Single Nucleotide Polymorphisms in Oxidative Stress-Related Genes Are Associated with Autism Spectrum Disorders
Abstract
1. Introduction
2. Results
2.1. Genotyping for SNPs in Antioxidant Defense Enzymes
2.2. Genotyping of Polymorphisms in Enzymes and Proteins Involved in Xenobiotic Metabolism
2.3. Evaluation of Oxidative Stress Markers
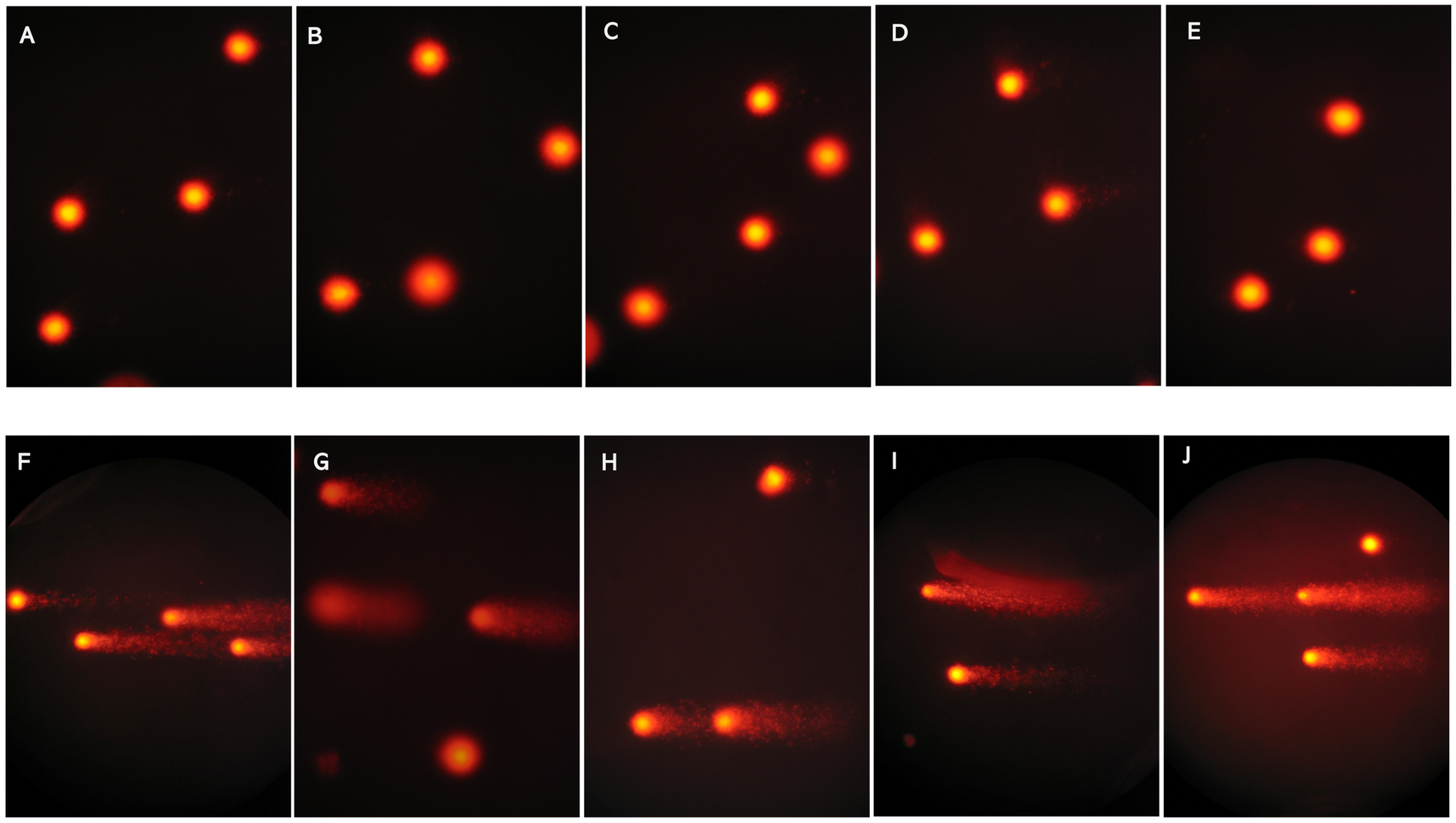
2.4. Comet Assay DNA Damage
3. Discussion
4. Materials and Methods
4.1. Study Cohorts
4.2. Genotyping by Real-Time PCR-Based Allelic Discrimination
4.3. Genotyping by PCR and Electrophoresis of GST Deletion Variants
4.4. Assessment of Oxidative Stress Levels
4.5. Assessment of Oxidative DNA Damage by Single Cell Gel Electrophoresis (Comet Assay)
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| GST | Glutathione S-transferase |
| dROMs | Reactive oxygen metabolites |
| BAP | Biological Antioxidant Potential |
| AOPP | Advanced oxidation protein products |
| GSH | Glutathione |
| GSSG | Glutathione disulfide |
| DSM | Diagnostic and Statistical Manual of Mental Disorders |
| MRI | Magnetic Resonance Imaging |
| EEG | Electroencephalogram |
| ADOS-2 | Autism Diagnostic Observation Schedule—Second Edition |
| ADI-R | Autism Diagnostic Interview—Revised |
| CARS | Children Autism Rating Scales |
| VABS | Vineland Adaptive Behavior Scales |
| SCGE | Single cell gel electrophoresis |
| SNPs | Single nucleotide polymorphisms |
| OR | Odds Ratio |
| PON1 | Paraoxonase 1 |
| SOD | Superoxide Dismutase |
| CAT | Catalase |
| GPX1 | Glutathione peroxidase 1 |
| CYP | Cytochrome P450 |
| AHR | Aryl hydrocarbon receptor |
| NATs | N-acetyl transferases |
| UGTs | UDP-glucuronosyl transferases |
| GSTs GWAS | Glutathione-S-transferases Genome-Wide Association Studies |
References
- American Psychiatric Association. American Psychiatric Association DSM-5 Task Force Diagnostic and Statistical Manual of Mental Disorders: DSM-5, 5th ed.; American Psychiatric Association: Washington, DC, USA, 2013; ISBN 978-0-89042-554-1. [Google Scholar]
- Maenner, M.J.; Shaw, K.A.; Bakian, A.V.; Bilder, D.A.; Durkin, M.S.; Esler, A.; Furnier, S.M.; Hallas, L.; Hall-Lande, J.; Hudson, A.; et al. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years-Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2018. Morb. Mortal. Wkly. Rep. Surveill. Summ. Wash. DC 2002 2021, 70, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, A.; Schiavi, S.; La Rosa, P.; Rossi-Espagnet, M.C.; Petrillo, S.; Bottino, F.; Tagliente, E.; Longo, D.; Lupi, E.; Casula, L.; et al. Sex Differences in Autism Spectrum Disorder: Diagnostic, Neurobiological, and Behavioral Features. Front. Psychiatry 2022, 13, 889636. [Google Scholar] [CrossRef]
- Tick, B.; Bolton, P.; Happé, F.; Rutter, M.; Rijsdijk, F. Heritability of Autism Spectrum Disorders: A Meta-Analysis of Twin Studies. J. Child Psychol. Psychiatry 2016, 57, 585–595. [Google Scholar] [CrossRef]
- Ronald, A.; Hoekstra, R.A. Autism Spectrum Disorders and Autistic Traits: A Decade of New Twin Studies. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2011, 156B, 255–274. [Google Scholar] [CrossRef]
- Havdahl, A.; Niarchou, M.; Starnawska, A.; Uddin, M.; van der Merwe, C.; Warrier, V. Genetic Contributions to Autism Spectrum Disorder. Psychol. Med. 2021, 51, 2260–2273. [Google Scholar] [CrossRef]
- Lim, H.K.; Yoon, J.H.; Song, M. Autism Spectrum Disorder Genes: Disease-Related Networks and Compensatory Strategies. Front. Mol. Neurosci. 2022, 15, 922840. [Google Scholar] [CrossRef]
- Genovese, A.; Butler, M.G. The Autism Spectrum: Behavioral, Psychiatric and Genetic Associations. Genes 2023, 14, 677. [Google Scholar] [CrossRef]
