Genome-Wide Isoform Switching Reveals SR45-Mediated Splicing Control of Arabidopsis Leaf Senescence
Abstract
1. Introduction
2. Results
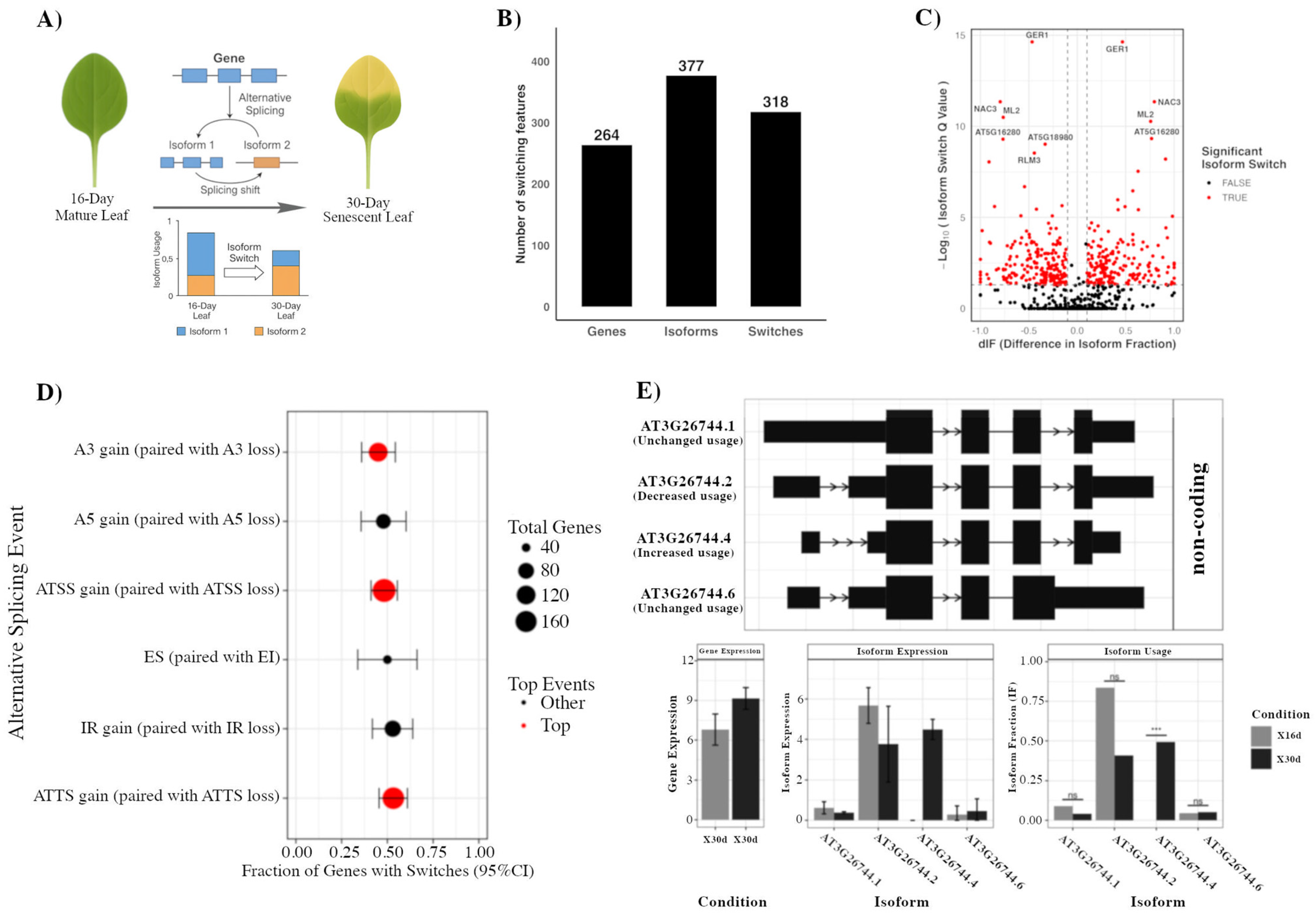
2.1. Isoform Switching During Arabidopsis Leaf Senescence
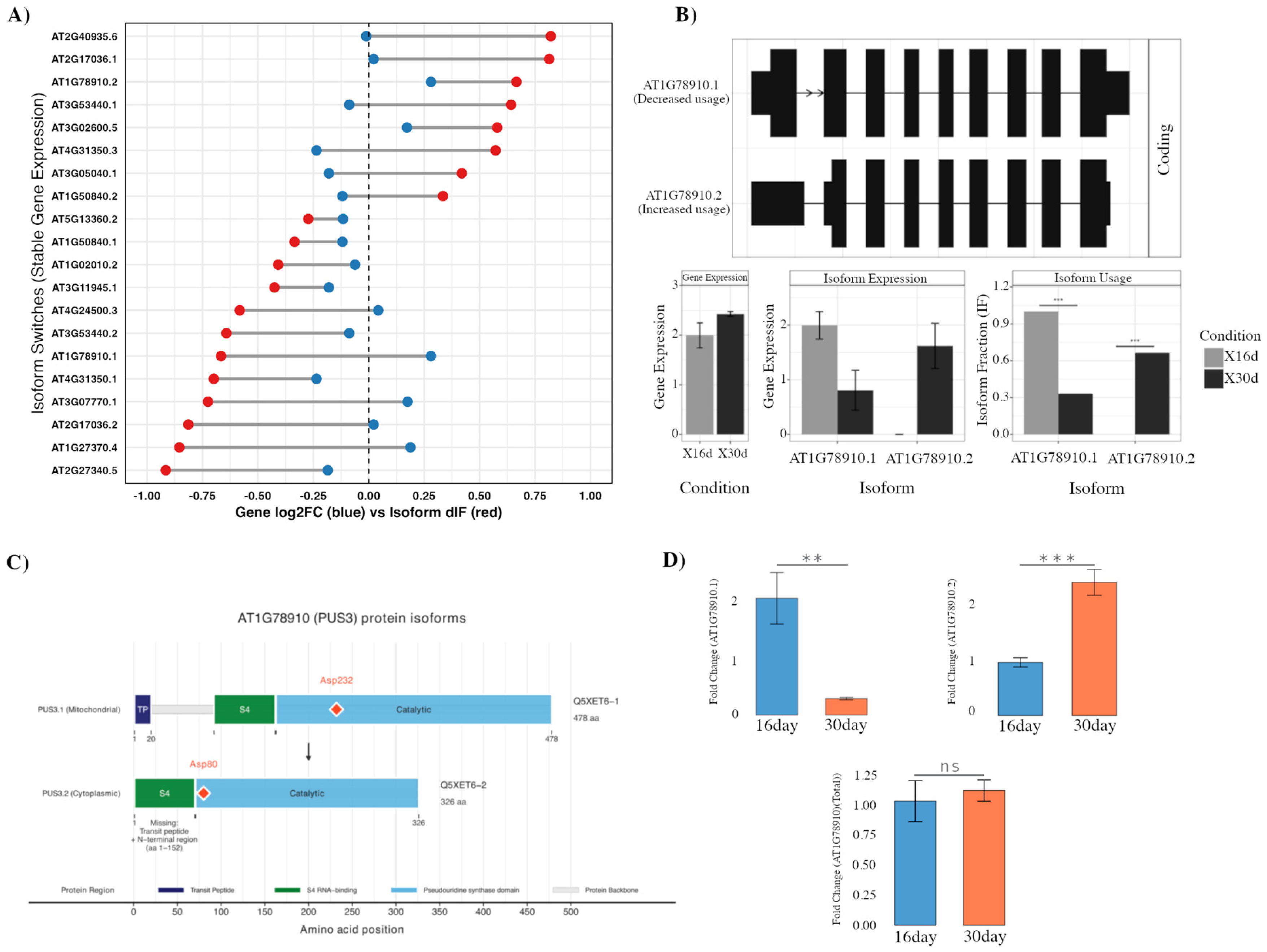
2.2. Isoform Switching Uncoupled from Gene Expression Alters Protein Localization
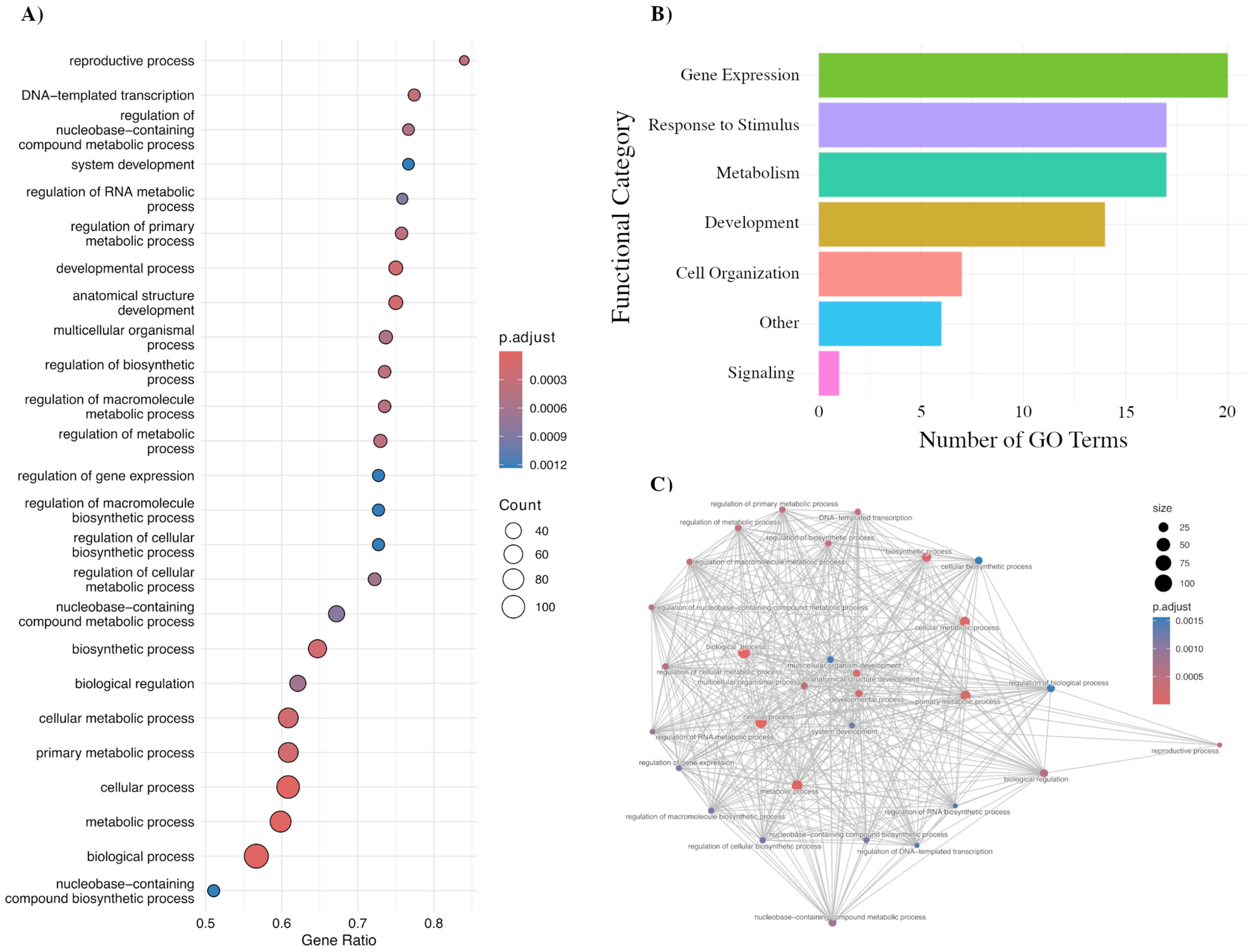
2.3. Functional Enrichment of Isoform Switching Genes
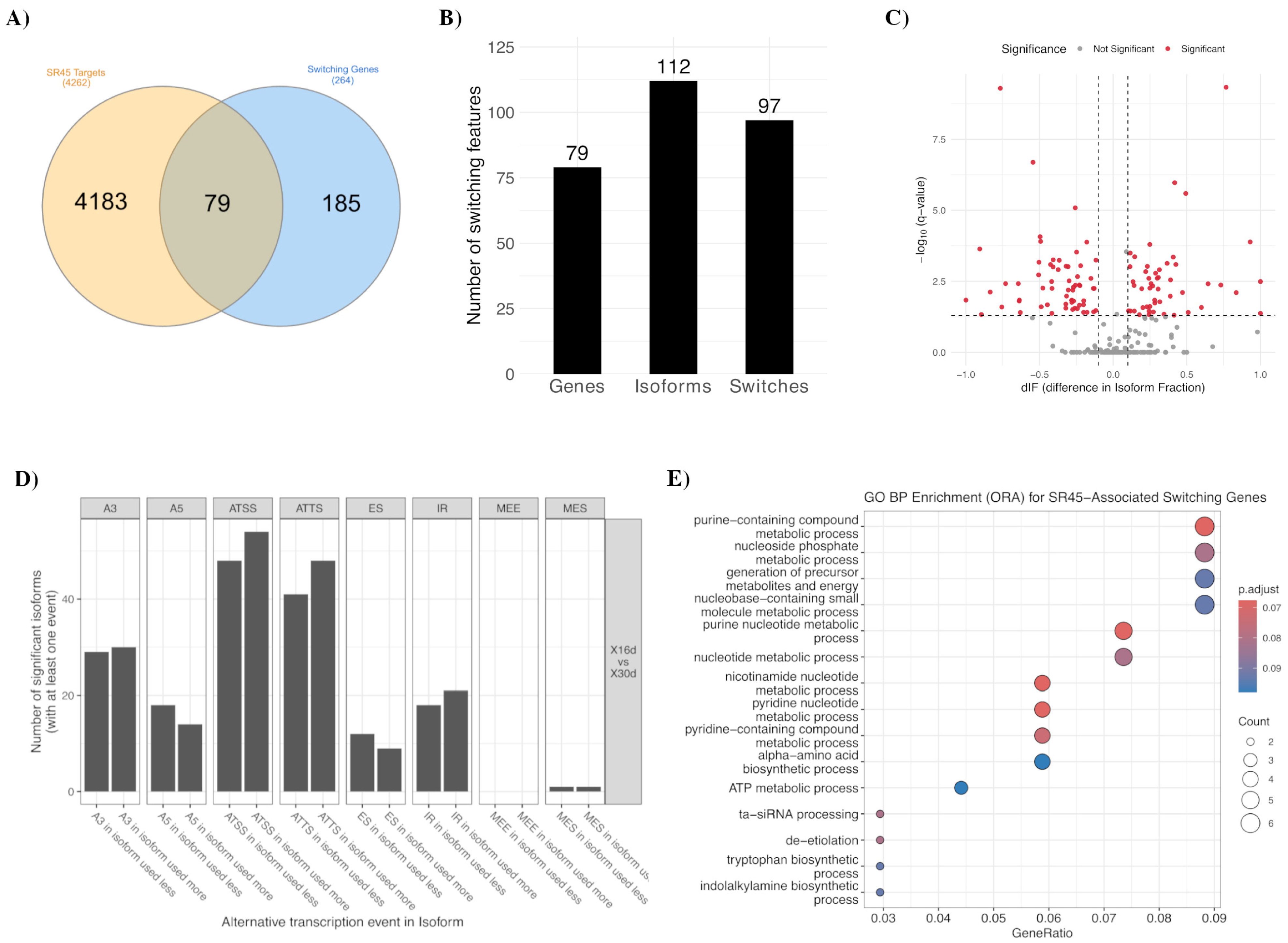
2.4. SR45 Targets Overlap with Isoform-Switching Genes
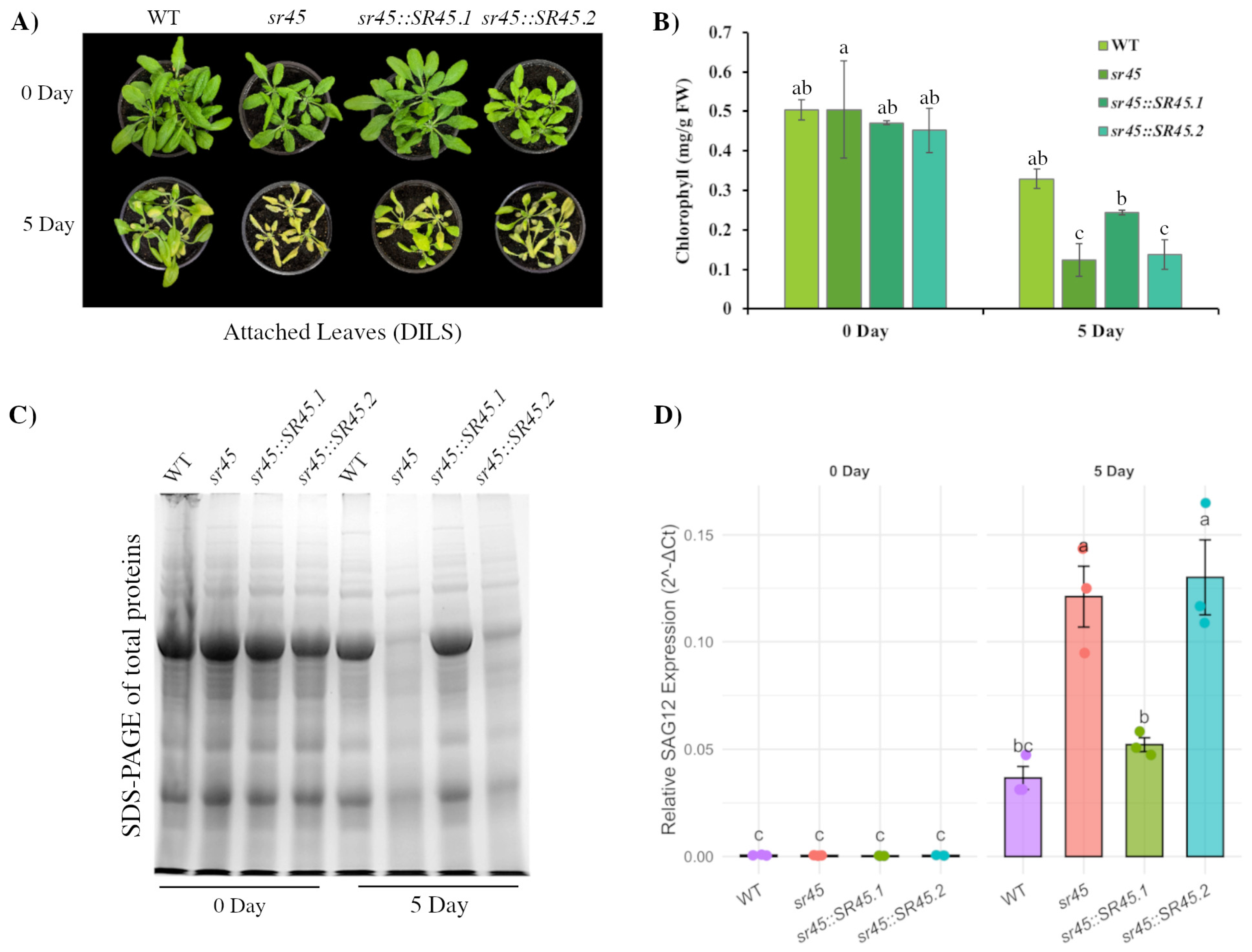
2.5. SR45 Regulates DILS in an Isoform-Specific Manner
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA-Seq Data Acquisition and Processing
4.3. Isoform Switching Analysis
4.4. Gene Ontology (GO) Enrichment Analysis
4.5. SR45 Target Overlap Analysis
4.6. Dark-Induced Leaf Senescence (DILS) and Chlorophyll Content Analysis
4.7. Total RNA Extraction and RT-PCR
4.8. Protein Extraction and SDS-PAGE
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dodge, J.D. Changes in chloroplast fine structure during the autumnal senescence of Betula leaves. Ann. Bot. 1970, 34, 817–824. [Google Scholar] [CrossRef]
