Significance of Epigenetic Alteration in Cancer-Associated Fibroblasts on the Development of Carcinoma
Abstract
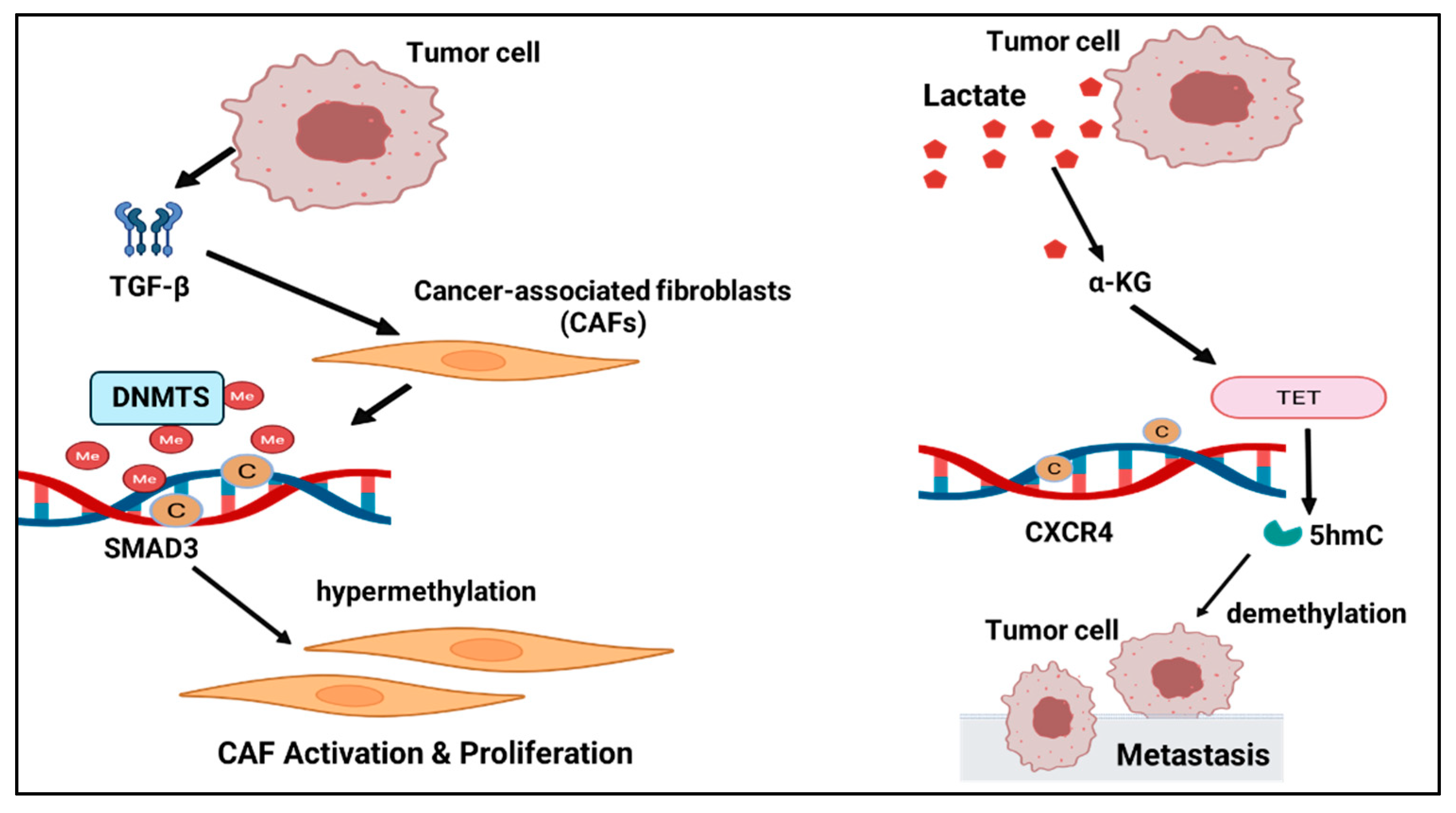
1. Introduction
1.1. DNA Methylation
1.2. Tumors and DNA Methylation (Table 1)
2. Histone Modifications in CAFs
2.1. Enhancer Reprogramming and AP-1/JUN
2.2. BET Inhibition Remodels Tumor Stroma/CAFs
2.3. Effects of HDACs
3. Non-Coding RNAs (miRNA/lncRNA/circRNA)
3.1. miRNA (miR-21) and CAFs
3.2. lncRNA and CAFs
3.3. circRNA and CAFs
4. Epigenetic Features of CAF Subsets
5. Spatial Epigenomics in CAFs
6. Conclusions
7. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Dzobo, K.; Senthebane, D.A.; Dandara, C. The Tumor Microenvironment in Tumorigenesis and Therapy Resistance Revisited. Cancers 2023, 15, 376. [Google Scholar] [CrossRef]
- Baghy, K.; Ladányi, A.; Reszegi, A.; Kovalszky, I. Insights into the Tumor Microenvironment—Components, Functions and Therapeutics. Int. J. Mol. Sci. 2023, 24, 17536. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.L.; Puré, E. Cancer-associated fibroblasts and their influence on tumor immunity and immunotherapy. eLife 2020, 9, e57243. [Google Scholar] [CrossRef] [PubMed]
- Asif, P.J.; Longobardi, C.; Hahne, M.; Medema, J.P. The Role of Cancer-Associated Fibroblasts in Cancer Invasion and Metastasis. Cancers 2021, 13, 4720. [Google Scholar] [CrossRef]
- Tao, L.; Huang, G.; Song, H.; Chen, Y.; Chen, L. Cancer associated fibroblasts: An essential role in the tumor microenvironment. Oncol. Lett. 2017, 14, 2611–2620. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Ma, W.; Wu, J.; Wu, Q.; Sun, C. Paracrine signaling in cancer-associated fibroblasts: Central regulators of the tumor immune microenvironment. J. Transl. Med. 2025, 23, 697. [Google Scholar] [CrossRef] [PubMed]
- Nissen, N.I.; Karsdal, M.; Willumsen, N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 2019, 38, 115. [Google Scholar] [CrossRef]
- Flynn, J.M.; Thadani, N.; Gallagher, E.E.; Azzaro, I.; Bodnar, C.M.; McCarty, C.P.; Romano, G.; Webster, M.R.; Capparelli, C. Plasticity and Functional Heterogeneity of Cancer-Associated Fibroblasts. Cancer Res. 2025, 85, 3378–3398. [Google Scholar] [CrossRef]
- Kehrberg, R.J.; Bhyravbhatla, N.; Batra, S.K.; Kumar, S. Epigenetic regulation of cancer-associated fibroblast heterogeneity. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188901. [Google Scholar] [CrossRef]
- Öhlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A.S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T.E.; Hearn, S.A.; Lee, E.J.; et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017, 214, 579–596. [Google Scholar] [CrossRef]
- Li, Q.; Lv, X.; Han, C.; Kong, Y.; Dai, Z.; Huo, D.; Li, T.; Li, D.; Li, W.; Wang, X.; et al. Enhancer reprogramming promotes the activation of cancer-associated fibroblasts and breast cancer metastasis. Theranostics 2022, 12, 7491–7508. [Google Scholar] [CrossRef]
- Ors Kumoglu, G.; Sendemir, A.; Tanyolac, M.B.; Bilir, B.; Kucuk, O.; Missirlis, Y.F. Epigenetic mechanisms in cancer. Longhua Chin. Med. 2022, 5, 4. [Google Scholar] [CrossRef]
- Sherif, Z.A.; Ogunwobi, O.O.; Ressom, H.W. Mechanisms and technologies in cancer epigenetics. Front. Oncol. 2025, 14, 1513654. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Halperin, C.; Hey, J.; Weichenhan, D.; Stein, Y.; Mayer, S.; Lutsik, P.; Plass, C.; Scherz-Shouval, R. Global DNA Methylation Analysis of Cancer-Associated Fibroblasts Reveals Extensive Epigenetic Rewiring Linked with RUNX1 Upregulation in Breast Cancer Stroma. Cancer Res. 2022, 82, 4139–4152. [Google Scholar] [CrossRef]
- Qiu, W.; Hu, M.; Sridhar, A.; Opeskin, K.; Fox, S.; Shipitsin, M.; Trivett, M.; Thompson, E.R.; Ramakrishna, M.; Gorringe, K.L.; et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat. Genet. 2008, 40, 650–655. [Google Scholar] [CrossRef]
- Ali, H.A.; Li, Y.; Bilal, A.H.M.; Qin, T.; Yuan, Z.; Zhao, W. A Comprehensive Review of BET Protein Biochemistry, Physiology, and Pathological Roles. Front. Pharmacol. 2022, 13, 818891. [Google Scholar] [CrossRef]
