Neoadjuvant Chemotherapy for T3 Tumors in the Era of Precision Medicine—Biology Is Still King
Abstract
1. Introduction
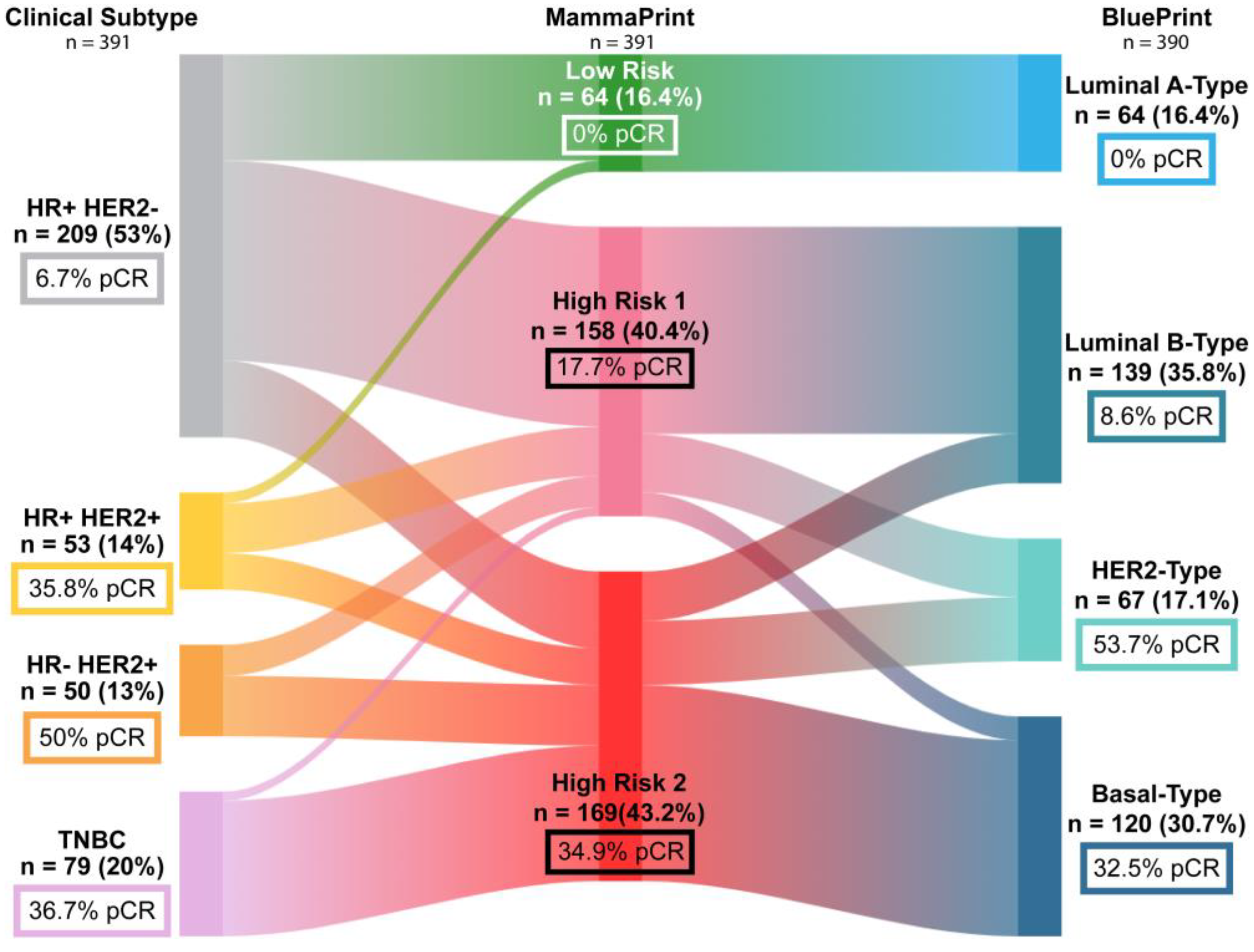
2. Results
3. Discussion
4. Materials and Methods
4.1. Patient Cohort
4.2. Molecular Subtyping
4.3. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Shelley Hwang, E.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef] [PubMed]
- Rashmi Kumar, N.; Schonfeld, R.; Gradishar, W.J.; Lurie, R.H.; Moran, M.S.; Abraham, J.; Abramson, V.; Aft, R.; Agnese, D.; Allison, K.H.; et al. NCCN Guidelines Version 6.2024 Breast Cancer; NCCN: Plymouth Meeting, PA, USA, 2024. [Google Scholar]
- Boughey, J.C.; Suman, V.J.; Mittendorf, E.A.; Ahrendt, G.M.; Wilke, L.G.; Taback, B.; Leitch, A.M.; Kuerer, H.M.; Bowling, M.; Flippo-Morton, T.S.; et al. Sentinel Lymph Node Surgery After Neoadjuvant Chemotherapy in Patients With Node-Positive Breast Cancer: The ACOSOG Z1071 (Alliance) Clinical Trial. JAMA 2013, 310, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive Molecular Portraits of Human Breast Tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The Genomic and Transcriptomic Architecture of 2,000 Breast Tumours Reveals Novel Subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.; Surujballi, J.; Awan, A.A.; Hutton, B.; Arnaout, A.; Shorr, R.; Vandermeer, L.; Alzahrani, M.J.; Clemons, M. A Scoping Review Characterizing “Choosing Wisely®” Recommendations for Breast Cancer Management. Breast Cancer Res Treat 2021, 185, 533–547. [Google Scholar] [CrossRef]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast Cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef]
- Beitsch, P.D.; Pellicane, J.V.; Pusztai, L.; Baron, P.; Cobain, E.F.; Murray, M.K.; Ashikari, A.; Kelemen, P.R.; Mislowsky, A.M.; Barone, J.; et al. MammaPrint Index as a Predictive Biomarker for Neoadjuvant Chemotherapy Response and Outcome in Patients with HR + HER2- Breast Cancer in NBRST. J. Clin. Oncol. 2023, 41, 521. [Google Scholar] [CrossRef]
- Blumencranz, P.; Habibi, M.; Shivers, S.; Acs, G.; Blumencranz, L.E.; Yoder, E.B.; van der Baan, B.; Menicucci, A.R.; Dauer, P.; Audeh, W.; et al. The Predictive Utility of MammaPrint and BluePrint in Identifying Patients with Locally Advanced Breast Cancer Who Are Most Likely to Have Nodal Downstaging and a Pathologic Complete Response After Neoadjuvant Chemotherapy. Ann. Surg. Oncol. 2023, 30, 8353–8361. [Google Scholar] [CrossRef]
- Gold, L.P.; Gray, C.R.; Marks, D.K.; Thomas, V.T.; Santillan, A.A.; Singleton, C.S.; Audeh, M.W.W.; Group, F.I. FLEX, a Real-World Evidence Full Transcriptome Study of 30,000 Patients with Early-Stage Breast Cancer. J. Clin. Oncol. 2023, 41, TPS621. [Google Scholar] [CrossRef]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.-Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef]