- Girondé, A.; Etienne, P.; Trouverie, J.; Bouchereau, A.; Le Cahérec, F.; Leport, L.; Avice, J.C. The contrasting N management of two oilseed rape genotypes reveals the mechanisms of proteolysis associated with leaf N remobilization and the respective contributions of leaves and stems to N storage and remobilization during seed filling. BMC Plant Biol. 2015, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef] [PubMed]
- Schippers, J.H.; Jing, H.C.; Hille, J.; Dijkwel, P.P. Developmental and hormonal control of leaf senescence. In Senescence Processes in Plants; Gan, S., Ed.; Blackwell Publishing: Oxford, UK, 2007; pp. 145–170. [Google Scholar]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef]
- Sobieszczuk-Nowicka, E.; Wrzesiński, T.; Bagniewska-Zadworna, A.; Kubala, S.; Rucińska-Sobkowiak, R.; Polcyn, W.; Mattoo, A.K. Physio-genetic dissection of dark-induced leaf senescence and timing its reversal in barley. Plant Physiol. 2018, 178, 654–671. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Buchanan-Wollaston, V. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC transcription factors in senescence: From molecular structure to function in crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Cao, J.; Liu, H.; Tan, S.; Li, Z. Transcription factors-regulated leaf senescence: Current knowledge, challenges and approaches. Int. J. Mol. Sci. 2023, 24, 9245. [Google Scholar] [CrossRef]