- Yamamoto, K.; Tateishi, K.; Kudo, Y.; Hoshikawa, M.; Tanaka, M.; Nakatsuka, T.; Fujiwara, H.; Miyabayashi, K.; Takahashi, R.; Tanaka, Y.; et al. Stromal remodeling by the BET bromodomain inhibitor JQ1 suppresses the progression of human pancreatic cancer. Oncotarget 2016, 7, 61469–61484. [Google Scholar] [CrossRef]
- Schmidt, M.; Maié, T.; Cramer, T.; Costa, I.G.; Wagner, W. Cancer-associated fibroblasts reveal aberrant DNA methylation across different types of cancer. Clin. Epigenetics 2024, 16, 164. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, T.D.; Von Ahrens, D.; Dawlaty, M.; Zou, Y.; Baddour, J.; Achreja, A.; Zhao, H.; Yang, L.; Patel, B.; Kwak, C.; et al. Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. eLife 2019, 8, e50663. [Google Scholar] [CrossRef]
- Li, C.; Teixeira, A.F.; Zhu, H.-J.; ten Dijke, P. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol. Cancer 2021, 20, 154. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Biffi, G.; Oni, T.E.; Spielman, B.; Hao, Y.; Elyada, E.; Park, Y.; Preall, J.; Tuveson, D.A. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFβ to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2019, 9, 282–301. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef]
- Li, B.L.; Lu, W.; Qu, J.J.; Ye, L.; Du, G.Q.; Wan, X.P. Loss of exosomal miR-148b from cancer-associated fibroblasts promotes endometrial cancer cell invasion and cancer metastasis. J. Cell. Physiol. 2019, 234, 2943–2953. [Google Scholar] [CrossRef]
- Xue, P.; Huang, S.; Han, X.; Zhang, C.; Yang, L.; Xiao, W.; Fu, J.; Li, H.; Zhou, Y. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 2022, 29, 82–95. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef]
- Shi, D.Q.; Ali, I.; Tang, J.; Yang, W.C. New Insights into 5hmC DNA Modification: Generation, Distribution and Function. Front. Genet. 2017, 8, 100. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Sato, T.; Issa, J.J.; Kropf, P. DNA Hypomethylating Drugs in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2017, 7, a026948. [Google Scholar] [CrossRef]
- Kang, G.H.; Lee, S.; Kim, J.-S.; Jung, H.-Y. Profile of Aberrant CpG Island Methylation along Multistep Gastric Carcinogenesis. Lab. Investig. 2003, 83, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA Methyltransferases in Cancer: Biology, Paradox, Aberrations, and Targeted Therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Wu, X.; Du, Q.; Lin, J.; Ma, W.; Liu, J. Systematic Characterization of DNA Methyltransferases Family in Tumor Progression and Antitumor Immunity. Technol. Cancer Res. Treat. 2024, 23, 15330338241260658. [Google Scholar] [CrossRef] [PubMed]
- Albrengues, J.; Bertero, T.; Grasset, E.; Bonan, S.; Maiel, M.; Bourget, I.; Philippe, C.; Herraiz Serrano, C.; Benamar, S.; Croce, O.; et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat. Commun. 2015, 6, 10204. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhou, D.; Rucki, A.A.; Williams, J.; Zhou, J.; Mo, G.; Murphy, A.; Fujiwara, K.; Kleponis, J.; Salman, B.; et al. Cancer-Associated Fibroblasts in Pancreatic Cancer Are Reprogrammed by Tumor-Induced Alterations in Genomic DNA Methylation. Cancer Res. 2016, 76, 5395–5404. [Google Scholar] [CrossRef] [PubMed]
- Neveu, W.A.; Mills, S.T.; Staitieh, B.S.; Sueblinvong, V. TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am. J. Physiol. Cell Physiol. 2015, 309, C616–C626. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharashi, L.A.; Al-Mohanna, F.H.; Tulbah, A.; Aboussekhra, A. The DNA methyl-transferase protein DNMT1 enhances tumor-promoting properties of breast stromal fibroblasts. Oncotarget 2018, 9, 2329–2343. [Google Scholar] [CrossRef]
- Cannon, A.; Thompson, C.; Hall, B.R.; Jain, M.; Kumar, S.; Batra, S.K. Desmoplasia in pancreatic ductal adenocarcinoma: Insight into pathological function and therapeutic potential. Genes Cancer 2018, 9, 78–86. [Google Scholar] [CrossRef]
