Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes
Abstract
1. Introduction
2. Results
2.1. Aggregated Networks Disruption Across Breast Cancer Subtypes
2.2. Intrachromosomal and Intracytoband Interaction Dynamics in Aggregated Networks
2.3. Breast Cancer Heterogeneity in Genomic Interactions According to Co-Expression Aggregated Networks
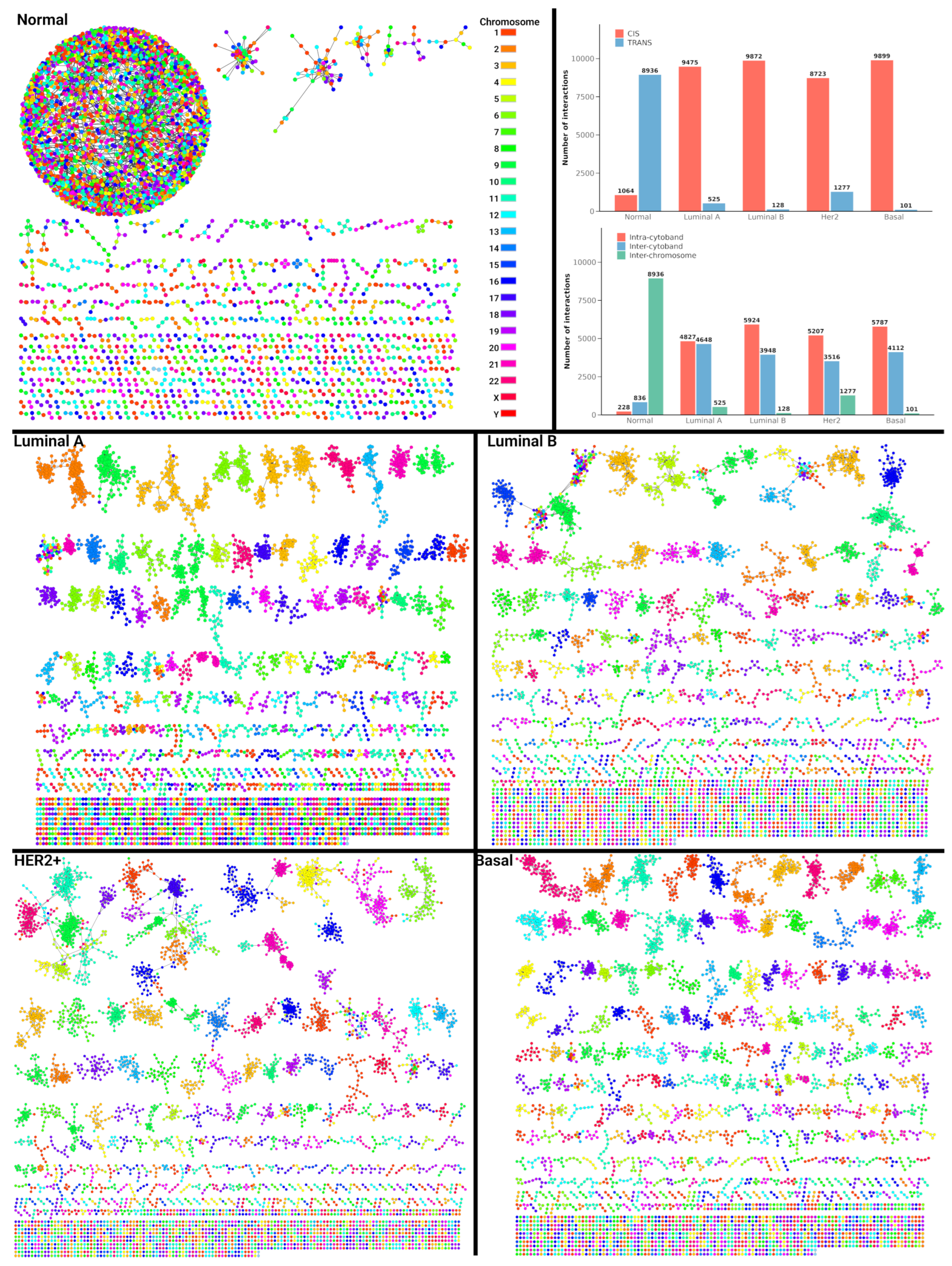
2.4. Interaction Patterns in Single-Sample Co-Expression Networks
2.5. Chromosomal and Cytoband Interaction Patterns in Single-Sample Networks
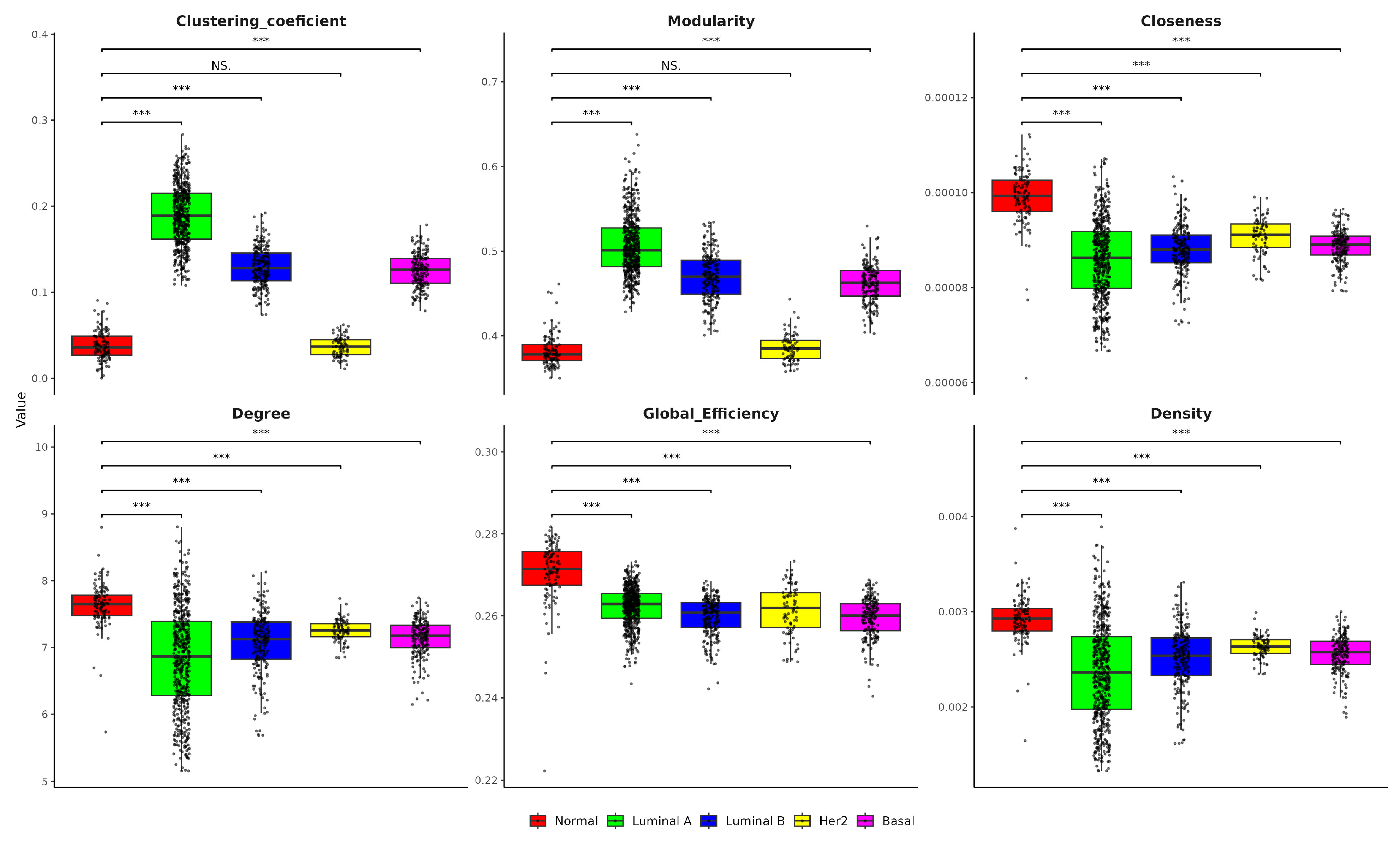
2.6. Single-Sample Network Analysis Reveals Key Topological Differences in Breast Cancer Subtypes
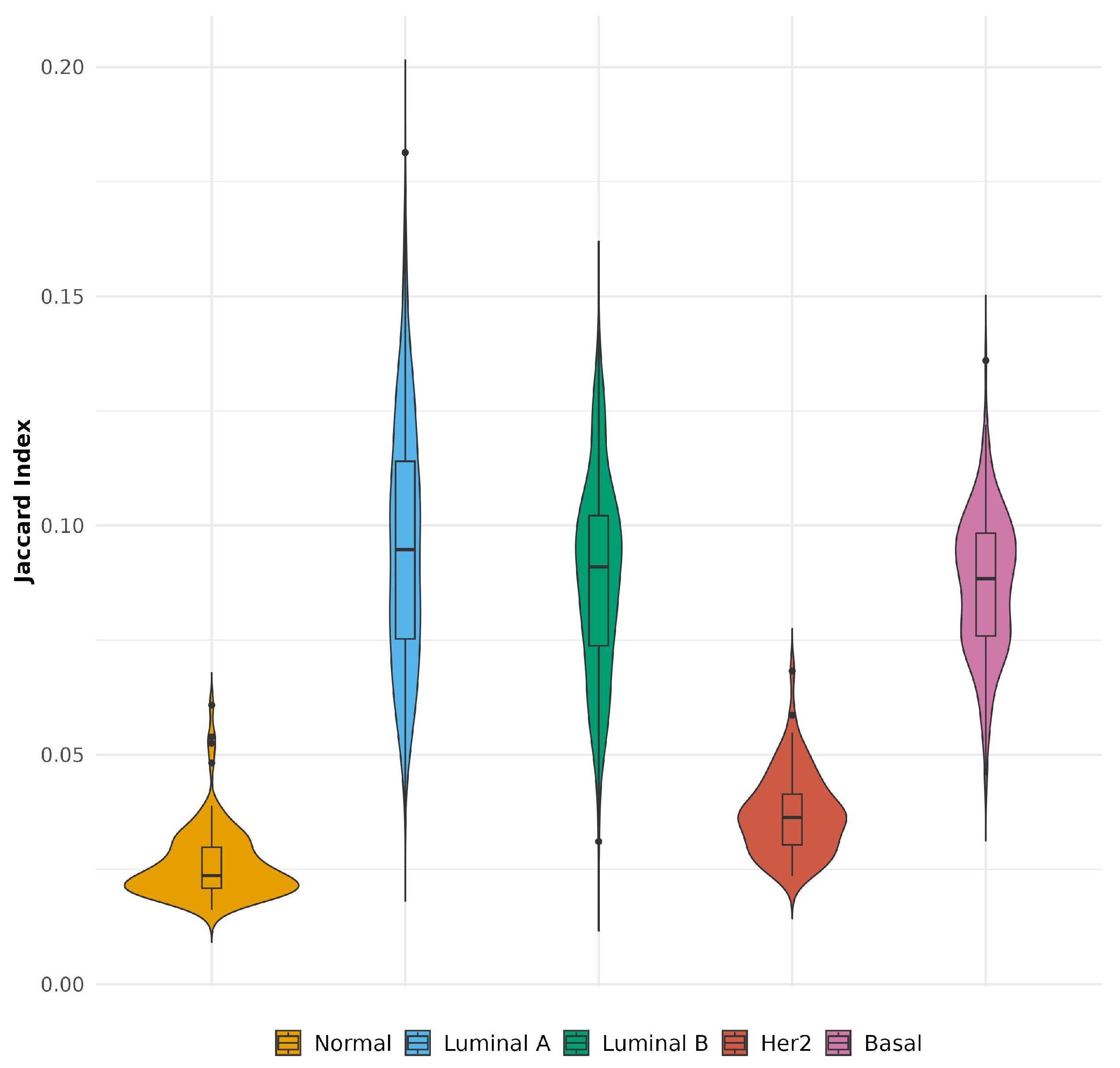
2.7. Comparative Co-Expression Patterns Between Aggregated and Single-Sample Networks
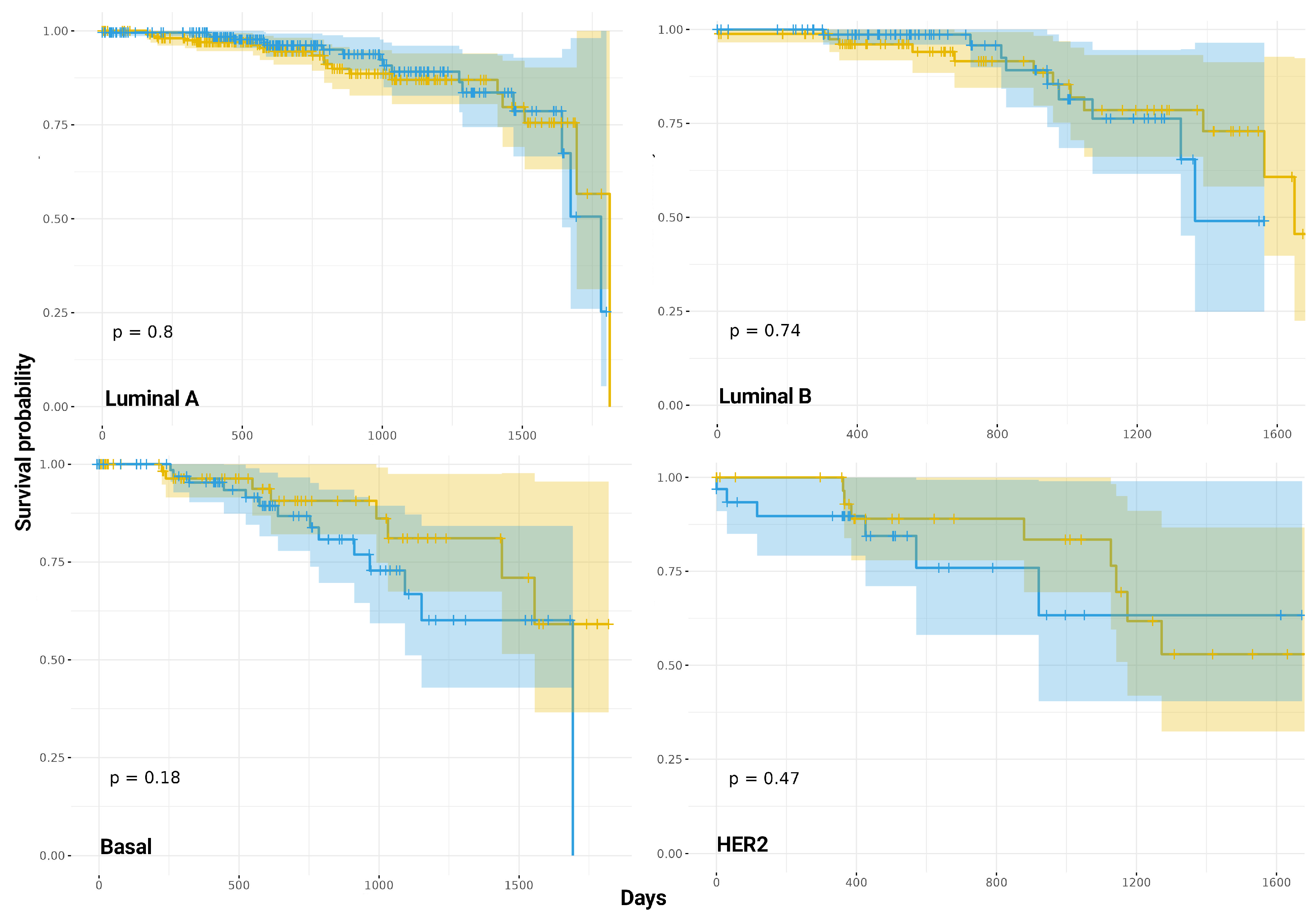
2.8. The Proportion of CIS Interactions Occurring in Single-Sample Co-Expression Networks and Survival Outcomes
2.9. High-Degree Gene Distribution and Cytoband Localization in Single-Sample Networks
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Pre-Processing
4.2. Gene Co-Expression Network Inference
4.3. Inference of Single-Sample Co-Expression Networks
4.4. Intra-Chromosomal Proportion of Co-Expression Networks
4.5. Survival Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Prim. 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Albert, R.; Barabasi, A.L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002, 74, 47–97. [Google Scholar] [CrossRef]
- Espinal-Enríquez, J.; Fresno, C.; Anda-Jáuregui, G.; Hernández-Lemus, E. RNA-Seq based genome-wide analysis reveals loss of inter-chromosomal regulation in breast cancer. Sci. Rep. 2017, 7, 1760. [Google Scholar] [CrossRef]
- García-Cortés, D.; de Anda-Jáuregui, G.; Fresno, C.; Hernández-Lemus, E.; Espinal-Enríquez, J. Gene Co-expression Is Distance-Dependent in Breast Cancer. Front. Oncol. 2020, 10, 1232. [Google Scholar] [CrossRef]
- Marcotte, R.; Sayad, A.; Brown, K.R.; Sanchez-Garcia, F.; Reimand, J.; Haider, M.; Virtanen, C.; Bradner, J.E.; Bader, G.D.; Mills, G.B.; et al. Functional genomic Landscape of Human Breast Cancer drivers, vulnerabilities, and resistance. Cell 2016, 164, 293–309. [Google Scholar] [CrossRef]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Sanchez-Garcia, F.; Villagrasa, P.; Matsui, J.; Kotliar, D.; Castro, V.; Akavia, U.D.; Chen, B.J.; Saucedo-Cuevas, L.; Rodriguez Barrueco, R.; Llobet-Navas, D.; et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell 2014, 159, 1461–1475. [Google Scholar] [CrossRef]