- Chaste, P.; Leboyer, M. Autism Risk Factors: Genes, Environment, and Gene-Environment Interactions. Dialogues Clin. Neurosci. 2012, 14, 281–292. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, X.; Zhong, L.; Zeng, L.; Li, L.; Yao, P. Understanding Autism: Causes, Diagnosis, and Advancing Therapies. Brain Res. Bull. 2025, 227, 111411. [Google Scholar] [CrossRef]
- Klei, L.; Sanders, S.J.; Murtha, M.T.; Hus, V.; Lowe, J.K.; Willsey, A.J.; Moreno-De-Luca, D.; Yu, T.W.; Fombonne, E.; Geschwind, D.; et al. Common Genetic Variants, Acting Additively, Are a Major Source of Risk for Autism. Mol. Autism 2012, 3, 9. [Google Scholar] [CrossRef]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of Common Genetic Risk Variants for Autism Spectrum Disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Bragg, M.; Chavarro, J.E.; Hamra, G.B.; Hart, J.E.; Tabb, L.P.; Weisskopf, M.G.; Volk, H.E.; Lyall, K. Prenatal Diet as a Modifier of Environmental Risk Factors for Autism and Related Neurodevelopmental Outcomes. Curr. Environ. Health Rep. 2022, 9, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Usui, N.; Shimada, S. Prenatal Environment and Neurodevelopmental Disorders. Front. Endocrinol. 2022, 13, 860110. [Google Scholar] [CrossRef]
- Khogeer, A.A.; AboMansour, I.S.; Mohammed, D.A. The Role of Genetics, Epigenetics, and the Environment in ASD: A Mini Review. Epigenomes 2022, 6, 15. [Google Scholar] [CrossRef]
- Mouat, J.S.; LaSalle, J.M. The Promise of DNA Methylation in Understanding Multigenerational Factors in Autism Spectrum Disorders. Front. Genet. 2022, 13, 831221. [Google Scholar] [CrossRef]
- Volk, H.E.; Ames, J.L.; Chen, A.; Fallin, M.D.; Hertz-Picciotto, I.; Halladay, A.; Hirtz, D.; Lavin, A.; Ritz, B.; Zoeller, T.; et al. Considering Toxic Chemicals in the Etiology of Autism. Pediatrics 2022, 149, e2021053012. [Google Scholar] [CrossRef]
- Welch, C.; Mulligan, K. Does Bisphenol A Confer Risk of Neurodevelopmental Disorders? What We Have Learned from Developmental Neurotoxicity Studies in Animal Models. Int. J. Mol. Sci. 2022, 23, 2894. [Google Scholar] [CrossRef]
- Rossignol, D.A.; Frye, R.E. Evidence Linking Oxidative Stress, Mitochondrial Dysfunction, and Inflammation in the Brain of Individuals with Autism. Front. Physiol. 2014, 5, 150. [Google Scholar] [CrossRef]
- Xu, N.; Li, X.; Zhong, Y. Inflammatory Cytokines: Potential Biomarkers of Immunologic Dysfunction in Autism Spectrum Disorders. Mediators Inflamm. 2015, 2015, 531518. [Google Scholar] [CrossRef]
- Pangrazzi, L.; Balasco, L.; Bozzi, Y. Oxidative Stress and Immune System Dysfunction in Autism Spectrum Disorders. Int. J. Mol. Sci. 2020, 21, 3293. [Google Scholar] [CrossRef]
- Waligóra, A.; Waligóra, S.; Kozarska, M.; Damasiewicz-Bodzek, A.; Gorczyca, P.; Tyrpień-Golder, K. Autism Spectrum Disorder (ASD)-Biomarkers of Oxidative Stress and Methylation and Transsulfuration Cycle. Psychiatr. Pol. 2019, 53, 771–788. [Google Scholar] [CrossRef]
- Spoto, G.; Butera, A.; Albertini, M.L.; Consoli, C.; Ceraolo, G.; Nicotera, A.G.; Rosa, G.D. The Ambiguous Role of Growth Factors in Autism: What Do We Really Know? Int. J. Mol. Sci. 2025, 26, 1607. [Google Scholar] [CrossRef]
- Zawadzka, A.; Cieślik, M.; Adamczyk, A. The Role of Maternal Immune Activation in the Pathogenesis of Autism: A Review of the Evidence, Proposed Mechanisms and Implications for Treatment. Int. J. Mol. Sci. 2021, 22, 11516. [Google Scholar] [CrossRef] [PubMed]
- McLellan, J.; Kim, D.H.J.; Bruce, M.; Ramirez-Celis, A.; Van de Water, J. Maternal Immune Dysregulation and Autism-Understanding the Role of Cytokines, Chemokines and Autoantibodies. Front. Psychiatry 2022, 13, 834910. [Google Scholar] [CrossRef] [PubMed]