- Oyon, D.; Lopez-Pascual, A.; Castello-Uribe, B.; Uriarte, I.; Orsi, G.; Llorente, S.; Elurbide, J.; Adan-Villaescusa, E.; Valbuena-Goiricelaya, E.; Irigaray-Miramon, A.; et al. Targeting of the G9a, DNMT1 and UHRF1 epigenetic complex as an effective strategy against pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2025, 44, 13. [Google Scholar] [CrossRef]
- Manoukian, P.; Kuhnen, L.C.; van Laarhoven, H.W.M.; Bijlsma, M.F. Association of epigenetic landscapes with heterogeneity and plasticity in pancreatic cancer. Crit. Rev. Oncol./Hematol. 2025, 206, 104573. [Google Scholar] [CrossRef]
- Gautam, S.K.; Batra, S.K.; Jain, M. Molecular and metabolic regulation of immunosuppression in metastatic pancreatic ductal adenocarcinoma. Mol. Cancer 2023, 22, 118. [Google Scholar] [CrossRef]
- Pan, X.; Zhou, J.; Xiao, Q.; Fujiwara, K.; Zhang, M.; Mo, G.; Gong, W.; Zheng, L. Cancer-associated fibroblast heterogeneity is associated with organ-specific metastasis in pancreatic ductal adenocarcinoma. J. Hematol. Oncol. 2021, 14, 184. [Google Scholar] [CrossRef]
- Mathot, P.; Grandin, M.; Devailly, G.; Souaze, F.; Cahais, V.; Moran, S.; Campone, M.; Herceg, Z.; Esteller, M.; Juin, P.; et al. DNA methylation signal has a major role in the response of human breast cancer cells to the microenvironment. Oncogenesis 2017, 6, e390. [Google Scholar] [CrossRef]
- Kadel, D.; Zhang, Y.; Sun, H.R.; Zhao, Y.; Dong, Q.Z.; Qin, L.X. Current perspectives of cancer-associated fibroblast in therapeutic resistance: Potential mechanism and future strategy. Cell Biol. Toxicol. 2019, 35, 407–421. [Google Scholar] [CrossRef]
- Wu, Y.; Li, N.; Shang, J.; Jiang, J.; Liu, X. Identification of cancer-associated fibroblast subtypes and prognostic model development in breast cancer: Role of the RUNX1/SDC1 axis in promoting invasion and metastasis. Cell Biol. Toxicol. 2025, 41, 21. [Google Scholar] [CrossRef]
- Wang, C.; Dong, D.; Zhao, N.; Liu, Y.; Bai, C.; Hua, J.; Cui, R.; Wei, X.; Zhao, T.; Ji, N.; et al. Tumor-derived CCL15 regulates RNA m6A methylation in cancer-associated fibroblasts to promote hepatocellular carcinoma growth. Cancer Lett. 2024, 611, 217420. [Google Scholar] [CrossRef] [PubMed]
- Su, S.-F.; Ho, H.; Li, J.-H.; Wu, M.-F.; Wang, H.-C.; Yeh, H.-Y.; Kuo, S.-W.; Chen, H.-W.; Ho, C.-C.; Li, K.-C. DNA methylome and transcriptome landscapes of cancer-associated fibroblasts reveal a smoking-associated malignancy index. J. Clin. Investig. 2021, 131, e139552. [Google Scholar] [CrossRef]
- Han, F.; Chen, S.; Zhang, K.; Wang, M.; Wang, P. Single-cell transcriptomic sequencing data reveal aberrant DNA methylation in SMAD3 promoter region in tumor-associated fibroblasts affecting molecular mechanism of radiosensitivity in non-small cell lung cancer. J. Transl. Med. 2024, 22, 288. [Google Scholar] [CrossRef] [PubMed]
- Papavassiliou, K.A.; Sofianidi, A.A.; Gogou, V.A.; Papavassiliou, A.G. Targeting Epigenetic Alterations Linked to Cancer-Associated Fibroblast Phenotypes in Lung Cancer. Cancers 2024, 16, 3976. [Google Scholar] [CrossRef] [PubMed]
- Juergens, R.A.; Wrangle, J.; Vendetti, F.P.; Murphy, S.C.; Zhao, M.; Coleman, B.; Sebree, R.; Rodgers, K.; Hooker, C.M.; Franco, N.; et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011, 1, 598–607. [Google Scholar] [CrossRef]
- Levy, B.P.; Giaccone, G.; Besse, B.; Felip, E.; Garassino, M.C.; Domine Gomez, M.; Garrido, P.; Piperdi, B.; Ponce-Aix, S.; Menezes, D.; et al. Randomised phase 2 study of pembrolizumab plus CC-486 versus pembrolizumab plus placebo in patients with previously treated advanced non-small cell lung cancer. Eur. J. Cancer 2019, 108, 120–128. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Jänne, P.A.; Opyrchal, M.; Hafez, N.; Raez, L.E.; Gabrilovich, D.I.; Wang, F.; Trepel, J.B.; Lee, M.J.; Yuno, A.; et al. Entinostat plus Pembrolizumab in Patients with Metastatic NSCLC Previously Treated with Anti-PD-(L)1 Therapy. Clin. Cancer Res. 2021, 27, 1019–1028. [Google Scholar] [CrossRef]
