Transcriptomic and Metabolomic Insights into Benzylisoquinoline Alkaloid Biosynthesis in Goldthread (Coptis trifolia)
Abstract
1. Introduction
2. Results
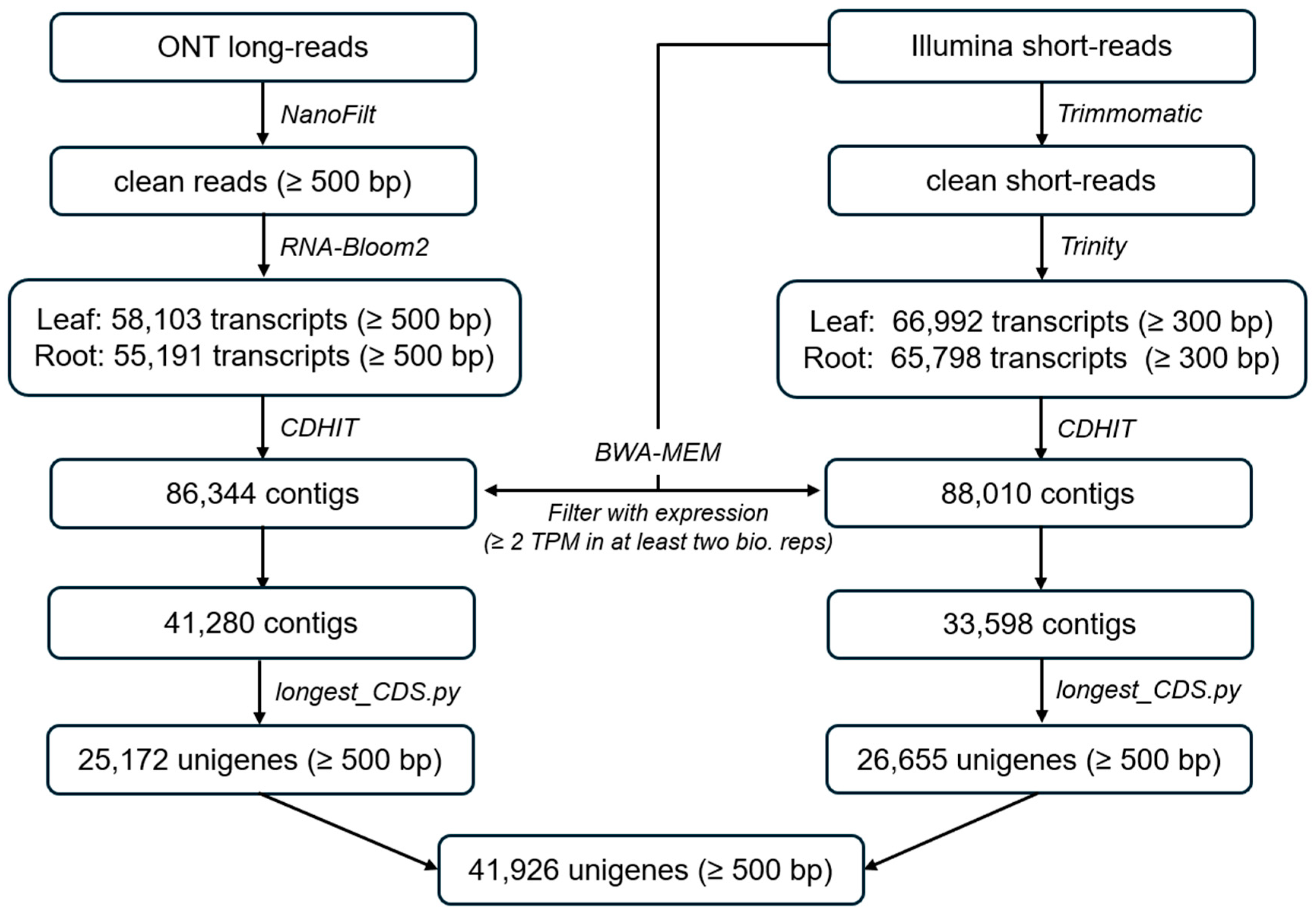
2.1. Transcriptome Assembly
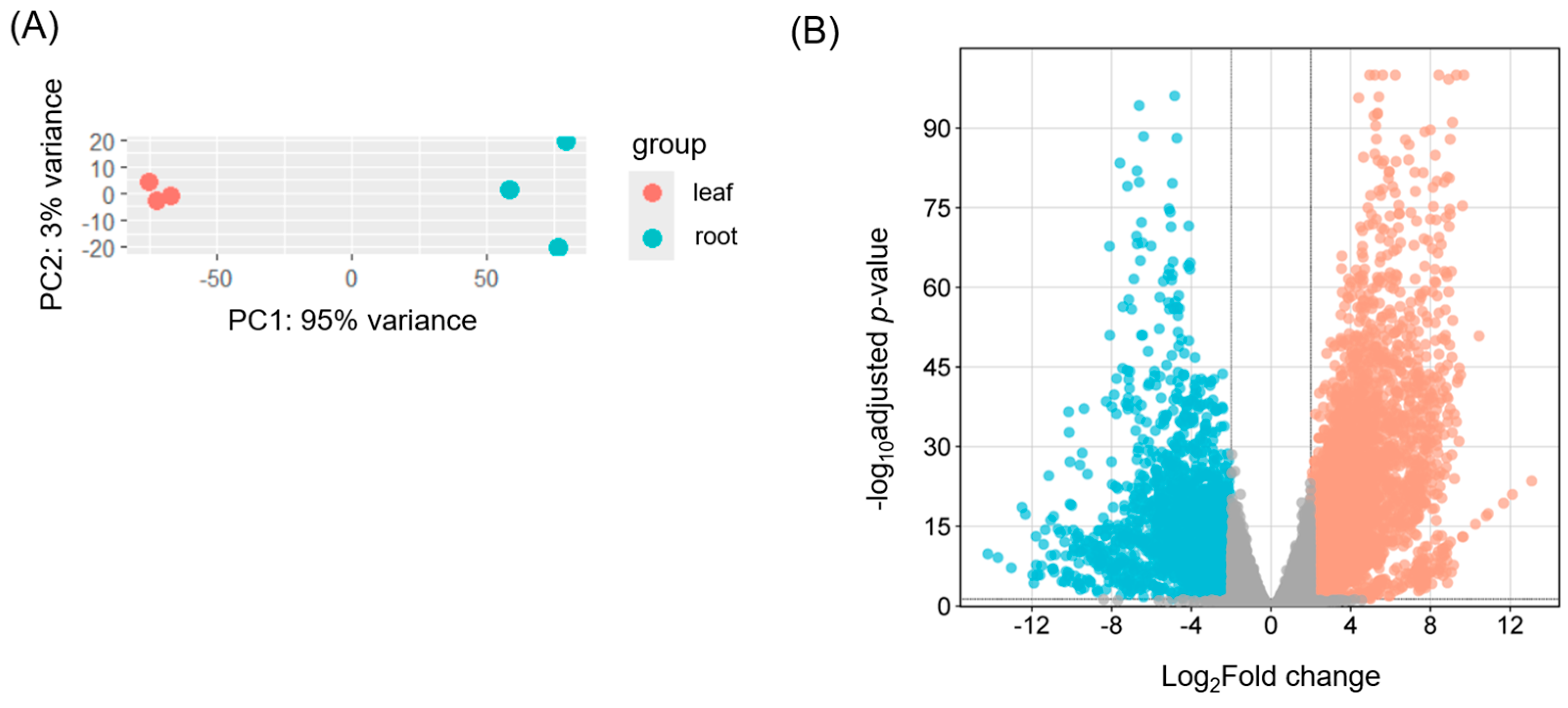
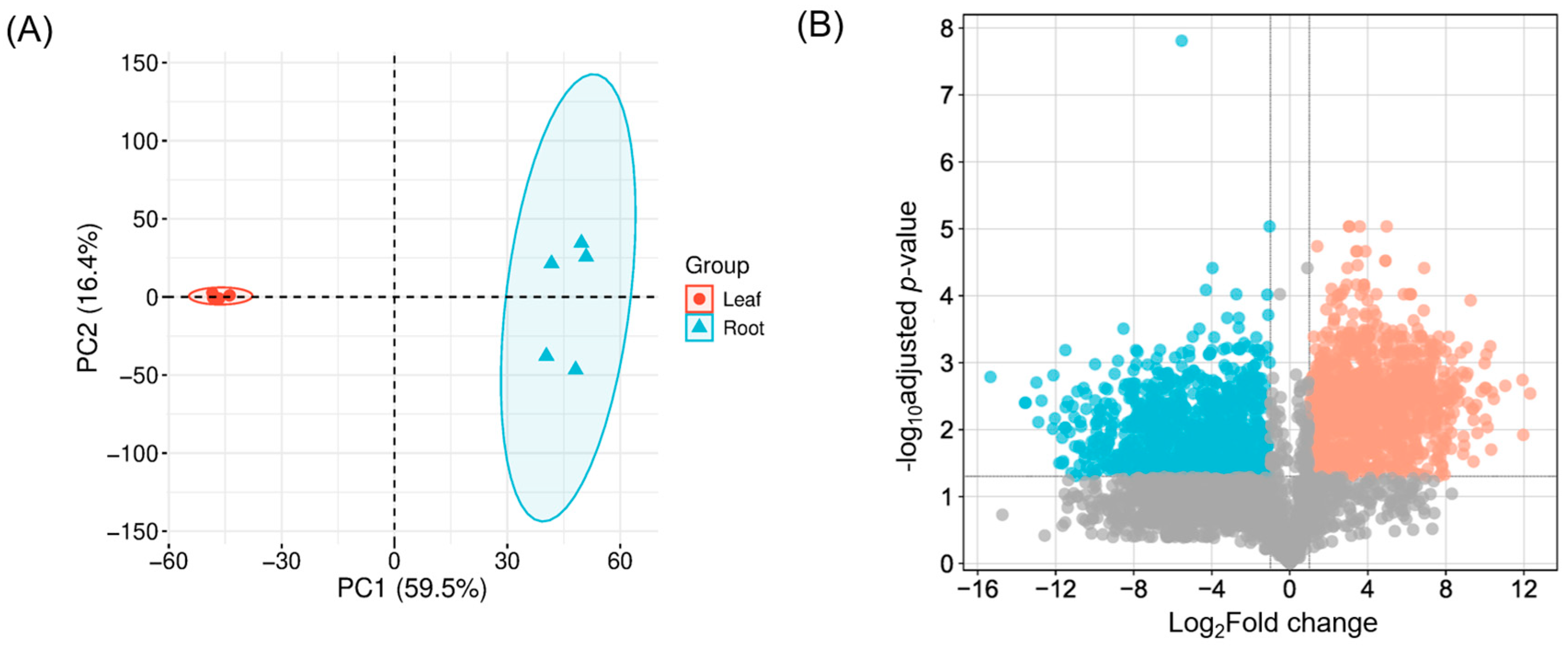
2.2. Transcriptome Analysis Using RNA-Seq Data
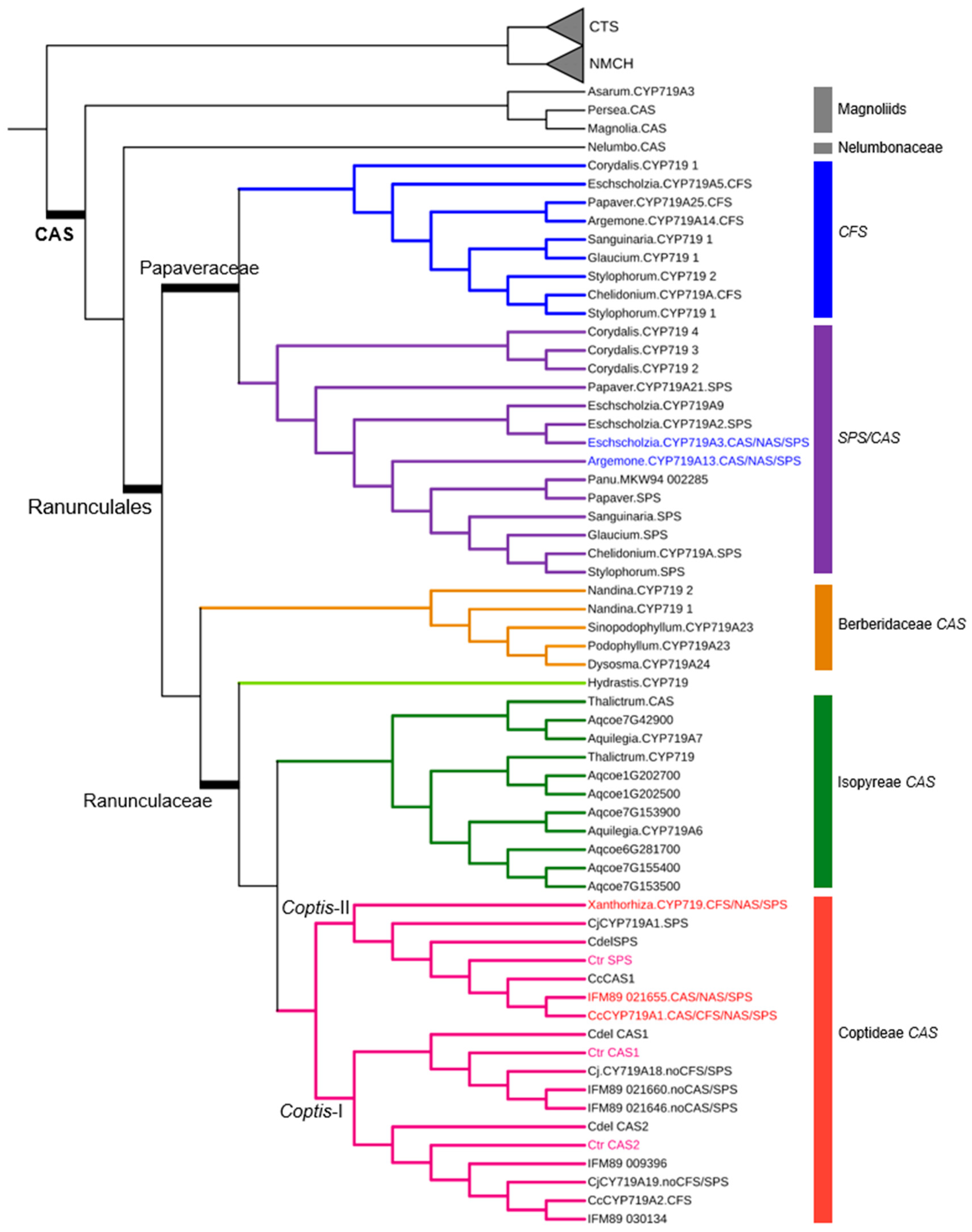
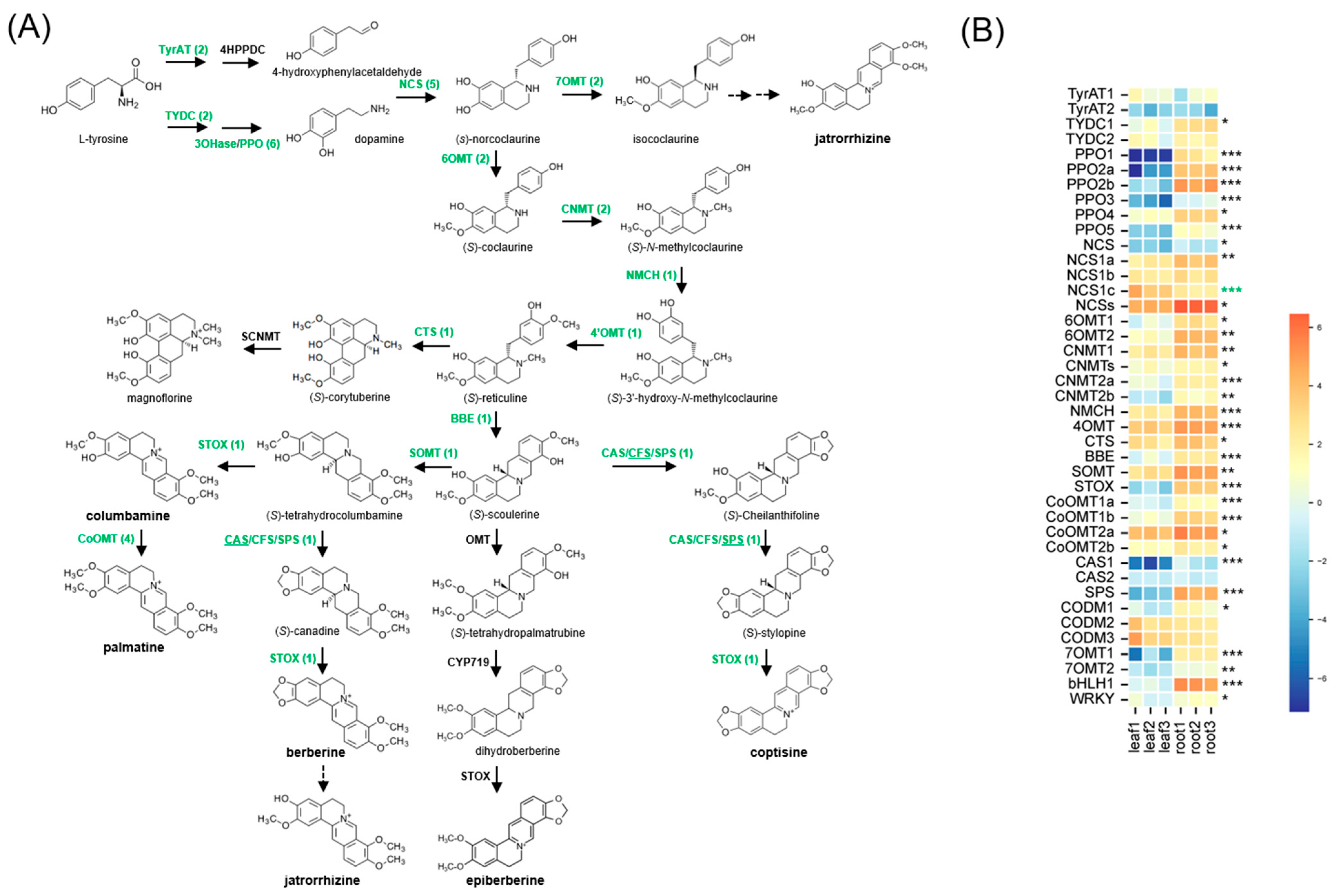
2.3. Identification of Full-Length Transcripts Involved in BIA Biosynthesis
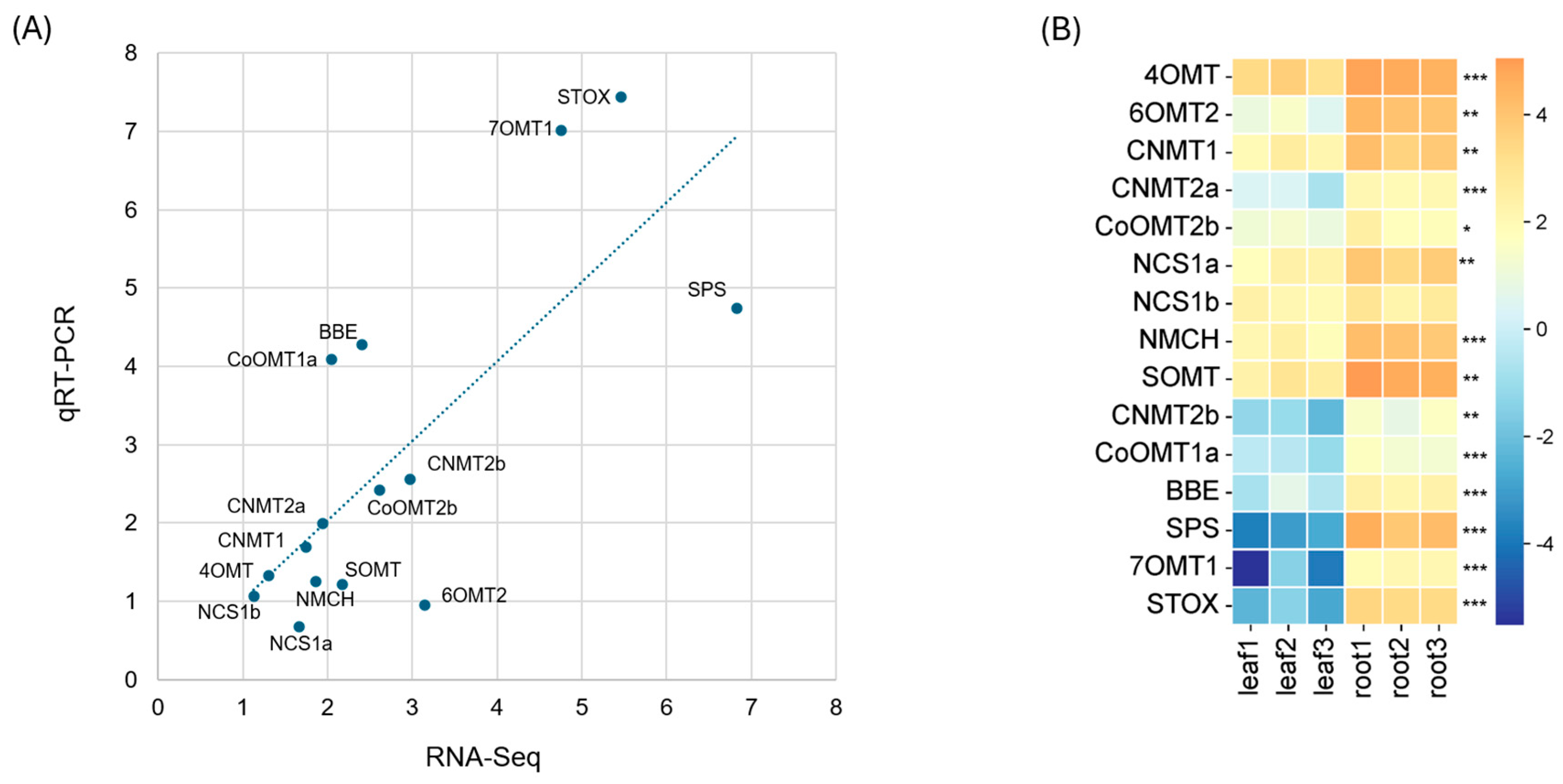
2.4. qRT-PCR Validation of RNA-Seq Results for 15 BIA Biosynthetic Genes
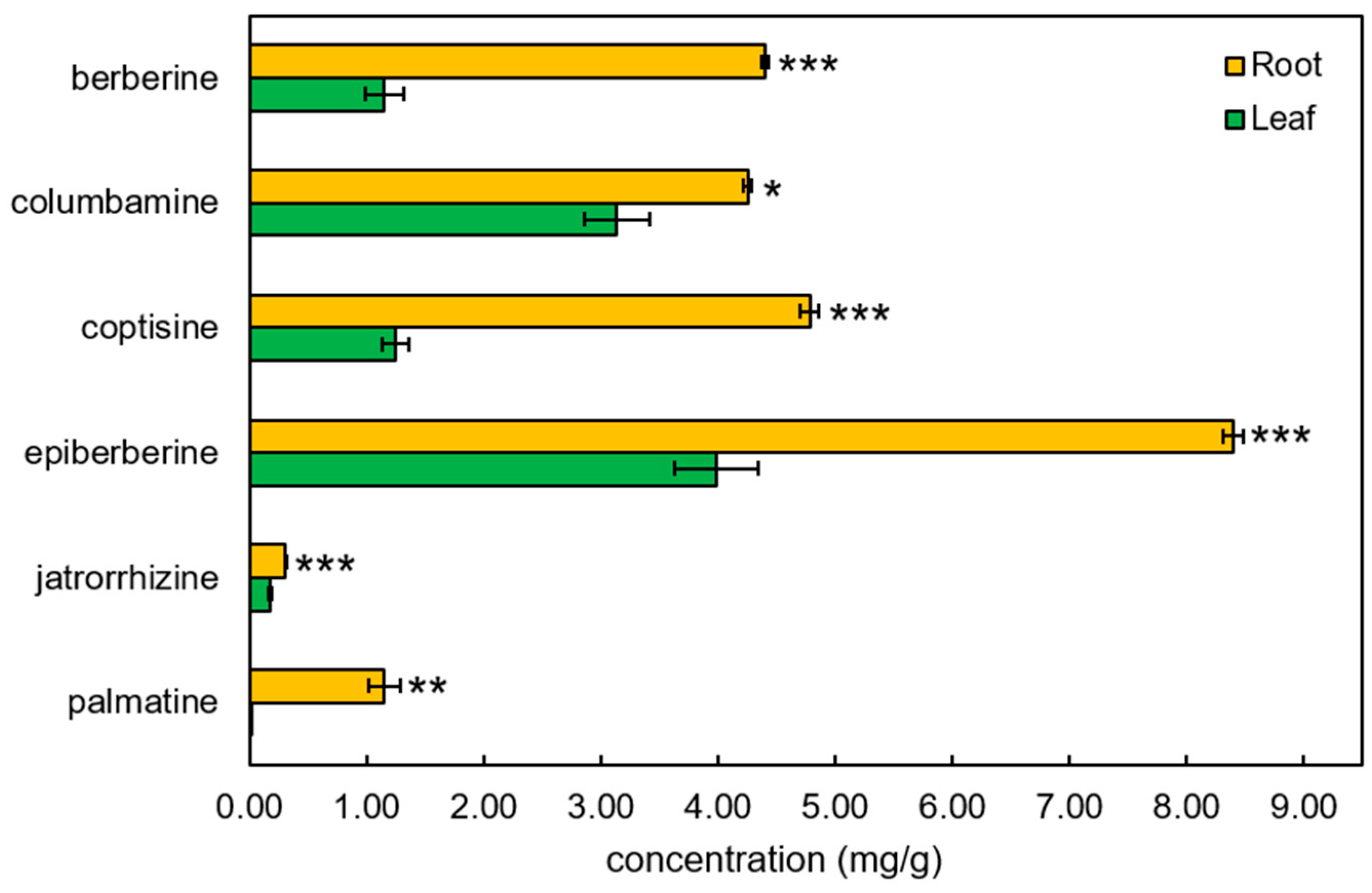
2.5. Untargeted and Targeted Metabolite Profiling
3. Discussion
3.1. High-Quality Coptis trifolia Transcriptome Assembly via Integrated ONT and Illumina Sequencing
3.2. Differential Gene Expression Between Leaves and Roots
3.3. Accumulation of BIA and Their Biosynthetic Genes in C. trifolia
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction, Library Construction, and RNA-Sequencing
4.3. Analysis of RNA-Seq Data
4.4. Identification of Genes Involved in BIA Biosynthesis
4.5. Validation of RNA-Seq Results Using qRT-PCR
4.6. Metabolite Profiling
4.6.1. Targeted Analysis of Six BIA-Related Metabolites
4.6.2. Untargeted Metabolomics
4.6.3. Quality Control and Method Validation