- Kim, J.H.; Chung, K.M.; Woo, H.R. Three positive regulators of leaf senescence in Arabidopsis, ORE1, ORE3 and ORE9, play roles in crosstalk among multiple hormone-mediated senescence pathways. Genes Genom. 2011, 33, 373–381. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, C.; Li, K.; Li, X.; Xu, M.; Guo, Y. CLE14 functions as a “brake signal” to suppress age-dependent and stress-induced leaf senescence by promoting JUB1-mediated ROS scavenging in Arabidopsis. Mol. Plant 2022, 15, 179–188. [Google Scholar] [CrossRef]
- Kim, H.J.; Park, J.H.; Kim, J.; Kim, J.J.; Hong, S.; Kim, J.; Hwang, D. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4930–E4939. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Xie, D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Wang, H.L.; Zhang, Y.; Wang, T.; Yang, Q.; Yang, Y.; Li, Z. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Koyama, T.; Nii, H.; Mitsuda, N.; Ohta, M.; Kitajima, S.; Ohme-Takagi, M.; Sato, F. A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 2013, 162, 991–1005. [Google Scholar] [CrossRef]
- Riester, L.; Köster-Hofmann, S.; Doll, J.; Berendzen, K.W.; Zentgraf, U. Impact of alternatively polyadenylated isoforms of ETHYLENE RESPONSE FACTOR4 with activator and repressor function on senescence in Arabidopsis thaliana L. Genes 2019, 10, 91. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Han, S.H.; Kim, S.H.; Piao, W.; Yanagisawa, S.; Paek, N.C. Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef]