- Piccart, M.; van ’t Veer, L.J.; Poncet, C.; Lopes Cardozo, J.M.N.; Delaloge, S.; Pierga, J.Y.; Vuylsteke, P.; Brain, E.; Vrijaldenhoven, S.; Neijenhuis, P.A.; et al. 70-Gene Signature as an Aid for Treatment Decisions in Early Breast Cancer: Updated Results of the Phase 3 Randomised MINDACT Trial with an Exploratory Analysis by Age. Lancet Oncol. 2021, 22, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene Expression and Benefit of Chemotherapy in Women with Node-Negative, Estrogen Receptor-Positive Breast Cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørile, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Ress, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular Portraits of Human Breast Tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Glück, S.; De Snoo, F.; Peeters, J.; Stork-Sloots, L.; Somlo, G. Molecular Subtyping of Early-Stage Breast Cancer Identifies a Group of Patients Who Do Not Benefit from Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 2013, 139, 759–767. [Google Scholar] [CrossRef]
- Whitworth, P.; Stork-Sloots, L.; de Snoo, F.A.; Richards, P.; Rotkis, M.; Beatty, J.; Mislowsky, A.; Pellicane, J.V.; Nguyen, B.; Lee, L.; et al. Chemosensitivity Predicted by BluePrint 80-Gene Functional Subtype and MammaPrint in the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST). Ann. Surg. Oncol. 2014, 21, 3261–3267. [Google Scholar] [CrossRef]
- Rios-Hoyo, A.; Xiong, K.; Marczyk, M.; García-Millán, R.; Wolf, D.M.; Huppert, L.A.; Nanda, R.; Yau, C.; Hirst, G.L.; van ’t Veer, L.; et al. Correlation of Hormone Receptor Positive HER2-Negative/MammaPrint High-2 Breast Cancer with Triple Negative Breast Cancer: Results from Gene Expression Data from the ISPY2 Trial. J. Clin. Oncol. 2024, 42, 573. [Google Scholar] [CrossRef]
- Lannin, D.R.; Wang, S. Are Small Breast Cancers Good Because They Are Small or Small Because They Are Good? N. Engl. J. Med. 2017, 376, 2286–2291. [Google Scholar] [CrossRef]
- Gao, J.J.; Swain, S.M. Luminal A Breast Cancer and Molecular Assays: A Review. Oncologist 2018, 23, 556–565. [Google Scholar] [CrossRef]
- Prat, A.; Fan, C.; Fernández, A.; Hoadley, K.A.; Martinello, R.; Vidal, M.; Viladot, M.; Pineda, E.; Arance, A.; Muñoz, M.; et al. Response and Survival of Breast Cancer Intrinsic Subtypes Following Multi-Agent Neoadjuvant Chemotherapy. BMC Med. 2015, 13, 303. [Google Scholar] [CrossRef]