4.6.4. Data Processing, Compound Detection, Grouping, Alignment, and Background Subtraction
4.6.5. Metabolite Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heinrich, M.; Mah, J.; Amirkia, V. Alkaloids Used as Medicines: Structural Phytochemistry Meets Biodiversity-An Update and Forward Look. Molecules 2021, 26, 1836. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.R.; Tay, K.C.; Su, Y.X.; Wong, C.K.; Tan, W.N.; Khaw, K.Y. Potential of Naturally Derived Alkaloids as Multi-Targeted Therapeutic Agents for Neurodegenerative Diseases. Molecules 2021, 26, 728. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Lou, G.H.; Zeng, H.R.; Hu, J.; Huang, Q.W.; Peng, W.; Yang, X.B. Coptidis Rhizoma: A comprehensive review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. Pharm. Biol. 2019, 57, 193–225. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, Y.; Ren, D.; Li, J.; Yuan, G.; An, L.; Du, P.; Ma, J. Antidiabetic mechanism of Coptis chinensis polysaccharide through its antioxidant property involving the JNK pathway. Pharm. Biol. 2015, 53, 1022–1029. [Google Scholar] [CrossRef] [PubMed]
- Kamath, S.; Skeels, M.; Pai, A. Significant differences in alkaloid content of Coptis chinensis (Huanglian), from its related American species. Chin. Med. 2009, 4, 17. [Google Scholar] [CrossRef]