- Zanin, M.; Tuñas, J.M.; Menasalvas, E. Understanding diseases as increased heterogeneity: A complex network computational framework. J. R. Soc. Interface 2018, 15, 20180405. [Google Scholar] [CrossRef] [PubMed]
- Kuijjer, M.L.; Tung, M.G.; Yuan, G.; Quackenbush, J.; Glass, K. Estimating Sample-Specific Regulatory Networks. iScience 2019, 14, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Li, L.; Zeng, T.; Chen, L. Cell-specific network constructed by single-cell RNA sequencing data. Nucleic Acids Res. 2019, 47, e62. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.; Ji, H.; Aihara, K.; Chen, L. Personalized characterization of diseases using sample-specific networks. Nucleic Acids Res. 2016, 44, e164. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Ramos, C.M.; Belova, T.; Brunner, T.H.; Ben Guebila, M.; Osorio, D.; Quackenbush, J.; Kuijjer, M.L. Regulatory Network of PD1 Signaling Is Associated with Prognosis in Glioblastoma Multiforme. Cancer Res. 2021, 81, 5401–5412. [Google Scholar] [CrossRef]
- Kuijjer, M.L.; Hsieh, P.H.; Quackenbush, J.; Glass, K. lionessR: Single sample network inference in R. BMC Cancer 2019, 19, 1003. [Google Scholar] [CrossRef]
- Zamora-Fuentes, J.M.; Hernández-Lemus, E.; Espinal-Enríquez, J. Gene Expression and Co-expression Networks Are Strongly Altered Through Stages in Clear Cell Renal Carcinoma. Front. Genet. 2020, 11, 578679. [Google Scholar] [CrossRef]
- Andonegui-Elguera, S.D.; Zamora-Fuentes, J.M.; Espinal-Enríquez, J.; Hernández-Lemus, E. Loss of Long Distance Co-Expression in Lung Cancer. Front. Genet. 2021, 12, 625741. [Google Scholar] [CrossRef]
- Hernández-Gómez, C.; Hernández-Lemus, E.; Espinal-Enríquez, J. CNVs in 8q24. 3 do not influence gene co-expression in breast cancer subtypes. Front. Genet. 2023, 14, 1141011. [Google Scholar] [CrossRef]
- Nakamura-García, A.K.; Espinal-Enríquez, J. The network structure of hematopoietic cancers. Sci. Rep. 2023, 13, 19837. [Google Scholar] [CrossRef]
- García-Cortés, D.; Hernández-Lemus, E.; Espinal-Enríquez, J. Luminal A Breast Cancer Co-expression Network: Structural and Functional Alterations. Front. Genet. 2021, 12, 629475. [Google Scholar] [CrossRef] [PubMed]