- Manti, S.; Spoto, G.; Nicotera, A.G.; Di Rosa, G.; Piedimonte, G. Impact of Respiratory Viral Infections during Pregnancy on the Neurological Outcomes of the Newborn: Current Knowledge. Front. Neurosci. 2023, 17, 1320319. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Kanda, Y.; Sone, H.; Aoyama, H. Oxidative Stress as a Common Key Event in Developmental Neurotoxicity. Oxid. Med. Cell. Longev. 2021, 2021, 6685204. [Google Scholar] [CrossRef]
- Frustaci, A.; Neri, M.; Cesario, A.; Adams, J.B.; Domenici, E.; Dalla Bernardina, B.; Bonassi, S. Oxidative Stress-Related Biomarkers in Autism: Systematic Review and Meta-Analyses. Free Radic. Biol. Med. 2012, 52, 2128–2141. [Google Scholar] [CrossRef]
- Liu, X.; Lin, J.; Zhang, H.; Khan, N.U.; Zhang, J.; Tang, X.; Cao, X.; Shen, L. Oxidative Stress in Autism Spectrum Disorder-Current Progress of Mechanisms and Biomarkers. Front. Psychiatry 2022, 13, 813304. [Google Scholar] [CrossRef]
- Thorsen, M. Oxidative Stress, Metabolic and Mitochondrial Abnormalities Associated with Autism Spectrum Disorder. Prog. Mol. Biol. Transl. Sci. 2020, 173, 331–354. [Google Scholar] [CrossRef]
- Gevezova, M.; Minchev, D.; Pacheva, I.; Sbirkov, Y.; Yordanova, R.; Timova, E.; Kotetarov, V.; Ivanov, I.; Sarafian, V. Cellular Bioenergetic and Metabolic Changes in Patients with Autism Spectrum Disorder. Curr. Top. Med. Chem. 2021, 21, 985–994. [Google Scholar] [CrossRef]
- Bjørklund, G.; Tinkov, A.A.; Hosnedlová, B.; Kizek, R.; Ajsuvakova, O.P.; Chirumbolo, S.; Skalnaya, M.G.; Peana, M.; Dadar, M.; El-Ansary, A.; et al. The Role of Glutathione Redox Imbalance in Autism Spectrum Disorder: A Review. Free Radic. Biol. Med. 2020, 160, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Iyanagi, T. Molecular Mechanism of Phase I and Phase II Drug-Metabolizing Enzymes: Implications for Detoxification. Int. Rev. Cytol. 2007, 260, 35–112. [Google Scholar] [CrossRef] [PubMed]
- Spoto, G.; Valentini, G.; Saia, M.C.; Butera, A.; Amore, G.; Salpietro, V.; Nicotera, A.G.; Di Rosa, G. Synaptopathies in Developmental and Epileptic Encephalopathies: A Focus on Pre-Synaptic Dysfunction. Front. Neurol. 2022, 13, 826211. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.E.; Rossignol, D.A. Treatments for Biomedical Abnormalities Associated with Autism Spectrum Disorder. Front. Pediatr. 2014, 2, 66. [Google Scholar] [CrossRef]
- Kałużna-Czaplińska, J.; Jóźwik-Pruska, J. Chromatographic and Mass Spectrometric Techniques in Studies on Oxidative Stress in Autism. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2016, 1019, 4–14. [Google Scholar] [CrossRef]
- Tanner, S.; Thomson, S.; Drummond, K.; O’Hely, M.; Symeonides, C.; Mansell, T.; Saffery, R.; Sly, P.D.; Collier, F.; Burgner, D.; et al. A Pathway-Based Genetic Score for Oxidative Stress: An Indicator of Host Vulnerability to Phthalate-Associated Adverse Neurodevelopment. Antioxid. Basel Switz. 2022, 11, 659. [Google Scholar] [CrossRef]
- Chen, L.; Shi, X.-J.; Liu, H.; Mao, X.; Gui, L.-N.; Wang, H.; Cheng, Y. Oxidative Stress Marker Aberrations in Children with Autism Spectrum Disorder: A Systematic Review and Meta-Analysis of 87 Studies (N = 9109). Transl. Psychiatry 2021, 11, 15. [Google Scholar] [CrossRef]
- Di Rosa, G.; Lenzo, P.; Parisi, E.; Neri, M.; Guerrera, S.; Nicotera, A.; Alibrandi, A.; Germanò, E.; Caccamo, D.; Spanò, M.; et al. Role of Plasma Homocysteine Levels and MTHFR Polymorphisms on IQ Scores in Children and Young Adults with Epilepsy Treated with Antiepileptic Drugs. Epilepsy Behav. EB 2013, 29, 548–551. [Google Scholar] [CrossRef]