- Yao, K.; Goldschmidt, R.; Turk, M.; Wesseling, J.; Stork-Sloots, L.; de Snoo, F.; Cristofanilli, M. Molecular Subtyping Improves Diagnostic Stratification of Patients with Primary Breast Cancer into Prognostically Defined Risk Groups. Breast Cancer Res. Treat. 2015, 154, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Symmans, W.F.; Wei, C.; Gould, R.; Yu, X.; Zhang, Y.; Liu, M.; Walls, A.; Bousamra, A.; Ramineni, M.; Sinn, B.; et al. Long-Term Prognostic Risk after Neoadjuvant Chemotherapy Associated with Residual Cancer Burden and Breast Cancer Subtype. J. Clin. Oncol. 2017, 35, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- van Olmen, J.P.; Jacobs, C.F.; Bartels, S.A.L.; Loo, C.E.; Sanders, J.; Vrancken Peeters, M.J.T.F.D.; Drukker, C.A.; van Duijnhoven, F.H.; Kok, M. Radiological, Pathological and Surgical Outcomes after Neoadjuvant Endocrine Treatment in Patients with ER-Positive/HER2-Negative Breast Cancer with a Clinical High Risk and a Low-Risk 70-Gene Signature. Breast 2024, 75, 103726. [Google Scholar] [CrossRef] [PubMed]
- Van’t Veer, L.J.; Dai, H.; Van de Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene Expression Profiling Predicts Clinical Outcome of Breast Cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.M.; Yau, C.; Sanil, A.; Glas, A.; Petricoin, E.; Wulfkuhle, J.; Severson, T.M.; Linn, S.; Brown-Swigart, L.; Hirst, G.; et al. DNA Repair Deficiency Biomarkers and the 70-Gene Ultra-High Risk Signature as Predictors of Veliparib/Carboplatin Response in the I-SPY 2 Breast Cancer Trial. NPJ Breast Cancer 2017, 3, 31. [Google Scholar] [CrossRef]
- Haan, J.C.; Bhaskaran, R.; Ellappalayam, A.; Bijl, Y.; Griffioen, C.J.; Lujinovic, E.; Audeh, W.M.; Penault-Llorca, F.; Mittempergher, L.; Glas, A.M. MammaPrint and BluePrint Comprehensively Capture the Cancer Hallmarks in Early-Stage Breast Cancer Patients. Genes Chromosomes Cancer 2022, 61, 148–160. [Google Scholar] [CrossRef]
- Krijgsman, O.; Roepman, P.; Zwart, W.; Carroll, J.S.; Tian, S.; De Snoo, F.A.; Bender, R.A.; Bernards, R.; Glas, A.M. A Diagnostic Gene Profile for Molecular Subtyping of Breast Cancer Associated with Treatment Response. Breast Cancer Res. Treat. 2012, 133, 37–47. [Google Scholar] [CrossRef]

| Clinical Characteristics | No. Patients (%) (n = 404) |
|---|---|
| Age in years—Mean (SD) | 52 (±12) |
| Menopausal Status | |
| Pre | 186 (51.7) |
| Post | 209 (46.04) |
| Unknown | 9 (2.2) |
| Race | |
| White | 293 (72.5) |
| Black | 67 (16.6) |
| Latin/Hispanic | 24 (5.9) |
| AAPI | 12 (2.97) |
| Other | 3 (0.7) |
| Unknown | 5 (1.2) |
| Histopatholgical Type | |
| IDC | 325 (80.5) |
| ILC | 49 (12.1) |
| Mixed IDC/ILC | 18 (4.5) |
| Other | 10 (2.5) |
| Unknown | 2 (0.5) |
| Nodal Status | |
| N0 | 104 (25.7) |
| N1 | 237 (58.7) |
| N2 | 38 (9.4) |
| N3 | 12 (3.0) |
| NX | 8 (2.0) |
| Unknown | 5 (1.2) |
| Grade | |
| G1 | 20 (5.0) |
| G2 | 150 (37.1) |