- Dorantes-Gilardi, R.; García-Cortés, D.; Hernández-Lemus, E.; Espinal-Enríquez, J. Multilayer approach reveals organizational principles disrupted in breast cancer co-expression networks. Appl. Netw. Sci. 2020, 5, 1–23. [Google Scholar] [CrossRef]
- Dorantes-Gilardi, R.; García-Cortés, D.; Hernández-Lemus, E.; Espinal-Enríquez, J. K-core genes underpin structural features of breast cancer. Sci. Rep. 2021, 11, 16284. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.Y.; Cao, W.M. The progress in our understanding of CIN in breast cancer research. Front. Oncol. 2023, 13, 1067735. [Google Scholar] [CrossRef]
- Duijf, P.H.G.; Nanayakkara, D.; Nones, K.; Srihari, S.; Kalimutho, M.; Khanna, K.K. Mechanisms of Genomic Instability in Breast Cancer. Trends Mol. Med. 2019, 25, 595–611. [Google Scholar] [CrossRef]
- Serrano-Carbajal, E.A.; Espinal-Enríquez, J.; Hernández-Lemus, E. Targeting metabolic deregulation landscapes in breast cancer subtypes. Front. Oncol. 2020, 10, 97. [Google Scholar] [CrossRef]
- Garcia-Cortes, D.; Hernandez-Lemus, E.; Espinal Enríquez, J. Loss of long-range co-expression is a common trait in cancer. bioRxiv 2022. [Google Scholar] [CrossRef]
- González-Espinoza, A.; Zamora-Fuentes, J.; Hernández-Lemus, E.; Espinal-Enríquez, J. Gene Co-Expression in Breast Cancer: A Matter of Distance. Front. Oncol. 2021, 11, 726493. [Google Scholar] [CrossRef]
- Hernández-Gómez, C.; Hernández-Lemus, E.; Espinal-Enríquez, J. The role of copy number variants in gene co-expression patterns for luminal b breast tumors. Front. Genet. 2022, 13, 806607. [Google Scholar] [CrossRef]
- Guo, L.; Kong, D.; Liu, J.; Zhan, L.; Luo, L.; Zheng, W.; Zheng, Q.; Chen, C.; Sun, S. Breast cancer heterogeneity and its implication in personalized precision therapy. Exp. Hematol. Oncol. 2023, 12, 1–27. [Google Scholar] [CrossRef]
- Lopes-Ramos, C.M.; Kuijjer, M.L.; Ogino, S.; Fuchs, C.S.; DeMeo, D.L.; Glass, K.; Quackenbush, J. Gene Regulatory Network Analysis Identifies Sex-Linked Differences in Colon Cancer Drug Metabolism. Cancer Res. 2018, 78, 5538–5547. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.G.; Barlow, K.F.; McLay, K.E.; Kaul, R.; Swarbreck, D.; Dunham, A.; Scott, C.E.; Howe, K.L.; Woodfine, K.; Spencer, C.C.A.; et al. The DNA sequence and biological annotation of human chromosome 1. Nature 2006, 441, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Newsham, I.F. The Long and Short of Chromosome 11 in Breast Cancer. Am. J. Pathol. 1998, 153, 5–9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reinholz, M.M.; Bruzek, A.K.; Visscher, D.W.; Lingle, W.L.; Schroeder, M.J.; Perez, E.A.; Jenkins, R.B. Breast cancer and aneusomy 17: Implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009, 10, 267–277. [Google Scholar] [CrossRef]