- Yang, Q.; Tan, S.; Wang, H.L.; Wang, T.; Cao, J.; Liu, H.; Li, Z. Spliceosomal protein U2B″ delays leaf senescence by enhancing splicing variant JAZ9β expression to attenuate jasmonate signaling in Arabidopsis. New Phytol. 2023, 240, 1116–1133. [Google Scholar] [CrossRef] [PubMed]
- Vitting-Seerup, K.; Sandelin, A. IsoformSwitchAnalyzeR: Analysis of changes in genome-wide patterns of alternative splicing and its functional consequences. Bioinformatics 2019, 35, 4469–4471. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, J.; Li, L.F.; Guo, P.; Wang, Y.; Cushman, S.A.; Shang, F.D. Roles and regulatory patterns of protein isoforms in plant adaptation and development. New Phytol. 2025, 245, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Kashkan, I.; Timofeyenko, K.; Růžička, K. How alternative splicing changes the properties of plant proteins. Quant. Plant Biol. 2022, 3, e14. [Google Scholar] [CrossRef]
- Wu, H.Y.L.; Jen, J.; Hsu, P.Y. What, where, and how: Regulation of translation and the translational landscape in plants. Plant Cell 2024, 36, 1540–1564. [Google Scholar] [CrossRef]
- Bernardes, W.S.; Menossi, M. Plant 3′ regulatory regions from mRNA-encoding genes and their uses to modulate expression. Front. Plant Sci. 2020, 11, 1252. [Google Scholar] [CrossRef]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative splicing and protein diversity: Plants versus animals. Front. Plant Sci. 2019, 10, 708. [Google Scholar] [CrossRef]