- Dan, Y.; Qian, Z.; Peng, Y.; Chen, C.; Liu, Y.; Tai, W.; Qi, J. Revision and Improvement of Criterion on Traditional Chinese Medicines in Chinese Pharmacopoeia 2015. Chin. Herb. Med. 2016, 8, 196–208. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, J.; Chen, Y. Advances in the biosynthesis of naturally occurring benzylisoquinoline alkaloids. Front. Plant Sci. 2025, 16, 1548471. [Google Scholar] [CrossRef]
- He, S.M.; Liang, Y.L.; Cong, K.; Chen, G.; Zhao, X.; Zhao, Q.M.; Zhang, J.J.; Wang, X.; Dong, Y.; Yang, J.L.; et al. Identification and Characterization of Genes Involved in Benzylisoquinoline Alkaloid Biosynthesis in Coptis Species. Front. Plant Sci. 2018, 9, 731. [Google Scholar] [CrossRef]
- Zhong, F.; Huang, L.; Qi, L.; Ma, Y.; Yan, Z. Full-length transcriptome analysis of Coptis deltoidea and identification of putative genes involved in benzylisoquinoline alkaloids biosynthesis based on combined sequencing platforms. Plant Mol. Biol. 2020, 102, 477–499. [Google Scholar] [CrossRef]
- Chen, D.X.; Pan, Y.; Wang, Y.; Cui, Y.Z.; Zhang, Y.J.; Mo, R.Y.; Wu, X.L.; Tan, J.; Zhang, J.; Guo, L.A.; et al. The chromosome-level reference genome of Coptis chinensis provides insights into genomic evolution and berberine biosynthesis. Hortic. Res. 2021, 8, 121. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Shu, S.; Li, Z.; Song, C.; Liu, D.; Niu, Y.; Liu, J.; Zhang, J.; Liu, H.; et al. Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids. Nat. Commun. 2021, 12, 3276. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; De Luca, V. Opium poppy and Madagascar periwinkle: Model non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 2008, 54, 763–784. [Google Scholar] [CrossRef] [PubMed]
- Eljounaidi, K.; Lichman, B.R. Nature’s Chemists: The Discovery and Engineering of Phytochemical Biosynthesis. Front. Chem. 2020, 8, 596479. [Google Scholar] [CrossRef] [PubMed]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Filsinger Interrante, M.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2015, 349, 1095–1100. [Google Scholar] [CrossRef]
- Ford, B.A. Coptis. In Flora of North America North of Mexico; Oxford University Press: Oxford, UK, 1993; Volume 3. [Google Scholar]
- Xiang, K.L.; Wu, S.D.; Yu, S.X.; Liu, Y.; Jabbour, F.; Erst, A.S.; Zhao, L.; Wang, W.; Chen, Z.D. The First Comprehensive Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Classification. PLoS ONE 2016, 11, e0153127. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Qiao, P.; Wang, J.; Wang, M.; Du, Y.; Xiong, F.; Luo, J.; Yuan, Q.; Dong, W.; et al. Evolutionary history of genus Coptis and its dynamic changes in the potential suitable distribution area. Front. Plant Sci. 2022, 13, 1003368. [Google Scholar] [CrossRef]