- Hall, J.M.; Friedman, L.; Guenther, C.; Lee, M.K.; Weber, J.L.; Black, D.M.; King, M.C. Closing in on a breast cancer gene on chromosome 17q. Am. J. Hum. Genet. 1992, 50, 1235–1242. [Google Scholar]
- Al-Kuraya, K.; Schraml, P.; Torhorst, J.; Tapia, C.; Zaharieva, B.; Novotny, H.; Spichtin, H.; Maurer, R.; Mirlacher, M.; Köchli, O.; et al. Prognostic Relevance of Gene Amplifications and Coamplifications in Breast Cancer. Cancer Res. 2004, 64, 8534–8540. [Google Scholar] [CrossRef]
- Valla, M.; Klæstad, E.; Ytterhus, B.; Bofin, A.M. CCND1 Amplification in Breast Cancer -associations With Proliferation, Histopathological Grade, Molecular Subtype and Prognosis. J. Mammary Gland. Biol. Neoplasia 2022, 27, 67–77. [Google Scholar] [CrossRef]
- Zeitz, M.J.; Ay, F.; Heidmann, J.D.; Lerner, P.L.; Noble, W.S.; Steelman, B.N.; Hoffman, A.R. Genomic Interaction Profiles in Breast Cancer Reveal Altered Chromatin Architecture. PLoS ONE 2013, 8, e73974. [Google Scholar] [CrossRef]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef]
- Arnold, A.; Papanikolaou, A. Cyclin D1 in Breast Cancer Pathogenesis. J. Clin. Oncol. 2005, 23, 4215–4224. [Google Scholar] [CrossRef]
- Qie, S.; Diehl, J.A. Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. 2016, 94, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; O’Grady, S.; Tang, M.; Crown, J. MYC as a target for cancer treatment. Cancer Treat. Rev. 2021, 94, 102154. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, Y.; Olopade, O.I. MYC and Breast Cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yu, Y. The Important Molecular Markers on Chromosome 17 and Their Clinical Impact in Breast Cancer. Int. J. Mol. Sci. 2011, 12, 5672–5683. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.M.; Shastry, M.; Hamilton, E. Targeting HER2-positive breast cancer: Advances and future directions. Nat. Rev. Drug Discov. 2023, 22, 101–126. [Google Scholar] [CrossRef]