- Alcaraz, J.; Ikemori, R.; Llorente, A.; Díaz-Valdivia, N.; Reguart, N.; Vizoso, M. Epigenetic Reprogramming of Tumor-Associated Fibroblasts in Lung Cancer: Therapeutic Opportunities. Cancers 2021, 13, 3782. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, H.; Luo, X.; Chen, Y.; Shi, C.; Wang, Y.; Bai, J.; Shao, Z.; Shang, Z. NNMT switches the proangiogenic phenotype of cancer-associated fibroblasts via epigenetically regulating ETS2/VEGFA axis. Oncogene 2024, 43, 2647–2660. [Google Scholar] [CrossRef]
- Wang, P.; Wang, G.; Li, H.; Yuan, Y.; Chen, H.; Wang, S.; Sun, Z.; Meng, F.; Li, Y.; Yang, F.; et al. Nicotinamide N-methyltransferase negatively regulates metastasis-promoting property of cancer-associated fibroblasts in lung adenocarcinoma. Cancer Commun. 2025, 45, 110–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Qiu, M.; Chen, K. 231P Cancer-associated fibroblast-expressed nicotinamide N-methyltransferase is negatively associated with metastasis of lung adenocarcinomas. ESMO Open 2024, 9, 102881. [Google Scholar] [CrossRef]
- Mrazek, A.A.; Carmical, J.R.; Wood, T.G.; Hellmich, M.R.; Eltorky, M.; Bohanon, F.J.; Chao, C. Colorectal Cancer-Associated Fibroblasts are Genotypically Distinct. Curr. Cancer Ther. Rev. 2014, 10, 97–218. [Google Scholar] [CrossRef]
- Niell, N.; Larriba, M.J.; Ferrer-Mayorga, G.; Sánchez-Pérez, I.; Cantero, R.; Real, F.X.; Del Peso, L.; Muñoz, A.; González-Sancho, J.M. The human PKP2/plakophilin-2 gene is induced by Wnt/β-catenin in normal and colon cancer-associated fibroblasts. Int. J. Cancer 2018, 142, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Swets, M.; Zaalberg, A.; Boot, A.; van Wezel, T.; Frouws, M.A.; Bastiaannet, E.; Gelderblom, H.; van de Velde, C.J.; Kuppen, P.J. Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer. Int. J. Mol. Sci. 2016, 18, 36. [Google Scholar] [CrossRef]
- Ling, E.; Ringel, A.; Sigal-Batikoff, I.; Abu-Freha, N.; Vaknine, H.; Friah, W.; Reshef, A.; Pinsk, I.; Fich, A.; Lamprecht, S. Human Colorectal Cancer Stage-dependent Global DNA Hypomethylation of Cancer-associated Fibroblasts. Anticancer Res. 2016, 36, 4503–4507. [Google Scholar] [CrossRef]
- Najgebauer, H.; Liloglou, T.; Jithesh, P.V.; Giger, O.T.; Varro, A.; Sanderson, C.M. Integrated omics profiling reveals novel patterns of epigenetic programming in cancer-associated myofibroblasts. Carcinogenesis 2019, 40, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Takeshima, H.; Iida, N.; Hattori, N.; Yamashita, S.; Moro, H.; Yasukawa, Y.; Nishiyama, K.; Hashimoto, T.; Sekine, S.; et al. Cancer cell niche factors secreted from cancer-associated fibroblast by loss of H3K27me3. Gut 2020, 69, 243–251. [Google Scholar] [CrossRef]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Zhu, M.; Tan, X.; Zhang, Y.; Liu, X.; Yang, L. Super-enhancers and the super-enhancer reader BRD4: Tumorigenic factors and therapeutic targets. Cell Death Discov. 2023, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.-P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone Recognition and Large-Scale Structural Analysis of the Human Bromodomain Family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef]
- Samani, K.; Sharma, U.R.; Sharma, A.R.; Pm, M.; Surendra, V. Role of BRD4 in cancer—A review. IP J. Diagn. Pathol. Oncol. 2020, 5, 128–134. [Google Scholar] [CrossRef]
- Cheung, K.L.; Kim, C.; Zhou, M.-M. The Functions of BET Proteins in Gene Transcription of Biology and Diseases. Front. Mol. Biosci. 2021, 8, 728777. [Google Scholar] [CrossRef]
- Ember, S.W.J.; Zhu, J.-Y.; Olesen, S.H.; Martin, M.P.; Becker, A.; Berndt, N.; Georg, G.I.; Schönbrunn, E. Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors. ACS Chem. Biol. 2014, 9, 1160–1171. [Google Scholar] [CrossRef]