- Alhabsi, A.; Ling, Y.; Crespi, M.; Reddy, A.S.; Mahfouz, M. Alternative splicing dynamics in plant adaptive responses to stress. Annu. Rev. Plant Biol. 2025, 76. in press. [Google Scholar] [CrossRef]
- Muhammad, S.; Xu, X.; Zhou, W.; Wu, L. Alternative splicing: An efficient regulatory approach towards plant developmental plasticity. Wiley Interdiscip. Rev. RNA 2023, 14, e1758. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Jin, X. Regulatory network of serine/arginine-rich (SR) proteins: The molecular mechanism and physiological function in plants. Int. J. Mol. Sci. 2022, 23, 10147. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.C.; Das, D.; Zhang, Y.; Fernie, A.R.; Liu, Y.G.; Chen, M.; Zhang, J. Plant serine/arginine-rich proteins: Versatile players in RNA processing. Planta 2023, 257, 109. [Google Scholar] [CrossRef] [PubMed]
- Shepard, P.J.; Hertel, K.J. The SR protein family. Genome Biol. 2009, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Fanara, S.; Schloesser, M.; Joris, M.; De Franco, S.; Vandevenne, M.; Kerff, F.; Motte, P. The Arabidopsis SR45 splicing factor bridges the splicing machinery and the exon–exon junction complex. J. Exp. Bot. 2024, 75, 2280–2298. [Google Scholar] [CrossRef]