- Bates, J.P.; Derakhshandeh, R.; Jones, L.; Webb, T.J. Mechanisms of immune evasion in breast cancer. BMC Cancer 2018, 18, 556. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, C.; Song, Y.; Wang, Z.; Wang, Y.; Luo, F.; Xu, Y.; Zhao, Y.; Wu, Z.; Xu, Y. Mechanism of immune evasion in breast cancer. OncoTargets Ther. 2017, 10, 1561–1573. [Google Scholar] [CrossRef]
- Hao, D.; Ren, C.; Li, C. Revisiting the variation of clustering coefficient of biological networks suggests new modular structure. BMC Syst. Biol. 2012, 6, 34. [Google Scholar] [CrossRef]
- Zamora-Fuentes, J.M.; Hernández-Lemus, E.; Espinal-Enríquez, J. Oncogenic role of mir-217 during clear cell renal carcinoma progression. Front. Oncol. 2022, 12, 934711. [Google Scholar] [CrossRef]
- Zamora-Fuentes, J.M.; Hernández-Lemus, E.; Espinal-Enríquez, J. Methylation-related genes involved in renal carcinoma progression. Front. Genet. 2023, 14, 1225158. [Google Scholar] [CrossRef]
- Alcalá-Corona, S.A.; Sandoval-Motta, S.; Espinal-Enriquez, J.; Hernandez-Lemus, E. Modularity in biological networks. Front. Genet. 2021, 12, 701331. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Pian, C.; Xiang, Q.; Liu, X.; Chen, Y. Personalized analysis of breast cancer using sample-specific networks. PeerJ 2020, 8, e9161. [Google Scholar] [CrossRef] [PubMed]
- Alcalá-Corona, S.A.; de Anda-Jáuregui, G.; Espinal-Enríquez, J.; Hernández-Lemus, E. Network Modularity in Breast Cancer Molecular Subtypes. Front. Physiol. 2017, 8, 915. [Google Scholar] [CrossRef] [PubMed]
- Alcalá-Corona, S.A.; de Anda-Jáuregui, G.; Espinal-Enriquez, J.; Tovar, H.; Hernández-Lemus, E. Network modularity and hierarchical structure in breast cancer molecular subtypes. In Unifying Themes in Complex Systems IX: Proceedings of the Ninth International Conference on Complex Systems 9; Springer: Cham, Switzerland, 2018; pp. 352–358. [Google Scholar]
- Chen, Y.; Li, H.; Sun, X. Construction and analysis of sample-specific driver modules for breast cancer. BMC Genom. 2022, 23, 717. [Google Scholar] [CrossRef] [PubMed]
- De Marzio, M.; Glass, K.; Kuijjer, M.L. Single-sample network modeling on omics data. BMC Biol. 2023, 21, 296. [Google Scholar] [CrossRef]
- Wang, J.; Chen, G.; Li, M.; Pan, Y. Integration of breast cancer gene signatures based on graph centrality. BMC Syst. Biol. 2011, 5, 1–10. [Google Scholar] [CrossRef]