- Kumar, K.; DeCant, B.T.; Grippo, P.J.; Hwang, R.F.; Bentrem, D.J.; Ebine, K.; Munshi, H.G. BET inhibitors block pancreatic stellate cell collagen I production and attenuate fibrosis in vivo. JCI Insight 2017, 2, e88032. [Google Scholar] [CrossRef]
- Shorstova, T.; Foulkes, W.D.; Witcher, M. Achieving clinical success with BET inhibitors as anti-cancer agents. Br. J. Cancer 2021, 124, 1478–1490. [Google Scholar] [CrossRef]
- Wang, W.; Tang, Y.-A.; Xiao, Q.; Lee, W.C.; Cheng, B.; Niu, Z.; Oguz, G.; Feng, M.; Lee, P.L.; Li, B.; et al. Stromal induction of BRD4 phosphorylation Results in Chromatin Remodeling and BET inhibitor Resistance in Colorectal Cancer. Nat. Commun. 2021, 12, 4441. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Elliott, I.A.; Wu, N.; Matsumura, C.; Vogelauer, M.; Attar, N.; Dann, A.; Ghukasyan, R.; Toste, P.A.; Patel, S.G.; et al. Histone deacetylase inhibitors provoke a tumor supportive phenotype in pancreatic cancer associated fibroblasts. Oncotarget 2017, 8, 19074–19088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ge, L.; Zhang, Y.; Zhang, Z.; Song, F.; Huang, P.; Xu, T. Targeted intervention of tumor microenvironment with HDAC inhibitors and their combination therapy strategies. Eur. J. Med. Res. 2025, 30, 69. [Google Scholar] [CrossRef]
- Liang, G.; Oh, T.G.; Hah, N.; Tiriac, H.; Shi, Y.; Truitt, M.L.; Antal, C.E.; Atkins, A.R.; Li, Y.; Fraser, C.; et al. Inhibiting stromal Class I HDACs curbs pancreatic cancer progression. Nat. Commun. 2023, 14, 7791. [Google Scholar] [CrossRef]
- Li, A.; Chen, P.; Leng, Y.; Kang, J. Histone deacetylase 6 regulates the immunosuppressive properties of cancer-associated fibroblasts in breast cancer through the STAT3-COX2-dependent pathway. Oncogene 2018, 37, 5952–5966. [Google Scholar] [CrossRef]
- Bauer, N.; Balourdas, D.I.; Schneider, J.R.; Zhang, X.; Berger, L.M.; Berger, B.T.; Schwalm, M.P.; Klopp, N.A.; Siveke, J.T.; Knapp, S.; et al. Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization. ACS Chem. Biol. 2024, 19, 266–279. [Google Scholar] [CrossRef]
- Doskey, L.C.; Scholtz, C.R.; Vail, N.R.; Khanal, S.; Lee, A.L.; Kandanur, S.G.S.; Hoell, Z.J.; Huehls, A.M.; Issa, M.R.; Kostallari, E.; et al. Efficacy and Toxicity Analysis of Selective BET Bromodomain Inhibitors in Models of Inflammatory Liver Disease. J. Med. Chem. 2025, 68, 8091–8105. [Google Scholar] [CrossRef] [PubMed]
- Manzotti, G.; Ciarrocchi, A.; Sancisi, V. Inhibition of BET Proteins and Histone Deacetylase (HDACs): Crossing Roads in Cancer Therapy. Cancers 2019, 11, 304. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Ren, B.; Fang, Y.; Ren, J.; Liu, X.; Wang, X.; Zhou, F.; Xiao, R.; Luo, X.; You, L.; et al. Epigenetic regulation in cancer. MedComm 2024, 5, e495. [Google Scholar] [CrossRef]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: First Steps from Bench to Bedside. Target Oncol. 2020, 15, 261–278. [Google Scholar] [CrossRef]
- Zhang, Y.; Lan, M.; Chen, Y. Minimal Information for Studies of Extracellular Vesicles (MISEV): Ten-Year Evolution (2014–2023). Pharmaceutics 2024, 16, 1394. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Gu, J.; Zhang, J.; Shi, H.; Hou, S.; Xu, X.; Chen, Y.; Zhang, Y.; Mao, F.; Qian, H.; et al. Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 2020, 11, 215. [Google Scholar] [CrossRef]
- Dai, X.; Chen, C.; Yang, Q.; Xue, J.; Chen, X.; Sun, B.; Luo, F.; Liu, X.; Xiao, T.; Xu, H.; et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018, 9, 454. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Yang, M.; Hou, Y.; Chen, Y.; Bie, P. Exosomal miR-29b from cancer-associated fibroblasts inhibits the migration and invasion of hepatocellular carcinoma cells. Transl. Cancer Res. 2020, 9, 2576–2587. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Lei, J.H.; Wang, Q.; Qu, T.F.; Cha, L.C.; Zhan, H.X.; Liu, S.L.; Hu, X.; Sun, C.D.; Guo, W.D.; et al. Cancer-associated fibroblast-secreted miR-421 promotes pancreatic cancer by regulating the SIRT3/H3K9Ac/HIF-1α axis. Kaohsiung J. Med. Sci. 2022, 38, 1080–1092. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhong, X.; Li, J.; Hu, R.; Yi, J.; Sun, J.; Xu, Y.; Zhou, X. Exosomal ncRNAs: Multifunctional contributors to the immunosuppressive tumor microenvironment of hepatocellular carcinoma. Biomed. Pharmacother. 2024, 173, 116409. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef]
