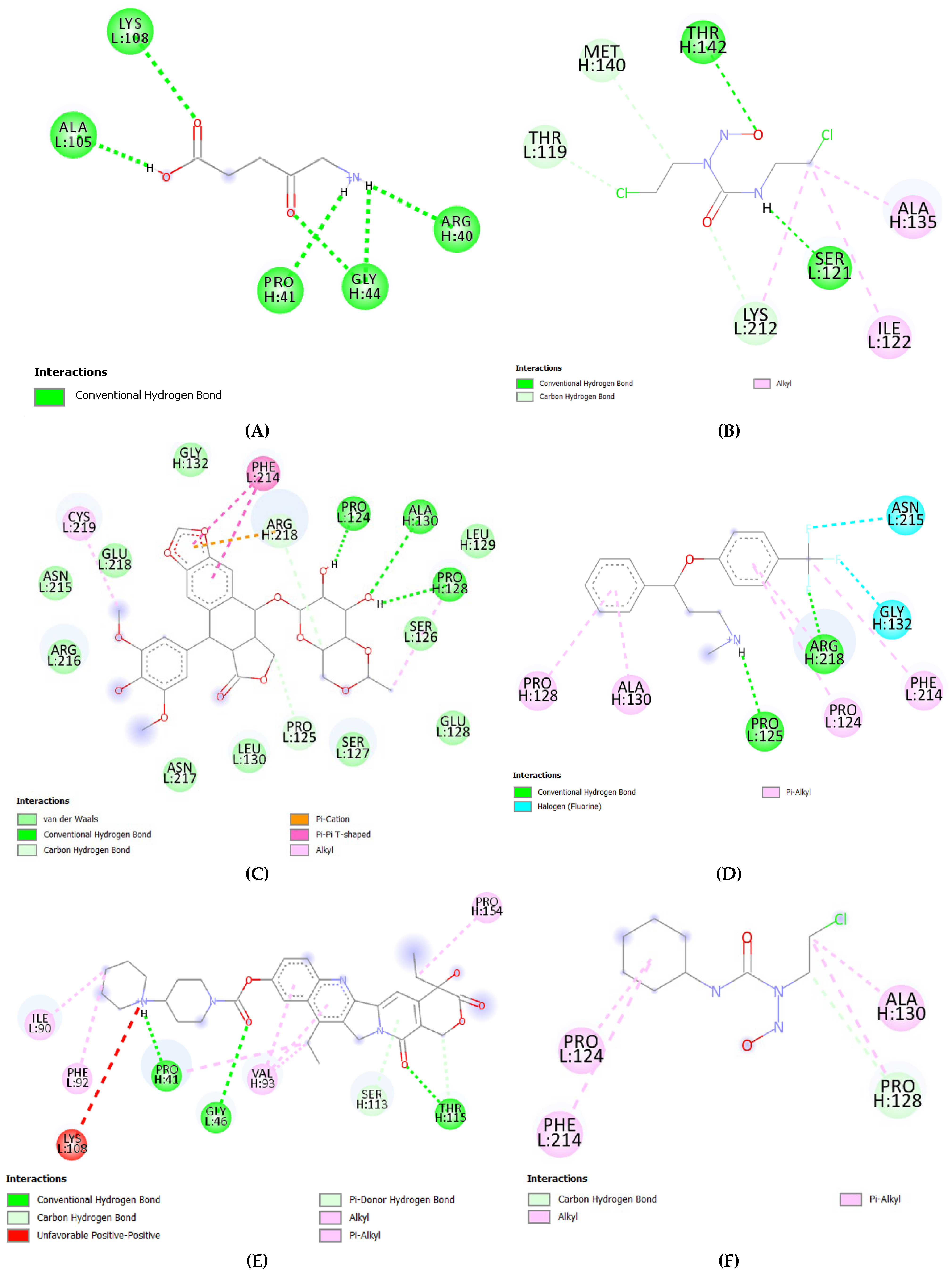

| S.No. | Receptor | PDB ID | Chain | Extracellular Domain |
|---|---|---|---|---|
| 1. | Beta-type platelet-derived growth factor receptor | 3MJG | Chain X, Y | 33–532 |
| 2. | TGF-beta type II receptor | 1KTZ | Chain B | 23–166 |
| 3. | Epidermal growth factor receptor | 1IVO | Chain A, B | 25–645 |
| 4. | Hepatocyte growth factor receptor | 1SHY | Chain B | 25–932 |
| 5. | Interleukin-4 receptor alpha chain | 1IAR | Chain B | 26–232 |
| 6. | Transferrin receptor protein 1 | 1SUV | Chain A | 89–760 |
| 7. | Vascular endothelial growth factor receptor 1 | 5T89 | Chain X, Y | 27–758 |
| 8. | Fibroblast growth factor receptor-1 | 1EVT | Chain C | 22–376 |
| 9. | Fibronectin receptor | 3VI4 | Chain L, H | 21–728 |
| 10. | Urokinase plasminogen activator surface receptor | 2I9B | Chain E, G | 23–305 |
| S.No. | Ligand | PDB ID | Chain |
|---|---|---|---|
| 1. | Platelet-derived growth factor subunit B | 3MJG | Chain A, B |
| 2. | Transforming growth factor beta 3 | 1KTZ | Chain A |
| 3. | Epidermal growth factor | 1IVO | Chain C |
| 4. | Hepatocyte growth factor | 1SHY | Chain A |
| 5. | Interleukin 4 | 1IAR | Chain B |
| 6. | Serotransferrin, N-lobe | 1SUV | Chain C, E |
| 7. | Vascular endothelial growth factor A | 5T89 | Chain V |
| 8. | Fibroblast growth factor 1 | 1EVT | Chain A |
| 9. | Fibronectin | 3VI4 | Chain B, D |
| 10. | Urokinase plasminogen activator | 2I9B | Chain A |
| S.No | Parameters | 3VI4-3MJG | 3VI4-1KTZ | 3VI4-1IVO | 3VI4-1SHY | 3VI4-1IAR | 3VI4-1SUV | 3VI4-5T89 | 3VI4-1EVT | 3VI4-3VI4 | 3VI4-2I9B |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | HADDOCK-v.2. Score | −57.3 ± 0.7 | 80.3 ± 14.2 | −24.4 ± 13.3 | 33.3 ± 2.0 | −42.7 ± 4.5 | 31.8 ± 6.9 | 47.9 ± 7.3 | 102.9 ± 3.1 | 178.4 ± 6.3 | 186.1 ± 9.9 |
| 2. | Cluster Size | 43 | 6 | 11 | 80 | 18 | 15 | 25 | 24 | 45 | 6 |
| 3. | RMSD from the Overall Lowest Energy Structure | 6.6 ± 0.2 | 0.8 ± 0.5 | 0.6 ± 0.4 | 35.3 ± 0.1 | 1.1 ± 0.6 | 3.6 ± 0.3 | 0.7 ± 0.4 | 13.0 ± 0.1 | 0.4 ± 0.2 | 20.3 ± 0.1 |
| 4. | Van der Waals Energy | −36.8 ± 4.6 | −83.3 ± 13.2 | −67.4 ± 8.0 | −93.2 ± 4.0 | −83.0 ± 7.2 | −54.0 ± 2.6 | −65.2 ± 8.9 | −75.5 ± 5.8 | −62.8 ± 9.8 | −58.0 ± 4.2 |
