Genomic Signatures of Adaptive Evolution in Taenioides sp. During Northward Invasion
Abstract
1. Introduction
2. Results
2.1. SNP and Genetic Diversity
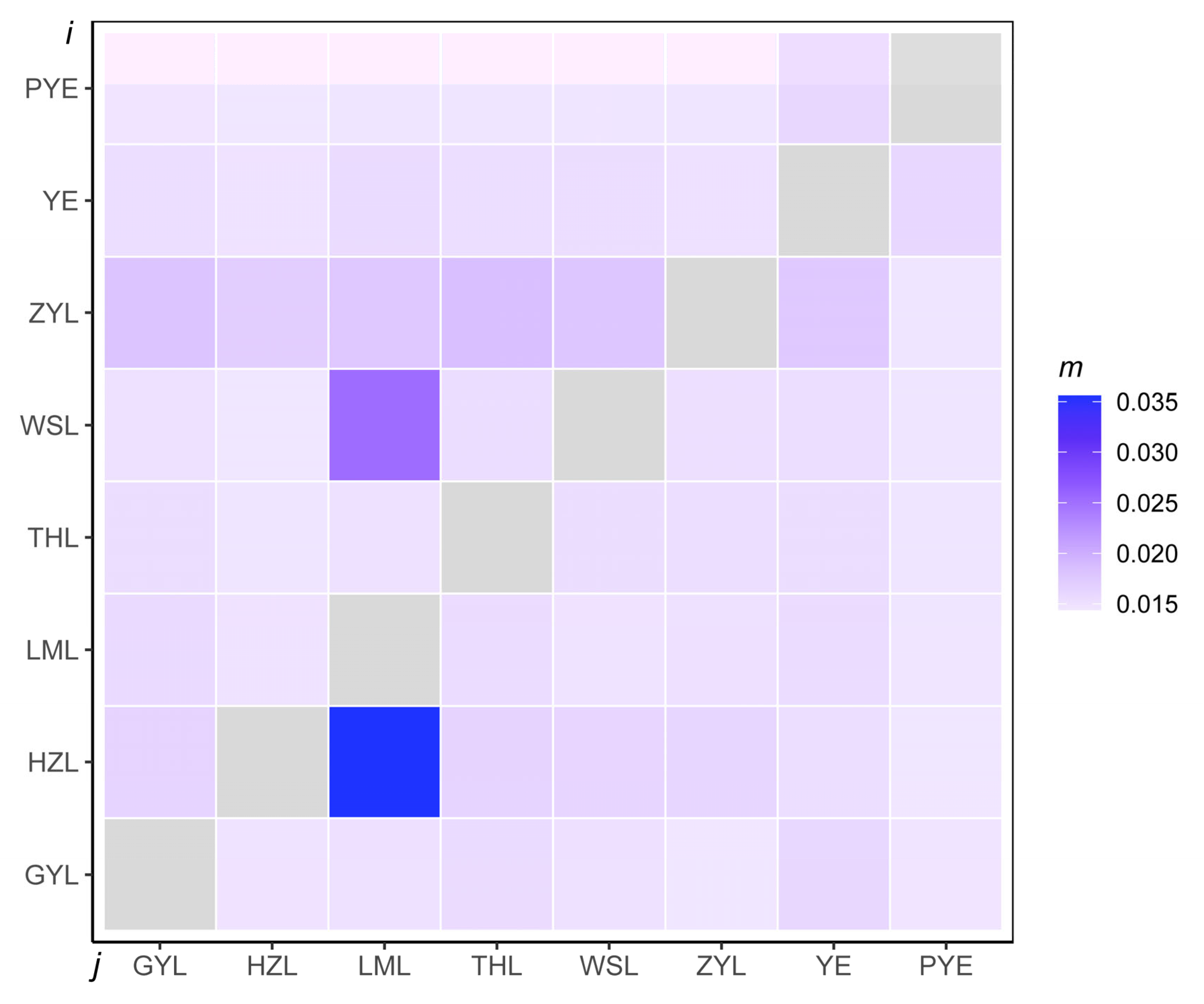
2.2. Population Genetic Structure
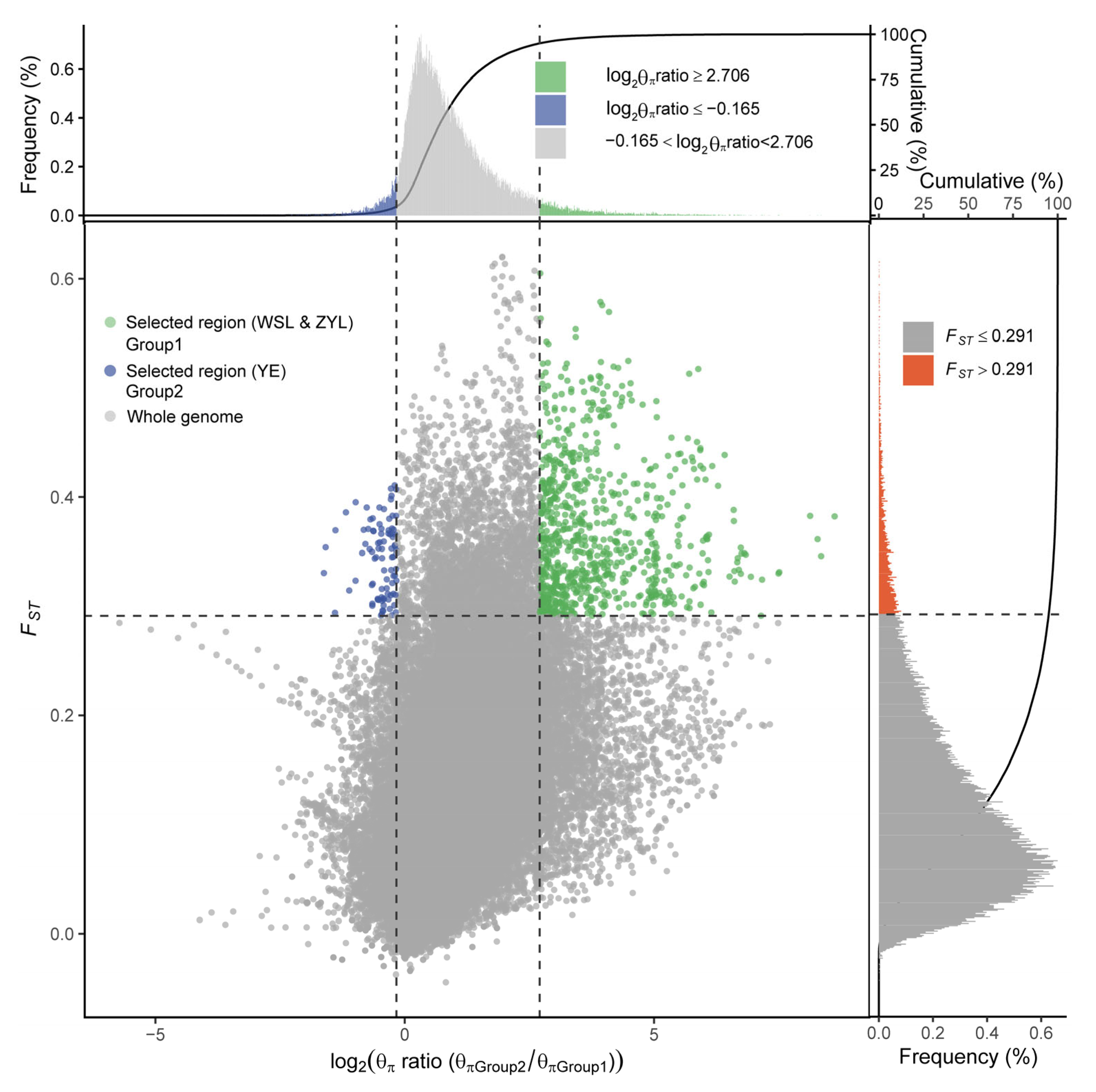
2.3. Candidate Loci Under Selection
2.4. Gene–Environment Associations
2.5. Functional Annotation and Enrichment
3. Discussion
3.1. Genetic Diversity and Large Bottleneck at the Invasion Front
3.2. Genetic Structure and the Invasion Source Inference
3.3. Adaptive Evolution Underlying the Northward Invasions
4. Materials and Methods
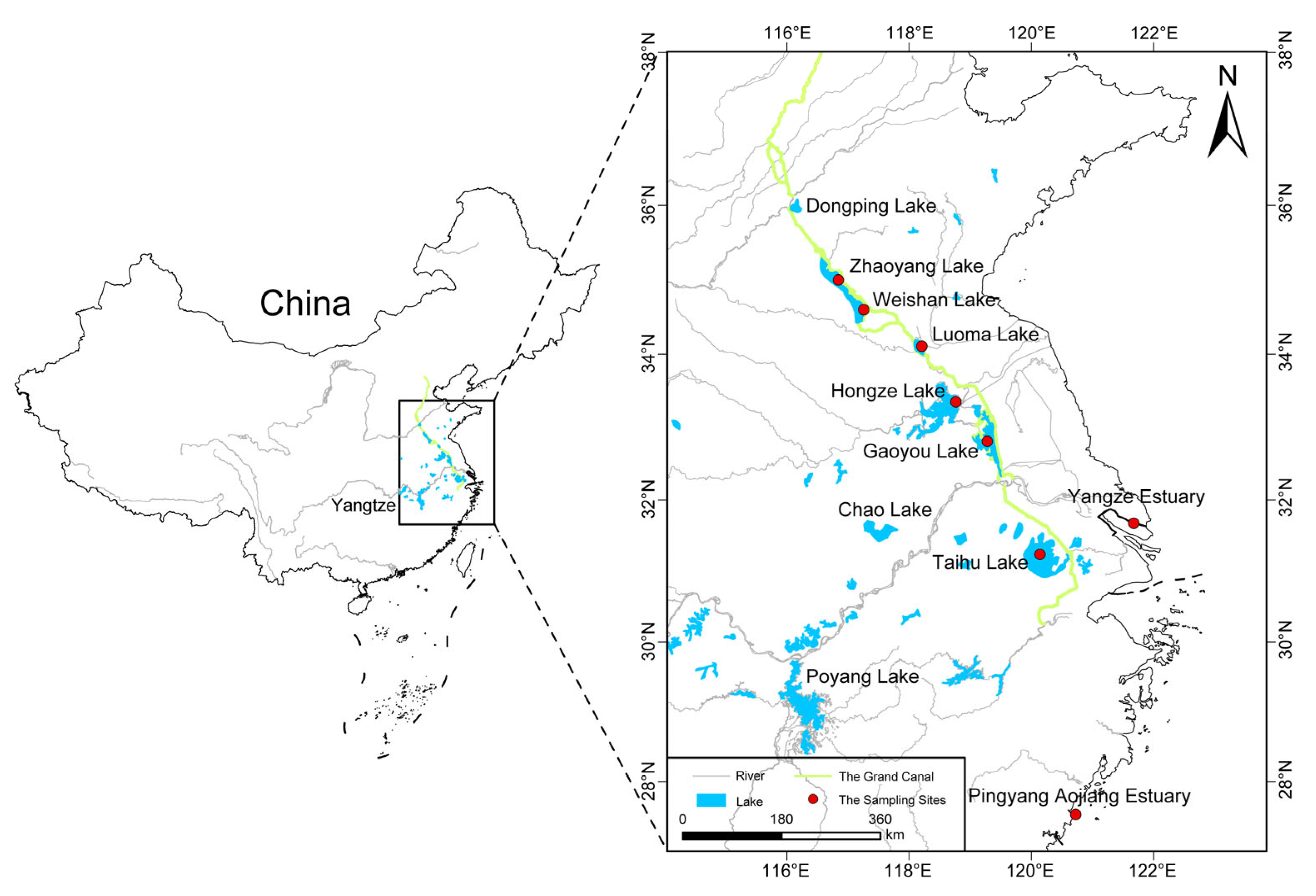
4.1. Sample Collection and Genomic DNA Extraction
4.2. Whole-Genome Resequencing and SNP Detection
4.3. Population Genetic Structure Inference
4.4. Genome-Wide Scan of Adaptive Signals
4.5. Genotype–Environment Association Analyses
4.6. Functional Annotation of Adaptive Loci