- Postorino, V.; Fatta, L.M.; Sanges, V.; Giovagnoli, G.; De Peppo, L.; Vicari, S.; Mazzone, L. Intellectual Disability in Autism Spectrum Disorder: Investigation of Prevalence in an Italian Sample of Children and Adolescents. Res. Dev. Disabil. 2016, 48, 193–201. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, X.; Jiao, J.; Yuan, D.; Li, S.; Luo, T.; Wang, M.; Situ, M.; Sun, X.; Huang, Y. Brain White Matter Microstructure Abnormalities in Children with Optimal Outcome from Autism: A Four-Year Follow-up Study. Sci. Rep. 2022, 12, 20151. [Google Scholar] [CrossRef]
- Marseglia, L.M.; Nicotera, A.; Salpietro, V.; Giaimo, E.; Cardile, G.; Bonsignore, M.; Alibrandi, A.; Caccamo, D.; Manti, S.; D’Angelo, G.; et al. Hyperhomocysteinemia and MTHFR Polymorphisms as Antenatal Risk Factors of White Matter Abnormalities in Two Cohorts of Late Preterm and Full Term Newborns. Oxid. Med. Cell. Longev. 2015, 2015, 543134. [Google Scholar] [CrossRef]
- Cannavò, L.; Perrone, S.; Viola, V.; Marseglia, L.; Di Rosa, G.; Gitto, E. Oxidative Stress and Respiratory Diseases in Preterm Newborns. Int. J. Mol. Sci. 2021, 22, 12504. [Google Scholar] [CrossRef] [PubMed]
- Usui, N.; Kobayashi, H.; Shimada, S. Neuroinflammation and Oxidative Stress in the Pathogenesis of Autism Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 5487. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, O.; Alzeer, S. Measuring Some Oxidative Stress Biomarkers in Autistic Syrian Children and Their Siblings: A Case-Control Study. Biomark. Insights 2022, 17, 11772719221123913. [Google Scholar] [CrossRef] [PubMed]
- Kern, J.K.; Jones, A.M. Evidence of Toxicity, Oxidative Stress, and Neuronal Insult in Autism. J. Toxicol. Environ. Health B Crit. Rev. 2006, 9, 485–499. [Google Scholar] [CrossRef]
- Amore, G.; Spoto, G.; Ieni, A.; Vetri, L.; Quatrosi, G.; Di Rosa, G.; Nicotera, A.G. A Focus on the Cerebellum: From Embryogenesis to an Age-Related Clinical Perspective. Front. Syst. Neurosci. 2021, 15, 646052. [Google Scholar] [CrossRef]
- Spoto, G.; Amore, G.; Vetri, L.; Quatrosi, G.; Cafeo, A.; Gitto, E.; Nicotera, A.G.; Di Rosa, G. Cerebellum and Prematurity: A Complex Interplay Between Disruptive and Dysmaturational Events. Front. Syst. Neurosci. 2021, 15, 655164. [Google Scholar] [CrossRef]
- Essa, M.M.; Guillemin, G.J.; Waly, M.I.; Al-Sharbati, M.M.; Al-Farsi, Y.M.; Hakkim, F.L.; Ali, A.; Al-Shafaee, M.S. Increased Markers of Oxidative Stress in Autistic Children of the Sultanate of Oman. Biol. Trace Elem. Res. 2012, 147, 25–27. [Google Scholar] [CrossRef]
- Ahmad, T.Y.; Tawfeeq, F.; Alameen, S. Biochemical Studies of Autism Spectrum Disorder Patients in Mosul City. Res. J. Chem. Sci. 2013, 3, 8–15. [Google Scholar]
- Yenkoyan, K.; Harutyunyan, H.; Harutyunyan, A. A Certain Role of SOD/CAT Imbalance in Pathogenesis of Autism Spectrum Disorders. Free Radic. Biol. Med. 2018, 123, 85–95. [Google Scholar] [CrossRef]
- Kovač, J.; Macedoni Lukšič, M.; Trebušak Podkrajšek, K.; Klančar, G.; Battelino, T. Rare Single Nucleotide Polymorphisms in the Regulatory Regions of the Superoxide Dismutase Genes in Autism Spectrum Disorder. Autism Res. Off. J. Int. Soc. Autism Res. 2014, 7, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Esparham, A.E.; Smith, T.; Belmont, J.M.; Haden, M.; Wagner, L.E.; Evans, R.G.; Drisko, J.A. Nutritional and Metabolic Biomarkers in Autism Spectrum Disorders: An Exploratory Study. Integr. Med. Encinitas Calif 2015, 14, 40–53. [Google Scholar]
- Morimoto, M.; Hashimoto, T.; Tsuda, Y.; Nakatsu, T.; Kitaoka, T.; Kyotani, S. Assessment of Oxidative Stress in Autism Spectrum Disorder Using Reactive Oxygen Metabolites and Biological Antioxidant Potential. PLoS ONE 2020, 15, e0233550. [Google Scholar] [CrossRef]