| G3 | 215 (53.2) |
| GX | 13 (3.2) |
| Unknown | 6 (1.5) |
| Receptor Status | |
| HR+HER2− | 209 (51.7) |
| HR+HER2+ | 53 (13.1) |
| HR−HER2+ | 50 (12.4) |
| TNBC | 79 (19.6) |
| Unknown | 13 (3.2) |
| MammaPrint | |
| Low Risk | 65 (16.1) |
| High Risk 1 | 167 (41.3) |
| High Risk 2 | 172 (42.6) |
| BluePrint | |
| Luminal A-Type | 64 (15.8) |
| Luminal B-Type | 150 (37.1) |
| HER2-Type | 68 (16.8) |
| Basal-Type | 121 (30.1) |
| Not Requested | 1 (0.3) |
| Characteristic | Odds Ratio | 95% CI | p-Value |
|---|---|---|---|
| BluePrint Subtype | |||
| Luminal (n = 214) | 1 | ||
| Basal (n = 121) | 3.06 | [1.15, 8.19] | 0.025 |
| HER2 (n = 68) | 6.27 | [2.19, 19.38] | 0.001 |
| Menopausal Status | |||
| Pre/Peri (n = 186) | 1 | ||
| Post (n = 209) | 0.66 | [0.36, 1.19] | 0.173 |
| Receptor Status | |||
| HR+HER2− (n = 209) | 1 | ||
| HR+HER2+ (n = 53) | 2.91 | [0.97, 8.23] | 0.048 |
| HR−HER2+ (n = 50) | 2.59 | [0.82, 8.05] | 0.101 |
| TNBC (n = 79) | 2.33 | [0.91, 6.34] | 0.085 |
| Lymph Node Stage | |||
| LN− (n = 104) | 1 | ||
| LN+ (n = 287) | 1.08 | [0.55, 2.18] | 0.816 |
| Grade | |||
| G1 (n = 20) | 1 | ||
| G2 (n = 150) | 2.77 | [0.39, 56.98] | 0.38 |
| G3 (n = 215) | 4.49 | [0.66, 91.11] | 0.191 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, R.L.; Santillan, A.; Habibi, M.; Beitsch, P.; Whitworth, P.; Ramaswamy, H.; Chmielewski-Stivers, N.; Menicucci, A.; Audeh, W.; O’Shaughnessy, J. Neoadjuvant Chemotherapy for T3 Tumors in the Era of Precision Medicine—Biology Is Still King. Int. J. Mol. Sci. 2025, 26, 491. https://doi.org/10.3390/ijms26020491
Rahman RL, Santillan A, Habibi M, Beitsch P, Whitworth P, Ramaswamy H, Chmielewski-Stivers N, Menicucci A, Audeh W, O’Shaughnessy J. Neoadjuvant Chemotherapy for T3 Tumors in the Era of Precision Medicine—Biology Is Still King. International Journal of Molecular Sciences. 2025; 26(2):491. https://doi.org/10.3390/ijms26020491
Chicago/Turabian StyleRahman, Rakhshanda Layeequr, Alfredo Santillan, Mehran Habibi, Peter Beitsch, Pat Whitworth, Harshini Ramaswamy, Nicole Chmielewski-Stivers, Andrea Menicucci, William Audeh, and Joyce O’Shaughnessy. 2025. "Neoadjuvant Chemotherapy for T3 Tumors in the Era of Precision Medicine—Biology Is Still King" International Journal of Molecular Sciences 26, no. 2: 491. https://doi.org/10.3390/ijms26020491
APA StyleRahman, R. L., Santillan, A., Habibi, M., Beitsch, P., Whitworth, P., Ramaswamy, H., Chmielewski-Stivers, N., Menicucci, A., Audeh, W., & O’Shaughnessy, J. (2025). Neoadjuvant Chemotherapy for T3 Tumors in the Era of Precision Medicine—Biology Is Still King. International Journal of Molecular Sciences, 26(2), 491. https://doi.org/10.3390/ijms26020491