- Ali, G.S.; Palusa, S.G.; Golovkin, M.; Prasad, J.; Manley, J.L.; Reddy, A.S. Regulation of plant developmental processes by a novel splicing factor. PLoS ONE 2007, 2, e471. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Carvalho, S.D.; Duque, P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol. 2010, 154, 772–783. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Szakonyi, D.; Simpson, C.G.; Barbosa, I.C.; Brown, J.W.; Baena-González, E.; Duque, P. The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 2016, 28, 1910–1925. [Google Scholar] [CrossRef]
- Albaqami, M.; Laluk, K.; Reddy, A.S. The Arabidopsis splicing regulator SR45 confers salt tolerance in a splice isoform-dependent manner. Plant Mol. Biol. 2019, 100, 379–390. [Google Scholar] [CrossRef]
- Albaqami, M. The splicing factor SR45 negatively regulates anthocyanin accumulation under high-light stress in Arabidopsis thaliana. Life 2023, 13, 1386. [Google Scholar] [CrossRef]
- Xing, D.; Wang, Y.; Hamilton, M.; Ben-Hur, A.; Reddy, A.S. Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 2015, 27, 3294–3308. [Google Scholar] [CrossRef]
- Zhang, X.N.; Mount, S.M. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009, 150, 1450–1458. [Google Scholar] [CrossRef]