| 5. | Electrostatic Energy | −115.6 ± 24.8 | −224.1 ± 48.0 | −332.4 ± 28.1 | −222.8 ± 20.8 | −271.9 ± 41.7 | −500.9 ± 51.5 | −401.5 ± 67.8 | −299.0 ± 70.1 | −711.4 ± 43.0 | −432.7 ± 43.8 |
| 6. | Desolvation Energy | 1.4 ± 0.6 | −16.1 ± 3.5 | −23.7 ± 3.6 | 22.5 ± 3.5 | −44.1 ± 4.2 | 23.7 ± 0.6 | 5.4 ± 4.6 | −3.3 ± 5.4 | 36.0 ± 3.3 | 16.9 ± 0.6 |
| 7. | Restraints Violation Energy | 11.8 ± 1.0 | 2245.6 ± 143.5 | 1332.8 ± 48.3 | 1484.8 ± 64.2 | 1388.4 ± 64.1 | 1623.2 ± 80.5 | 1879.9 ± 142.3 | 2414.5 ± 108.9 | 3474.8 ± 68.1 | 3136.9 ± 158.7 |
| 8. | Buried Surface Area | 1076.7 ± 24.2 | 2412.0 ± 60.4 | 2395.7 ± 79.7 | 2499.1 ± 84.6 | 2785.3 ± 66.8 | 2055.4 ± 136.6 | 2742.9 ± 100.4 | 2513.0 ± 57.7 | 2563.2 ± 47.4 | 2254.6 ± 48.7 |
| 9. | Z-Score | −1.2 | −1.5 | −1.8 | −2.2 | −2.6 | −1.3 | −2.1 | −1.4 | −2.3 | −2.5 |
| Ligand | Receptor | PDB ID | Binding Affinity |
| Fibronectin (3VI4) | Beta-type platelet-derived growth factor receptor | 3MJG | −21.3 |
| TGF-beta type II receptor | 1KTZ | −18.9 | |
| Epidermal growth factor receptor | 1IVO | −19.4 | |
| Hepatocyte growth factor receptor | 1SHY | −18.5 | |
| Iinterleukin-4 receptor alpha chain | 1IAR | −17.7 | |
| Transferrin receptor protein 1 | 1SUV | −17.3 | |
| Vascular endothelial growth factor receptor 1 | 5T89 | −17.7 | |
| Fibroblast growth factor receptor 1 | 1EVT | −20.8 | |
| Fibronectin receptor | 3VI4 | −17.2 | |
| Urokinase plasminogen activator surface receptor | 2I9B | −19.9 |
| Receptor | Receptor | PDB ID | Binding Affinity |
|---|---|---|---|
| Fibronectin receptor (3VI4) | Beta-type platelet-derived growth factor receptor | 3MJG | −19.5 |
| TGF-beta type II receptor | 1KTZ | −18.6 | |
| Epidermal growth factor receptor | 1IVO | −20.4 | |
| Hepatocyte growth factor receptor | 1SHY | −18.5 | |
| Interleukin-4 receptor alpha chain | 1IAR | −17.1 | |
| Transferrin receptor protein 1 | 1SUV | −18.5 | |
| Vascular endothelial growth factor receptor 1 | 5T89 | −16.2 | |
| Fibroblast growth factor receptor 1 | 1EVT | −19.6 | |
| Fibronectin receptor | 3VI4 | −18.2 | |
| Urokinase plasminogen activator surface receptor | 2I9B | −20.8 |
| S. No | Ligand | Drugs | Binding Energy | RMSD | Image (Drugs) |