- Kitaoka, T.; Morimoto, M.; Hashimoto, T.; Tsuda, Y.; Nakatsu, T.; Kyotani, S. Evaluation of the Efficacy of Drug Treatment Based on Measurement of the Oxidative Stress, Using Reactive Oxygen Metabolites and Biological Antioxidant Potential, in Children with Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder. J. Pharm. Health Care Sci. 2020, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Manivasagam, T.; Arunadevi, S.; Essa, M.M.; SaravanaBabu, C.; Borah, A.; Thenmozhi, A.J.; Qoronfleh, M.W. Role of Oxidative Stress and Antioxidants in Autism. Adv. Neurobiol. 2020, 24, 193–206. [Google Scholar] [CrossRef]
- Shao, X.; Yan, C.; Sun, D.; Fu, C.; Tian, C.; Duan, L.; Zhu, G. Association Between Glutathione Peroxidase-1 (GPx-1) Polymorphisms and Schizophrenia in the Chinese Han Population. Neuropsychiatr. Dis. Treat. 2020, 16, 2297–2305. [Google Scholar] [CrossRef]
- Banhela, N.; Naidoo, P.; Naidoo, S. Association between Pesticide Exposure and Paraoxonase-1 (PON1) Polymorphisms, and Neurobehavioural Outcomes in Children: A Systematic Review. Syst. Rev. 2020, 9, 109. [Google Scholar] [CrossRef]
- Gaita, L.; Manzi, B.; Sacco, R.; Lintas, C.; Altieri, L.; Lombardi, F.; Pawlowski, T.L.; Redman, M.; Craig, D.W.; Huentelman, M.J.; et al. Decreased Serum Arylesterase Activity in Autism Spectrum Disorders. Psychiatry Res. 2010, 180, 105–113. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Cole, T.B.; Marsillach, J.; Furlong, C.E. Paraoxonase 1 (PON1) as a Genetic Determinant of Susceptibility to Organophosphate Toxicity. Toxicology 2013, 307, 115–122. [Google Scholar] [CrossRef]
- Mandic-Maravic, V.; Mitkovic-Voncina, M.; Pljesa-Ercegovac, M.; Savic-Radojevic, A.; Djordjevic, M.; Ercegovac, M.; Pekmezovic, T.; Simic, T.; Pejovic-Milovancevic, M. Glutathione S-Transferase Polymorphisms and Clinical Characteristics in Autism Spectrum Disorders. Front. Psychiatry 2021, 12, 672389. [Google Scholar] [CrossRef]
- Morales, E.; Sunyer, J.; Castro-Giner, F.; Estivill, X.; Julvez, J.; Ribas-Fitó, N.; Torrent, M.; Grimalt, J.O.; de Cid, R. Influence of Glutathione S-Transferase Polymorphisms on Cognitive Functioning Effects Induced by p,p’-DDT among Preschoolers. Environ. Health Perspect. 2008, 116, 1581–1585. [Google Scholar] [CrossRef]
- Mandic-Maravic, V.; Coric, V.; Mitkovic-Voncina, M.; Djordjevic, M.; Savic-Radojevic, A.; Ercegovac, M.; Matic, M.; Simic, T.; Lecic-Tosevski, D.; Toskovic, O.; et al. Interaction of Glutathione S-Transferase Polymorphisms and Tobacco Smoking during Pregnancy in Susceptibility to Autism Spectrum Disorders. Sci. Rep. 2019, 9, 3206. [Google Scholar] [CrossRef]
- Lamberti, M.; Siracusano, R.; Italiano, D.; Alosi, N.; Cucinotta, F.; Di Rosa, G.; Germanò, E.; Spina, E.; Gagliano, A. Head-to-Head Comparison of Aripiprazole and Risperidone in the Treatment of ADHD Symptoms in Children with Autistic Spectrum Disorder and ADHD: A Pilot, Open-Label, Randomized Controlled Study. Paediatr. Drugs 2016, 18, 319–329. [Google Scholar] [CrossRef]
- Arranz, M.J.; Salazar, J.; Bote, V.; Artigas-Baleri, A.; Serra-LLovich, A.; Triviño, E.; Roige, J.; Lombardia, C.; Cancino, M.; Hernandez, M.; et al. Pharmacogenetic Interventions Improve the Clinical Outcome of Treatment-Resistant Autistic Spectrum Disorder Sufferers. Pharmaceutics 2022, 14, 999. [Google Scholar] [CrossRef]