- Zhang, X.N.; Mo, C.; Garrett, W.M.; Cooper, B. Phosphothreonine 218 is required for the function of SR45.1 in regulating flower petal development in Arabidopsis. Plant Signal. Behav. 2014, 9, e29134. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Köster, T.; Nolte, C.; Weinholdt, C.; Lewinski, M.; Grosse, I.; Staiger, D. Adaptation of iCLIP to plants determines the binding landscape of the clock-regulated RNA-binding protein AtGRP7. Genome Biol. 2017, 18, 204. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Day, I.S.; Golovkin, M.; Palusa, S.G.; Link, A.; Ali, G.S.; Thomas, J.; Reddy, A.S. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing. Plant J. 2012, 71, 936–947. [Google Scholar] [CrossRef]
- Chen, S.L.; Rooney, T.J.; Hu, A.R.; Beard, H.S.; Garrett, W.M.; Mangalath, L.M.; Zhang, X.N. Quantitative proteomics reveals a role for SERINE/ARGININE-rich 45 in regulating RNA metabolism and modulating transcriptional suppression via the ASAP complex in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1116. [Google Scholar] [CrossRef]
- Li, Y.; Xia, C.; Feng, J.; Yang, D.; Wu, F.; Cao, Y.; Ma, L. The SNW domain of SKIP is required for its integration into the spliceosome and its interaction with the Paf1 complex in Arabidopsis. Mol. Plant 2016, 9, 1040–1050. [Google Scholar] [CrossRef]