- He, Y.; Hou, P.; Fan, G.; Arain, S.; Peng, C. Comprehensive analyses of molecular phylogeny and main alkaloids for Coptis (Ranunculaceae) species identification. Biochem. Syst. Ecol. 2014, 56, 88–94. [Google Scholar] [CrossRef]
- Yamada, Y.; Kokabu, Y.; Chaki, K.; Yoshimoto, T.; Ohgaki, M.; Yoshida, S.; Kato, N.; Koyama, T.; Sato, F. Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in Coptis japonica. Plant Cell Physiol. 2011, 52, 1131–1141. [Google Scholar] [CrossRef]
- Liu, W.; Tian, X.; Feng, Y.; Hu, J.; Wang, B.; Chen, S.; Liu, D.; Liu, Y. Genome-wide analysis of bHLH gene family in Coptis chinensis provides insights into the regulatory role in benzylisoquinoline alkaloid biosynthesis. Plant Physiol. Biochem. 2023, 201, 107846. [Google Scholar] [CrossRef]
- Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J.G.; Yazaki, K.; Sato, F. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007, 48, 8–18. [Google Scholar] [CrossRef]
- Choi, K.B.; Morishige, T.; Shitan, N.; Yazaki, K.; Sato, F. Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 2002, 277, 830–835. [Google Scholar] [CrossRef]
- Ikezawa, N.; Iwasa, K.; Sato, F. Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 2007, 274, 1019–1035. [Google Scholar] [CrossRef]