- Eltalal, S.; El Ayouty, M.; El-Said, A.; Wahba, Y. CYP2C9 (*2&*3) and CYP2C19 (*2&*3) Polymorphisms among Children with Nonlesional Epilepsy: A Single-Center Study. Acta Neurol. Belg. 2021, 121, 1623–1631. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Dong, W.; Li, H.; Yang, X.; Jin, Y.; Zhang, Z.; Jiang, Y. CYP2C9*3/*3 Gene Expression Affects the Total and Free Concentrations of Valproic Acid in Pediatric Patients with Epilepsy. Pharmacogenomics Pers. Med. 2021, 14, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Nicotera, A.G.; Hagerman, R.J.; Catania, M.V.; Buono, S.; Di Nuovo, S.; Liprino, E.M.; Stracuzzi, E.; Giusto, S.; Di Vita, G.; Musumeci, S.A. EEG Abnormalities as a Neurophysiological Biomarker of Severity in Autism Spectrum Disorder: A Pilot Cohort Study. J. Autism Dev. Disord. 2019, 49, 2337–2347. [Google Scholar] [CrossRef] [PubMed]
- Kloosterboer, S.M.; de Winter, B.C.M.; Reichart, C.G.; Kouijzer, M.E.J.; de Kroon, M.M.J.; van Daalen, E.; Ester, W.A.; Rieken, R.; Dieleman, G.C.; van Altena, D.; et al. Risperidone Plasma Concentrations Are Associated with Side Effects and Effectiveness in Children and Adolescents with Autism Spectrum Disorder. Br. J. Clin. Pharmacol. 2021, 87, 1069–1081. [Google Scholar] [CrossRef]
- Youngster, I.; Zachor, D.A.; Gabis, L.V.; Bar-Chaim, A.; Benveniste-Levkovitz, P.; Britzi, M.; Soback, S.; Ziv-Baran, T.; Berkovitch, M. CYP2D6 Genotyping in Paediatric Patients with Autism Treated with Risperidone: A Preliminary Cohort Study. Dev. Med. Child Neurol. 2014, 56, 990–994. [Google Scholar] [CrossRef]
- Vanwong, N.; Ngamsamut, N.; Medhasi, S.; Puangpetch, A.; Chamnanphon, M.; Tan-Kam, T.; Hongkaew, Y.; Limsila, P.; Sukasem, C. Impact of CYP2D6 Polymorphism on Steady-State Plasma Levels of Risperidone and 9-Hydroxyrisperidone in Thai Children and Adolescents with Autism Spectrum Disorder. J. Child Adolesc. Psychopharmacol. 2017, 27, 185–191. [Google Scholar] [CrossRef]
- Biswas, M.; Vanwong, N.; Sukasem, C. Pharmacogenomics in Clinical Practice to Prevent Risperidone-Induced Hyperprolactinemia in Autism Spectrum Disorder. Pharmacogenomics 2022, 23, 493–503. [Google Scholar] [CrossRef]
- Horinouchi, T.; Maeyama, K.; Nagai, M.; Mizobuchi, M.; Takagi, Y.; Okada, Y.; Kato, T.; Nishimura, M.; Kawasaki, Y.; Yoshioka, M.; et al. Genetic Analysis of UGT1A1 Polymorphisms Using Preserved Dried Umbilical Cord for Assessing the Potential of Neonatal Jaundice as a Risk Factor for Autism Spectrum Disorder in Children. J. Autism Dev. Disord. 2022, 52, 483–489. [Google Scholar] [CrossRef]
- Spoto, G.; Di Rosa, G.; Nicotera, A.G. The Impact of Genetics on Cognition: Insights into Cognitive Disorders and Single Nucleotide Polymorphisms. J. Pers. Med. 2024, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, A.; Baago, I.B.; Rygner, Z.; Jorgensen, M.B.; Andersen, P.K.; Kessing, L.V.; Poulsen, H.E. Association of Oxidative Stress-Induced Nucleic Acid Damage With Psychiatric Disorders in Adults: A Systematic Review and Meta-Analysis. JAMA Psychiatry 2022, 79, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Steullet, P. Editorial for the Special Issue “Oxidative Stress, Inflammation and Antioxidant Defense System in Psychiatric Disorders” in Antioxidants (2022–2023). Antioxid. Basel Switz. 2024, 13, 1307. [Google Scholar] [CrossRef] [PubMed]
- Schröder, S.S.; Danner, U.N.; Spek, A.A.; van Elburg, A.A. Problematic Eating Behaviours of Autistic Women-A Scoping Review. Eur. Eat. Disord. Rev. J. Eat. Disord. Assoc. 2022, 30, 510–537. [Google Scholar] [CrossRef]
- Al-Beltagi, M.; Saeed, N.K.; Bediwy, A.S.; Elbeltagi, R.; Alhawamdeh, R. Role of Gastrointestinal Health in Managing Children with Autism Spectrum Disorder. World J. Clin. Pediatr. 2023, 12, 171–196. [Google Scholar] [CrossRef]