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, M.; North, H.L.; Peng, Y.; Liu, H.; Liu, B.; Pan, R.; Zhou, Y.; Zheng, W.; Liu, K.; Yang, B. Adaptive evolution to the natural and anthropogenic environment in a global invasive crop pest, the cotton bollworm. Innov. 2023, 4, 100454. [Google Scholar] [CrossRef]
- Bernardi, G.; Azzurro, E.; Golani, D.; Miller, M.R. Genomic signatures of rapid adaptive evolution in the bluespotted cornetfish, a Mediterranean Lessepsian invader. Mol. Ecol. 2016, 25, 3384–3396. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Peng, Y.; Guo, C.; Qu, W.; Yang, N.; Zhu, Y.; Jeong, I.; Li, X.; Ghanim, M. Invasion genomics uncover complex introduction patterns of the globally invasive whitefly, Bemisia tabaci MED. Divers. Distrib. 2023, 29, 1172–1189. [Google Scholar] [CrossRef]
- Baltazar-Soares, M.; Blanchet, S.; Cote, J.; Tarkan, A.S.; Záhorská, E.; Gozlan, R.E.; Eizaguirre, C. Genomic footprints of a biological invasion: Introduction from Asia and dispersal in Europe of the topmouth gudgeon (Pseudorasbora parva). Mol. Ecol. 2020, 29, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Britton, J.R.; Gozlan, R.E. How many founders for a biological invasion? Predicting introduction outcomes from propagule pressure. Ecology 2013, 94, 2558–2566. [Google Scholar] [CrossRef] [PubMed]