- Diaz Chavez, M.L.; Rolf, M.; Gesell, A.; Kutchan, T.M. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 2011, 507, 186–193. [Google Scholar] [CrossRef]
- Ikezawa, N.; Tanaka, M.; Nagayoshi, M.; Shinkyo, R.; Sakaki, T.; Inouye, K.; Sato, F. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003, 278, 38557–38565. [Google Scholar] [CrossRef]
- Wu, L.; Zhao, B.; Deng, Z.; Wang, B.; Yu, Y. A biosynthetic network for protoberberine production in Coptis chinensis. Hortic. Res. 2024, 11, uhad259. [Google Scholar] [CrossRef]
- Narcross, L.; Bourgeois, L.; Fossati, E.; Burton, E.; Martin, V.J. Mining Enzyme Diversity of Transcriptome Libraries through DNA Synthesis for Benzylisoquinoline Alkaloid Pathway Optimization in Yeast. ACS Synth. Biol. 2016, 5, 1505–1518. [Google Scholar] [CrossRef]
- Jackson, D.J.; Cerveau, N.; Posnien, N. De novo assembly of transcriptomes and differential gene expression analysis using short-read data from emerging model organisms—A brief guide. Front. Zool. 2024, 21, 17. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.N.; Takahashi, H.; Uchimiya, H. The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 2009, 103, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Shen, C.; Qi, L.; Ma, Y. A Multi-Level Strategy Based on Metabolic and Molecular Genetic Approaches for the Characterization of Different Coptis Medicines Using HPLC-UV and RAD-seq Techniques. Molecules 2018, 23, 3090. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Zhu, T.; Hu, X.; Hou, C.; He, J.; Liu, X. Transcriptome and Metabolome Analysis of Isoquinoline Alkaloid Biosynthesis of Coptis chinensis in Different Years. Genes 2023, 14, 2232. [Google Scholar] [CrossRef]
- Morishige, T.; Dubouzet, E.; Choi, K.B.; Yazaki, K.; Sato, F. Molecular cloning of columbamine O-methyltransferase from cultured Coptis japonica cells. Eur. J. Biochem. 2002, 269, 5659–5667. [Google Scholar] [CrossRef]
- Li, Y.; Winzer, T.; He, Z.; Graham, I.A. Over 100 Million Years of Enzyme Evolution Underpinning the Production of Morphine in the Papaveraceae Family of Flowering Plants. Plant Commun. 2020, 1, 100029. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Nip, K.M.; Hafezqorani, S.; Gagalova, K.K.; Chiu, R.; Yang, C.; Warren, R.L.; Birol, I. Reference-free assembly of long-read transcriptome sequencing data with RNA-Bloom2. Nat. Commun. 2023, 14, 2940. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Fiehn, O.; Wohlgemuth, G.; Scholz, M.; Kind, T.; Lee, D.Y.; Lu, Y.; Moon, S.; Nikolau, B. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008, 53, 691–704. [Google Scholar] [CrossRef]