- Taha, Z.; Abdalhai, K.A. A Review of the Efficacy of the Dietary Intervention in Autism Spectrum Disorder. Open Access Maced. J. Med. Sci. 2021, 9, 88–94. [Google Scholar] [CrossRef]
- Pangrazzi, L.; Balasco, L.; Bozzi, Y. Natural Antioxidants: A Novel Therapeutic Approach to Autism Spectrum Disorders? Antioxid. Basel Switz. 2020, 9, 1186. [Google Scholar] [CrossRef]
- Albertini, M.L.; Spoto, G.; Ceraolo, G.; Fichera, M.F.; Consoli, C.; Nicotera, A.G.; Di Rosa, G. Sleep Disorders in Children with Autism Spectrum Disorder: Developmental Impact and Intervention Strategies. Brain Sci. 2025, 15, 983. [Google Scholar] [CrossRef]
- Colak, H.; Sariyer, E.T.; Nogay, N.H. The Effect of Nutritional Interventions Reducing Oxidative Stress on Behavioural and Gastrointestinal Problems in Autism Spectrum Disorder. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 2023, 83, 135–164. [Google Scholar] [CrossRef]
- Hadders-Algra, M. Early Diagnostics and Early Intervention in Neurodevelopmental Disorders-Age-Dependent Challenges and Opportunities. J. Clin. Med. 2021, 10, 861. [Google Scholar] [CrossRef]
- Arand, M.; Mühlbauer, R.; Hengstler, J.; Jäger, E.; Fuchs, J.; Winkler, L.; Oesch, F. A Multiplex Polymerase Chain Reaction Protocol for the Simultaneous Analysis of the Glutathione S-Transferase GSTM1 and GSTT1 Polymorphisms. Anal. Biochem. 1996, 236, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Tanito, M.; Kaidzu, S.; Takai, Y.; Ohira, A. Status of Systemic Oxidative Stresses in Patients with Primary Open-Angle Glaucoma and Pseudoexfoliation Syndrome. PLoS ONE 2012, 7, e49680. [Google Scholar] [CrossRef] [PubMed]
- Alagozlu, H.; Gorgul, A.; Bilgihan, A.; Tuncer, C.; Unal, S. Increased Plasma Levels of Advanced Oxidation Protein Products (AOPP) as a Marker for Oxidative Stress in Patients with Active Ulcerative Colitis. Clin. Res. Hepatol. Gastroenterol. 2013, 37, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R. The Comet Assay for DNA Damage and Repair: Principles, Applications, and Limitations. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Olive, P.L.; Banáth, J.P. The Comet Assay: A Method to Measure DNA Damage in Individual Cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Gangemi, C.; Calabrò, C.; Vecchio, M.; Di Mauro, D.; Renis, M.; Ientile, R.; Currò, M.; Caccamo, D. Assessment of Glutathione Peroxidase-1 Polymorphisms, Oxidative Stress and DNA Damage in Sensitivity-Related Illnesses. Life Sci. 2016, 145, 27–33. [Google Scholar] [CrossRef]
| Genotype | ASD (n = 106) % | Controls (n = 90) % | p-Value | Odds Ratio (95% C.I.) |
|---|---|---|---|---|
| SOD2 A16V | ||||
| AA | 33.02 | 21.11 | 0.0777 | - |
| AV | 36.79 | 26.67 | 0.1671 | - |
| VV | 30.19 | 52.22 | 0.0021 | 0.3956 (0.2202 to 0.7107) |
| CAT -844C>T | ||||
| CC | 53.77 | 68.89 | 0.0398 | 0.5253 (0.2920 to 0.9453) |
| CT | 46.23 | 31.11 | 0.0398 | 1.904 (1.058 to 3.425) |
| TT | - | - | - | |
| GPx1 rs180668 C>T | ||||
| CC | 54.81 | 75 | 0.0028 | 0.3924 (0.2118 to 0.7270) |
| CT | 34.62 | 25 | 0.1569 | - |
| TT | 10.58 | 0 | 0.001 | 22.26 (1.292 to 383.6) |
| PON Q192R T>C | ||||
| TT | 47.62 | 63.33 | 0.0471 | 0.5263 (0.2871 to 0.9649) |
| TC | 38.10 | 36.67 | 0.8763 | - |
| CC | 14.29 | 0.00 | 0.0001 | 31.21 (1.816 to 536.4) |
| Genotype | ASD (n = 106) % | Controls (n = 90) % | p-Value | Odds Ratio (95% C.I.) |
|---|---|---|---|---|
| CYP2C9*2, *3 a | ||||
| *1/1 | 73.3 | 84.4 | 0.0524 | - |
| *1/*2 | 6.7 | 12.2 | 0.1058 | - |
| *1/*3 | 18.3 | 1.1 | 0.0011 | 18.88 (2.236–159.4) |
| *2/*2 | 1.7 | 2.2 | 1 | - |
| CYP2C19*2 b | ||||
| *1/*1 | 73.91 | 80.0 | 0.445 | - |
| *1/*2 | 21.74 | 16.7 | 0.4228 | - |
| *2/*2 | 4.35 | 3.33 | 1 | - |
| CYP2D6*41 c | ||||
| *1/*1 | 77.5 | 100 | <0.0001 | 0.01878 (0.001–0.340) |