- Kan, R.L.; Chen, J.; Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 2022, 38, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Savardashtaki, A.; Shabaninejad, Z.; Movahedpour, A.; Sahebnasagh, R.; Mirzaei, H.; Hamblin, M.R. miRNAs derived from cancer-associated fibroblasts in colorectal cancer. Epigenomics 2019, 11, 1627–1645. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, D.; Wang, Y.; Sun, P.; Hou, X.; Larner, J.; Xiong, W.; Mi, J. MiR-21/Smad 7 signaling determines TGF-β1-induced CAF formation. Sci. Rep. 2013, 3, 2038. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, J.; Li, W.; Zhang, C. Micro-RNA-21 Regulates Cancer-Associated Fibroblast-Mediated Drug Resistance in Pancreatic Cancer. Oncol. Res. 2018, 26, 827–835. [Google Scholar] [CrossRef]
- Kunita, A.; Morita, S.; Irisa, T.U.; Goto, A.; Niki, T.; Takai, D.; Nakajima, J.; Fukayama, M. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci. Rep. 2018, 8, 8838. [Google Scholar] [CrossRef]
- Shintani, Y.; Kimura, T.; Funaki, S.; Ose, N.; Kanou, T.; Fukui, E. Therapeutic Targeting of Cancer-Associated Fibroblasts in the Non-Small Cell Lung Cancer Tumor Microenvironment. Cancers 2023, 15, 335. [Google Scholar] [CrossRef]
- Ren, Y.; Jia, H.H.; Xu, Y.Q.; Zhou, X.; Zhao, X.H.; Wang, Y.F.; Song, X.; Zhu, Z.Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ß1 secretion. Mol. Cancer 2018, 17, 5. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, S.; Hu, C.; Li, G.; Lin, H.; Xia, R.; Ye, Y.; He, R.; Li, Z.; Lin, Q.; et al. Cancer-associated fibroblast-induced lncRNA UPK1A-AS1 confers platinum resistance in pancreatic cancer via efficient double-strand break repair. Oncogene 2022, 41, 2372–2389. [Google Scholar] [CrossRef]
- Deng, X.; Ruan, H.; Zhang, X.; Xu, X.; Zhu, Y.; Peng, H.; Kong, F.; Guan, M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int. J. Cancer 2020, 146, 1700–1716. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Li, G.; Jiang, K.; Li, Z.; Liu, T. Cancer therapy resistance mediated by cancer-associated fibroblast-derived extracellular vesicles: Biological mechanisms to clinical significance and implications. Mol. Cancer 2024, 23, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.; Fu, L.; Shi, M.; Hu, G.; Du, F.; Wang, Z.; Xiao, Y.; Zhang, Y.; Li, Y. Role of exosomal non-coding RNAs in cancer-associated fibroblast-mediated therapy resistance (Review). Int. J. Oncol. 2025, 67, 68. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhang, J.; Bao, C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer 2021, 21, 933. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W.F.; Courtois, E.T.; Burkhart, R.A.; Teinor, J.A.; Belleau, P.; Biffi, G.; Lucito, M.S.; et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019, 9, 1102–1123. [Google Scholar] [CrossRef]
- Han, X.; Li, Y.; Xu, Y.; Zhao, X.; Zhang, Y.; Yang, X.; Wang, Y.; Zhao, R.; Anderson, G.J.; Zhao, Y.; et al. Reversal of pancreatic desmoplasia by re-educating stellate cells with a tumour microenvironment-activated nanosystem. Nat. Commun. 2018, 9, 3390. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β signaling in health, disease and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Kurisaki, A.; Kose, S.; Yoneda, Y.; Heldin, C.-H.; Moustakas, A. Transforming Growth Factor-β Induces Nuclear Import of Smad3 in an Importin-β1 and Ran-dependent Manner. Mol. Biol. Cell 2001, 12, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Stratton, M.S.; Bagchi, R.A.; Felisbino, M.B.; Hirsch, R.A.; Smith, H.E.; Riching, A.S.; Enyart, B.Y.; Koch, K.A.; Cavasin, M.A.; Alexanian, M.; et al. Dynamic Chromatin Targeting of BRD4 Stimulates Cardiac Fibroblast Activation. Circ. Res. 2019, 125, 662–677. [Google Scholar] [CrossRef]