- Simoes, S.S.; Silva, I.; Ajenjo, A.C.; Dias, M.J. Validation and application of an UPLC-MS/MS method for the quantification of synthetic cannabinoids in urine samples and analysis of seized materials from the Portuguese market. Forensic Sci. Int. 2014, 243, 117–125. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koh, Y.-S.; Zhu, F.; Hwang, Y.; Yoo, M.-J. Transcriptomic and Metabolomic Insights into Benzylisoquinoline Alkaloid Biosynthesis in Goldthread (Coptis trifolia). Int. J. Mol. Sci. 2025, 26, 9704. https://doi.org/10.3390/ijms26199704
Koh Y-S, Zhu F, Hwang Y, Yoo M-J. Transcriptomic and Metabolomic Insights into Benzylisoquinoline Alkaloid Biosynthesis in Goldthread (Coptis trifolia). International Journal of Molecular Sciences. 2025; 26(19):9704. https://doi.org/10.3390/ijms26199704
Chicago/Turabian StyleKoh, Yoo-Shin, Fanchao Zhu, Yoojeong Hwang, and Mi-Jeong Yoo. 2025. "Transcriptomic and Metabolomic Insights into Benzylisoquinoline Alkaloid Biosynthesis in Goldthread (Coptis trifolia)" International Journal of Molecular Sciences 26, no. 19: 9704. https://doi.org/10.3390/ijms26199704
APA StyleKoh, Y.-S., Zhu, F., Hwang, Y., & Yoo, M.-J. (2025). Transcriptomic and Metabolomic Insights into Benzylisoquinoline Alkaloid Biosynthesis in Goldthread (Coptis trifolia). International Journal of Molecular Sciences, 26(19), 9704. https://doi.org/10.3390/ijms26199704