| *1/*41 | 20 | - | 0.0002 | 44.4 (2.420–814.6) |
| *41/*41 | 2.5 | - | 0.2623 | - |
| AHR Arg254Lys d | ||||
| Arg/Arg | 83.9 | 77.8 | 0.4109 | - |
| Arg/Lys | 16.1 | 16.7 | 1 | - |
| Lys/Lys | - | 5.5 | 0.0797 | - |
| NAT1 Arg187Gln f | ||||
| Arg/Arg | 97.26 | 96.67 | 1 | - |
| Arg/Gln | 2.74 | 3.33 | 1 | - |
| Gln/Gln | - | - | - | - |
| NAT2rs1799931 G>A g | ||||
| G/G | 100 | 96.67 | 0.2466 | - |
| G/A | - | 3.33 | 0.2466 | - |
| A/A | - | - | - | - |
| UGT1A1*6 G>A h | ||||
| WT 1/1 G/G | 89.09 | 85.56 | 0.6191 | |
| HT *1/*6 G/A | - | 14.44 | 0.0019 | 0.05172 (0.003–0.889) |
| MUT *6/*6 A/A | 10.91 | - | 0.0025 | 23.77 (1.310–431.1) |
| GSTP1 I105V/A114V | ||||
| *A/*A | 45.2 | 62.2 | 0.024 | 0.5025 (0.283–0.891) |
| *A/*B | 34.5 | 28.9 | 0.5378 | - |
| *B/*B | 8.3 | 4.4 | 0.3888 | - |
| *A/*C | 4.8 | 2.2 | 0.4558 | - |
| *A/*D | 1.2 | - | 1 | - |
| *B/*C | 5.9 | 2.2 | 0.2923 | - |
| GSTM1 Null (*0) e | ||||
| *1/*1+*1/*0 | 58.8 | 53.3 | 0.1494 | - |
| *0/*0 | 41.2 | 46.7 | 0.1494 | - |
| GSTT1 Null (*0) e | ||||
| *1/*1+*1/*0 | 64.7 | 80 | 0.0296 | 0.43 (0.210–0.878) |
| *0/*0 | 35.3 | 20 | 0.0296 | 2.326 (1.139–4.750) |
| GSTM1 Null/ GSTT1 Null e | ||||
| (*1/*1+*1/*0)/ (*1/*1+*1/*0) | 38.2 | 47 | 0.615 | - |
| (*1/*1+*1/*0)/*0/*0 | 16.2 | 15.5 | 0.915 | - |
| *0/*0/(*1/*1+*1/*0) | 22.05 | 33.3 | 0.12 | - |
| (*0/*0)/(*0/*0) | 19.1 | 4.44 | 0.0032 | 5.000 (1.64–15.24) |
| Redox Marker | ASD (n = 106) | Controls (n = 90) |
|---|---|---|
| AOPP | 329.1 ± 149.7 *** | 74.9 ± 15.1 |
| dROMs | 327.1 ± 115.9 | 319.3 ± 50.1 |
| BAP | 2428.0 ± 1479.3 | 2128.4 ± 1820.6 |
| AOPP | dROMs | BAP | |
|---|---|---|---|
| Group 1 (SOD2 Wt + 2-9 other SNPs) (n = 35) | 312.2 ± 111.9 | 357.3 ± 107.0 | 1694.0 ± 1061.9 |
| Group 2 (SOD2 Ht + 2-6 other SNPs) (n = 39) | 303.0 ± 134.3 | 347.3 ± 101.6 | 3008.8 ± 1396.2 ** |
| Group 3 (SOD2 Mut + 2-6 other SNPs) (n = 32) | 378.4 ± 151.0 | 401.7 ± 160.4 | 2392.5 ±1248.5 ** |
| Comet Parameters | ASD (n = 32) | Controls (n = 27) |
|---|---|---|
| % COMET + cells | 55.4 ± 4.66 *** | 4.5 ± 2.9 |
| Head length | 215.8 ± 15.3 | 209.5 ± 15.7 |
| Tail length | 64.5 ± 14.1 *** | 36.6 ± 9.5 |
| Comet length | 274.0 ± 20.3 *** | 252.5 ± 13.6 |
| Head DNA | 85.9 ± 4.5 | 112.4 ± 101.9 |
| Tail DNA | 14.04 ± 4.6 * | 11.6 ± 4.1 |
| Tail Moment | 14.8 ± 9.8 * | 9.3 ± 6.4 |
| Olive Tail Moment | 11.2 ± 6.2 *** | 4.0 ± 2.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spoto, G.; Bertuccio, M.P.; Visalli, G.; Currò, M.; Di Rosa, G.; Caccamo, D. Single Nucleotide Polymorphisms in Oxidative Stress-Related Genes Are Associated with Autism Spectrum Disorders. Int. J. Mol. Sci. 2025, 26, 9768. https://doi.org/10.3390/ijms26199768
Spoto G, Bertuccio MP, Visalli G, Currò M, Di Rosa G, Caccamo D. Single Nucleotide Polymorphisms in Oxidative Stress-Related Genes Are Associated with Autism Spectrum Disorders. International Journal of Molecular Sciences. 2025; 26(19):9768. https://doi.org/10.3390/ijms26199768
Chicago/Turabian StyleSpoto, Giulia, Maria Paola Bertuccio, Giuseppa Visalli, Monica Currò, Gabriella Di Rosa, and Daniela Caccamo. 2025. "Single Nucleotide Polymorphisms in Oxidative Stress-Related Genes Are Associated with Autism Spectrum Disorders" International Journal of Molecular Sciences 26, no. 19: 9768. https://doi.org/10.3390/ijms26199768
APA StyleSpoto, G., Bertuccio, M. P., Visalli, G., Currò, M., Di Rosa, G., & Caccamo, D. (2025). Single Nucleotide Polymorphisms in Oxidative Stress-Related Genes Are Associated with Autism Spectrum Disorders. International Journal of Molecular Sciences, 26(19), 9768. https://doi.org/10.3390/ijms26199768