- Niu, N.; Shen, X.; Wang, Z.; Chen, Y.; Weng, Y.; Yu, F.; Tang, Y.; Lu, P.; Liu, M.; Wang, L.; et al. Tumor cell-intrinsic epigenetic dysregulation shapes cancer-associated fibroblasts heterogeneity to metabolically support pancreatic cancer. Cancer Cell 2024, 42, 869–884.e9. [Google Scholar] [CrossRef]
- Noel, P.; Hussein, S.; Ng, S.; Antal, C.E.; Lin, W.; Rodela, E.; Delgado, P.; Naveed, S.; Downes, M.; Lin, Y.; et al. Triptolide targets super-enhancer networks in pancreatic cancer cells and cancer-associated fibroblasts. Oncogenesis 2020, 9, 100. [Google Scholar] [CrossRef]
- Soutto, M.; Bhat, N.; Khalafi, S.; Zhu, S.; Poveda, J.; Garcia-Buitrago, M.; Zaika, A.; El-Rifai, W. NF-kB-dependent activation of STAT3 by H. pylori is suppressed by TFF1. Cancer Cell Int. 2021, 21, 444. [Google Scholar] [CrossRef]
- Wojciak, J.M.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 2009, 28, 948–958. [Google Scholar] [CrossRef]
- Wang, R.; Cherukuri, P.; Luo, J. Activation of Stat3 Sequence-specific DNA Binding and Transcription by p300/CREB-binding Protein-mediated Acetylation. J. Biol. Chem. 2005, 280, 11528–11534. [Google Scholar] [CrossRef]
- Narita, T.; Ito, S.; Higashijima, Y.; Chu, W.K.; Neumann, K.; Walter, J.; Satpathy, S.; Liebner, T.; Hamilton, W.B.; Maskey, E.; et al. Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Mol. Cell 2021, 81, 2166–2182.e6. [Google Scholar] [CrossRef]
- Liu, Y.; Sinjab, A.; Min, J.; Han, G.; Paradiso, F.; Zhang, Y.; Wang, R.; Pei, G.; Dai, Y.; Cho, K.S.; et al. Conserved spatial subtypes and cellular neighborhoods of cancer-associated fibroblasts revealed by single-cell spatial multi-omics. Cancer Cell 2025, 43, 905–924.e6. [Google Scholar] [CrossRef]
- Song, J.; Wei, R.; Liu, C.; Zhao, Z.; Liu, X.; Wang, Y.; Liu, F. Antigen-presenting cancer associated fibroblasts enhance antitumor immunity and predict immunotherapy response. Nat. Commun. 2025, 16, 2175. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Z.; Zhang, Y.; Pradhan, R.N.; Ganguly, D.; Chandra, R.; Murimwa, G.; Wright, S.; Gu, X.; Maddipati, R.; et al. Mesothelial cell-derived antigen-presenting cancer-associated fibroblasts induce expansion of regulatory T cells in pancreatic cancer. Cancer Cell 2022, 40, 656–673.e7. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, W.; Xu, H.; Xu, J.; Li, J.; Liu, X.; Lu, X.; Dai, J.; Jiang, Y.; Wang, W.; et al. Cancer-associated fibroblasts promote EGFR-TKI resistance via the CTHRC1/glycolysis/H3K18la positive feedback loop. Oncogene 2025, 44, 1400–1414. [Google Scholar] [CrossRef]
- Zhou, S.; Xiao, L.; Hu, L.; Zuo, F.; Wang, Y.; Fei, B.; Dai, J.; Zhou, X. CAFs promote immune evasion in gastric cancer through histone lactylation-mediated suppression of NCAPG ubiquitination. J. Transl. Med. 2025, 23, 989. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liang, P.; Chen, Z.; Jin, T.; He, F.; Chen, X.; Yang, K. CAF-secreted LOX promotes PD-L1 expression via histone Lactylation and regulates tumor EMT through TGFβ/IGF1 signaling in gastric Cancer. Cell Signal. 2024, 124, 111462. [Google Scholar] [CrossRef]
- Deng, Y.; Bartosovic, M.; Ma, S.; Zhang, D.; Kukanja, P.; Xiao, Y.; Su, G.; Liu, Y.; Qin, X.; Rosoklija, G.B.; et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 2022, 609, 375–383. [Google Scholar] [CrossRef]
- Jing, S.-y.; Wang, H.-q.; Lin, P.; Yuan, J.; Tang, Z.-x.; Li, H. Quantifying and interpreting biologically meaningful spatial signatures within tumor microenvironments. npj Precis. Oncol. 2025, 9, 68. [Google Scholar] [CrossRef]