|---|---|---|---|---|---|
| 1. | Fibronectin | Temozolomide | −5.4 | 1.54 | 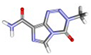 |
| 2. | Carmustine | −4.9 | 1.64 |  | |
| 3. | Lomustine | −5.4 | 1.25 |  | |
| 4. | Irinotecan | −9.4 | 1.81 |  | |
| 5. | Procarbazine | −5.8 | 3.22 |  | |
| 6. | Vincristine | −7.9 | 2.85 |  | |
| 7. | Fluoxetine | −6.2 | 1.79 |  | |
| 8. | Etoposide | −8.2 | 0.84 |  | |
| 9. | Vorasidenib | −7.9 | 4.25 |  | |
| 10. | 5-Aminolevulinic acid | −4.4 | 0.64 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.W.A.; Alam, M.J.; Sherwani, S.; Alouffi, S.; Al-Motair, K.; Khan, S.; Khan, S.N. Molecular Docking and Simulation Analysis of Glioblastoma Cell Surface Receptors and Their Ligands: Identification of Inhibitory Drugs Targeting Fibronectin Ligand to Potentially Halt Glioblastoma Pathogenesis. Int. J. Mol. Sci. 2025, 26, 10038. https://doi.org/10.3390/ijms262010038
Khan MWA, Alam MJ, Sherwani S, Alouffi S, Al-Motair K, Khan S, Khan SN. Molecular Docking and Simulation Analysis of Glioblastoma Cell Surface Receptors and Their Ligands: Identification of Inhibitory Drugs Targeting Fibronectin Ligand to Potentially Halt Glioblastoma Pathogenesis. International Journal of Molecular Sciences. 2025; 26(20):10038. https://doi.org/10.3390/ijms262010038
Chicago/Turabian StyleKhan, Mohd Wajid Ali, Mohammad Jahoor Alam, Subuhi Sherwani, Sultan Alouffi, Khalid Al-Motair, Saif Khan, and Shahper Nazeer Khan. 2025. "Molecular Docking and Simulation Analysis of Glioblastoma Cell Surface Receptors and Their Ligands: Identification of Inhibitory Drugs Targeting Fibronectin Ligand to Potentially Halt Glioblastoma Pathogenesis" International Journal of Molecular Sciences 26, no. 20: 10038. https://doi.org/10.3390/ijms262010038
APA StyleKhan, M. W. A., Alam, M. J., Sherwani, S., Alouffi, S., Al-Motair, K., Khan, S., & Khan, S. N. (2025). Molecular Docking and Simulation Analysis of Glioblastoma Cell Surface Receptors and Their Ligands: Identification of Inhibitory Drugs Targeting Fibronectin Ligand to Potentially Halt Glioblastoma Pathogenesis. International Journal of Molecular Sciences, 26(20), 10038. https://doi.org/10.3390/ijms262010038