- Kumar, S.; Vindal, V. Architecture and topologies of gene regulatory networks associated with breast cancer, adjacent normal, and normal tissues. Funct. Integr. Genom. 2023, 23, 324. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.X.; Li, Y.Y. Differential Regulatory Analysis Based on Coexpression Network in Cancer Research. BioMed Res. Int. 2016, 2016, 4241293. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Network, T.C.G.A. Comprehensive molecular portraits of human breast tumors. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Tijhuis, A.E.; Johnson, S.C.; McClelland, S.E. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol. Cytogenet. 2019, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bach, D.H.; Zhang, W.; Sood, A.K. Chromosomal Instability in Tumor Initiation and Development. Cancer Res. 2019, 79, 3995–4002. [Google Scholar] [CrossRef] [PubMed]
- Kwei, K.A.; Kung, Y.; Salari, K.; Holcomb, I.N.; Pollack, J.R. Genomic instability in breast cancer: Pathogenesis and clinical implications. Mol. Oncol. 2010, 4, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Hosea, R.; Hillary, S.; Naqvi, S.; Wu, S.; Kasim, V. The two sides of chromosomal instability: Drivers and brakes in cancer. Signal Transduct. Target. Ther. 2024, 9, 1–30. [Google Scholar] [CrossRef]
- Scully, O.J.; Shyamasundar, S.; Matsumoto, K.; Dheen, S.T.; Yip, G.W.; Bay, B.H. C1QBP Mediates Breast Cancer Cell Proliferation and Growth via Multiple Potential Signalling Pathways. Int. J. Mol. Sci. 2023, 24, 1343. [Google Scholar] [CrossRef]
- Dheeraj, A.; Marques, F.J.G.; Tailor, D.; Bermudez, A.; Resendez, A.; Pandrala, M.; Grau, B.; Kumar, P.; Haley, C.B.; Honkala, A.; et al. Inhibition of protein translational machinery in triple-negative breast cancer as a promising therapeutic strategy. Cell Rep. Med. 2024, 5, 101552. [Google Scholar] [CrossRef]
- Metro, G.; Zheng, Z.; Fabi, A.; Schell, M.; Antoniani, B.; Mottolese, M.; Monteiro, A.; Vici, P.; Rivera, S.L.; Boulware, D.; et al. In situ protein expression of RRM1, ERCC1 and BRCA1 in metastatic breast cancer patients treated with gemcitabine-based chemotherapy. Cancer Investig. 2010, 28, 172–180. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Li, X.; Hu, Y.; Liu, O.; Wang, Y.; Li, S.; Yang, Q.; Lin, B. PSMD8 can serve as potential biomarker and therapeutic target of the PSMD family in ovarian cancer: Based on bioinformatics analysis and in vitro validation. BMC Cancer 2023, 23, 573. [Google Scholar] [CrossRef]