- Grout, J.A.; Sirven, P.; Leader, A.M.; Maskey, S.; Hector, E.; Puisieux, I.; Steffan, F.; Cheng, E.; Tung, N.; Maurin, M.; et al. Spatial Positioning and Matrix Programs of Cancer-Associated Fibroblasts Promote T-cell Exclusion in Human Lung Tumors. Cancer Discov. 2022, 12, 2606–2625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Deng, Y.; Kukanja, P.; Agirre, E.; Bartosovic, M.; Dong, M.; Ma, C.; Ma, S.; Su, G.; Bao, S.; et al. Spatial epigenome–transcriptome co-profiling of mammalian tissues. Nature 2023, 616, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Jieun Kim, H.; Ruan, T.; Swarbrick, A. Pan-Cancer Spatial Profiling Reveals Conserved Subtypes and Niches of Cancer-Associated Fibroblasts. Cancer Res. 2025, 85, 2555–2557. [Google Scholar] [CrossRef]
- Carmona-Fontaine, C.; Deforet, M.; Akkari, L.; Thompson, C.B.; Joyce, J.A.; Xavier, J.B. Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2017, 114, 2934–2939. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.Y.; Yang, J.L.; Lai, R.; Zhou, Z.J.; Tang, D.; Hu, L.; Zhao, L.J. Impact of lactate on immune cell function in the tumor microenvironment: Mechanisms and therapeutic perspectives. Front. Immunol. 2025, 16, 1563303. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhao, R.; Shen, Y.; Guo, D.; Deng, L.; Cai, R.; Shen, Z.; Xie, Z.; Hang, N.; Fu, S.; et al. Lactylation in Tumor Immune Escape and Immunotherapy: Multifaceted Functions and Therapeutic Strategies. Research 2025, 8, 0793. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Chen, Y.; Mao, L.; Cardilla, A.; Lee, C.N.; Cui, Y.; Jin, D.; Hua, Y.; Xu, X.; Deng, Y. Spatial profiling of chromatin accessibility in formalin-fixed paraffin-embedded tissues. Nat. Commun. 2025, 16, 5945. [Google Scholar] [CrossRef]
- Zhang, H.; Polavarapu, V.K.; Xing, P.; Zhao, M.; Mathot, L.; Zhao, L.; Rosen, G.; Swartling, F.J.; Sjöblom, T.; Chen, X. Profiling chromatin accessibility in formalin-fixed paraffin-embedded samples. Genome Res. 2022, 32, 150–161. [Google Scholar] [CrossRef]
- Li, H.; Bao, S.; Farzad, N.; Qin, X.; Fung, A.A.; Zhang, D.; Bai, Z.; Tao, B.; Fan, R. Spatially resolved genome-wide joint profiling of epigenome and transcriptome with spatial-ATAC-RNA-seq and spatial-CUT&Tag-RNA-seq. Nat. Protoc. 2025, 20, 2383–2417. [Google Scholar] [CrossRef]
- Cords, L.; Tietscher, S.; Anzeneder, T.; Langwieder, C.; Rees, M.; de Souza, N.; Bodenmiller, B. Cancer-associated fibroblast classification in single-cell and spatial proteomics data. Nat. Commun. 2023, 14, 4294. [Google Scholar] [CrossRef]
| Cancer Type | Methylation Gene in CAF | Direction |
|---|---|---|
| Pancreatic cancer | SOCS1; STAT3/IGF-1 axis | Hyper |
| Breast cancer | RUNX1/Syndecan-1 (SDC1) | Hyper |
| Hepatocellular carcinoma | CCL15 | Hyper |
| Lung cancer | SMAD3; TGF-β-driven | Mixed/hyper (SMAD3) |
| Oral cancer | ||
| Colorectal cancer | LINE-1, COL1A1, GJA4 | Global hypo |
| Gastric cancer and Esophageal cancer | SMAD3, SPON2; H3K27me3 loss | Promoter hyper |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Nishikubo, H.; Ma, D.; Pan, J.; Sano, T.; Imanishi, D.; Sakuma, T.; Fan, C.; Yashiro, M. Significance of Epigenetic Alteration in Cancer-Associated Fibroblasts on the Development of Carcinoma. Int. J. Mol. Sci. 2025, 26, 9695. https://doi.org/10.3390/ijms26199695
Gao H, Nishikubo H, Ma D, Pan J, Sano T, Imanishi D, Sakuma T, Fan C, Yashiro M. Significance of Epigenetic Alteration in Cancer-Associated Fibroblasts on the Development of Carcinoma. International Journal of Molecular Sciences. 2025; 26(19):9695. https://doi.org/10.3390/ijms26199695
Chicago/Turabian StyleGao, Hongdong, Hinano Nishikubo, Dongheng Ma, Juncheng Pan, Tomoya Sano, Daiki Imanishi, Takashi Sakuma, Canfeng Fan, and Masakazu Yashiro. 2025. "Significance of Epigenetic Alteration in Cancer-Associated Fibroblasts on the Development of Carcinoma" International Journal of Molecular Sciences 26, no. 19: 9695. https://doi.org/10.3390/ijms26199695
APA StyleGao, H., Nishikubo, H., Ma, D., Pan, J., Sano, T., Imanishi, D., Sakuma, T., Fan, C., & Yashiro, M. (2025). Significance of Epigenetic Alteration in Cancer-Associated Fibroblasts on the Development of Carcinoma. International Journal of Molecular Sciences, 26(19), 9695. https://doi.org/10.3390/ijms26199695