- Woo, H.R.; Koo, H.J.; Kim, J.; Jeong, H.; Yang, J.O.; Lee, I.H.; Lim, P.O. Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol. 2016, 171, 452–467. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A. The landscape of isoform switches in human cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Yan, H.; Sheng, M.; Wang, C.; Liu, Y.; Yang, J.; Liu, F.; Su, Z. AtSPX1-mediated transcriptional regulation during leaf senescence in Arabidopsis thaliana. Plant Sci. 2019, 283, 238–246. [Google Scholar] [CrossRef]
- Wessel, D.M.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albaqami, M.; Almaghrabi, G.O. Genome-Wide Isoform Switching Reveals SR45-Mediated Splicing Control of Arabidopsis Leaf Senescence. Int. J. Mol. Sci. 2025, 26, 9784. https://doi.org/10.3390/ijms26199784
Albaqami M, Almaghrabi GO. Genome-Wide Isoform Switching Reveals SR45-Mediated Splicing Control of Arabidopsis Leaf Senescence. International Journal of Molecular Sciences. 2025; 26(19):9784. https://doi.org/10.3390/ijms26199784
Chicago/Turabian StyleAlbaqami, Mohammed, and Ghaydaa Osamah Almaghrabi. 2025. "Genome-Wide Isoform Switching Reveals SR45-Mediated Splicing Control of Arabidopsis Leaf Senescence" International Journal of Molecular Sciences 26, no. 19: 9784. https://doi.org/10.3390/ijms26199784
APA StyleAlbaqami, M., & Almaghrabi, G. O. (2025). Genome-Wide Isoform Switching Reveals SR45-Mediated Splicing Control of Arabidopsis Leaf Senescence. International Journal of Molecular Sciences, 26(19), 9784. https://doi.org/10.3390/ijms26199784