- Geng, X.; Yuan, J.; Xu, W.; Zou, D.; Sun, Y.; Li, J. YWHAB is regulated by IRX5 and inhibits the migration and invasion of breast cancer cells. Oncol. Lett. 2024, 28, 469. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, G.; Jiang, C.; Guo, T.; Tang, H.; Yuan, Z.; Wang, Y.; Tan, X.; Chen, J.; Zhang, E.; et al. USP31 serves as a potential biomarker for predicting prognosis and immune responses for clear cell renal cell carcinoma via single-cell and bulk RNA-sequencing. J. Gene Med. 2024, 26, e3594. [Google Scholar] [CrossRef]
- Shen, H.; Ma, W.; Hu, Y.; Liu, Y.; Song, Y.; Fu, L.; Qin, Z. Mitochondrial Sirtuins in Cancer: A Revisited Review from Molecular Mechanisms to Therapeutic Strategies. Theranostics 2024, 14, 2993. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, K.; Holm, K.; Nordenskjold, B.; Borg, A.; Landberg, G. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. BCR 2008, 10, R81. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, X.; Guo, H.; Tong, Y. Variants in the 8q24 region associated with risk of breast cancer: Systematic research synopsis and meta-analysis. Medicine 2020, 99, e19217. [Google Scholar] [CrossRef] [PubMed]
- Ahmadiyeh, N.; Pomerantz, M.M.; Grisanzio, C.; Herman, P.; Jia, L.; Almendro, V.; He, H.H.; Brown, M.; Liu, X.S.; Davis, M.; et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA 2010, 107, 9742–9746. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Ma, H.; Wu, C.; Dai, J.; Zhang, R.; Shi, Y.; Lu, J.; Miao, X.; Wang, M.; Zhou, Y.; et al. Genetic Variants at 6p21.1 and 7p15.3 Are Associated with Risk of Multiple Cancers in Han Chinese. Am. J. Hum. Genet. 2012, 91, 928–934. [Google Scholar] [CrossRef]
- Khan, S.; Fagerholm, R.; Rafiq, S.; Tapper, W.; Aittomäki, K.; Liu, J.; Blomqvist, C.; Eccles, D.; Nevanlinna, H. Polymorphism at 19q13.41 predicts breast cancer survival specifically after endocrine therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4086–4096. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-Seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Nueda, M.J.; Ferrer, A.; Conesa, A. ARSyN: A method for the identification and removal of systematic noise in multifactorial time course microarray experiments. Biostatistics 2012, 13, 553–566. [Google Scholar] [CrossRef] [PubMed]
- Margolin, A.A.; Nemenman, I.; Basso, K.; Wiggins, C.; Stolovitzky, G.; Dalla Favera, R.; Califano, A. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinform. 2006, 7 (Suppl. 1), S7. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Caldelas, T.E.; Alcalá-Corona, S.A.; Espinal-Enríquez, J.; Hernandez-Lemus, E. Unveiling the link between inflammation and adaptive immunity in breast cancer. Front. Immunol. 2019, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]



| Subtype | Chromosome | Frequency | Proportion |
|---|---|---|---|
| 1 | 223 | 0.000105 | |
| Healthy | 19 | 98 | 0.000090 |
| 2 | 89 | 0.000114 | |
| 17 | 1742 | 0.002479 | |
| Luminal A | 11 | 1601 | 0.001847 |
| 8 | 871 | 0.003520 | |
| 11 | 1612 | 0.001860 | |
| Luminal B | 17 | 1530 | 0.002177 |
| 8 | 792 | 0.003201 | |
| 17 | 2047 | 0.002913 | |
| Her2 | 8 | 754 | 0.003047 |
| 11 | 698 | 0.000805 | |
| 1 | 1013 | 0.000475 | |
| Basal | 19 | 749 | 0.000687 |
| 10 | 641 | 0.002396 |
| Subtype | Cytoband | Frecuency | Proportion |
|---|---|---|---|
| 6p21.32 | 17 | 0.010650 | |
| Healthy | 8p11.23 | 13 | 0.068420 |
| 8q24.3 | 7 | 0.001210 | |
| 11q13.1 | 264 | 0.051250 | |
| Luminal A | 8q24.3 | 206 | 0.035650 |
| 11q13.2 | 151 | 0.062530 | |
| 8q24.3 | 377 | 0.065247 | |
| Luminal B | 17q11.2 | 227 | 0.066706 |
| 11q13.1 | 205 | 0.039798 | |
| 17q11.2 | 367 | 0.107850 | |
| Her2 | 8q24.3 | 331 | 0.057290 |
| 17q25.3 | 325 | 0.061870 | |
| 6p21.1 | 272 | 0.066420 | |
| Basal | 8q24.3 | 224 | 0.038770 |
| 17q25.3 | 182 | 0.034650 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponce-Cusi, R.; López-Sánchez, P.; Maracaja-Coutinho, V.; Espinal-Enríquez, J. Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes. Int. J. Mol. Sci. 2024, 25, 12163. https://doi.org/10.3390/ijms252212163
Ponce-Cusi R, López-Sánchez P, Maracaja-Coutinho V, Espinal-Enríquez J. Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes. International Journal of Molecular Sciences. 2024; 25(22):12163. https://doi.org/10.3390/ijms252212163
Chicago/Turabian StylePonce-Cusi, Richard, Patricio López-Sánchez, Vinicius Maracaja-Coutinho, and Jesús Espinal-Enríquez. 2024. "Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes" International Journal of Molecular Sciences 25, no. 22: 12163. https://doi.org/10.3390/ijms252212163
APA StylePonce-Cusi, R., López-Sánchez, P., Maracaja-Coutinho, V., & Espinal-Enríquez, J. (2024). Single-Sample Networks Reveal Intra-Cytoband Co-Expression Hotspots in Breast Cancer Subtypes. International Journal of Molecular Sciences, 25(22), 12163. https://doi.org/10.3390/ijms252212163