- Simberloff, D. The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 81–102. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Huang, X.; Li, S.; Zhan, A. Local environment-driven adaptive evolution in a marine invasive ascidian (Molgula manhattensis). Ecol. Evol. 2021, 11, 4252–4266. [Google Scholar] [CrossRef]
- Qin, J.; Bjorn Victor, S.; Zhang, L.; Cheng, F.; Xie, S. Patterns of genetic diversity: Stepping-stone dispersal of an invasive fish introduced by an inter-basin water transfer project. Freshw. Biol. 2022, 67, 2078–2088. [Google Scholar] [CrossRef]
- Sun, C.; Lü, Z.; Fang, J.; Yao, C.; Zhao, S.; Liu, Y.; Gong, L.; Liu, B.; Liu, L.; Liu, J. Population structure of Taenioides sp. (Gobiiformes, Gobiidae) reveals their invasion history to inland waters of China based on mitochondrial DNA control region. ZooKeys 2024, 1203, 239. [Google Scholar] [CrossRef]
- Ni, Y.; Wu, H.L. Fishes in the Jiangsu Province; China Agriculture Press: Beijing, China, 2006; p. 699. [Google Scholar]
- Wu, H.; Zhong, J. Fauna Sinica: Osteichthyes Perciformes (V), Gobioidei; Science Press: Beijing, China, 2008; p. 747. [Google Scholar]
- Dlugosch, K.M.; Parker, I.M. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008, 17, 431–449. [Google Scholar] [CrossRef]
- Cui, M.; Roe, A.D.; Boyle, B.; Keena, M.; Wu, Y.; Braswell, W.E.; Smith, M.T.; Gasman, B.; Shi, J.; Javal, M. Tracking the North American Asian longhorned beetle invasion with genomics. Evol. Appl. 2024, 17, e70036. [Google Scholar] [CrossRef] [PubMed]
- Marchini, G.L.; Sherlock, N.C.; Ramakrishnan, A.P.; Rosenthal, D.M.; Cruzan, M.B. Rapid purging of genetic load in a metapopulation and consequences for range expansion in an invasive plant. Biol. Invasions 2016, 18, 183–196. [Google Scholar] [CrossRef]
- Teixeira, D.F.; Neto, F.R.A.; Gomes, L.C.; Beheregaray, L.B.; Carvalho, D.C. Invasion dynamics of the white piranha (Serrasalmus brandtii) in a Neotropical river basin. Biol. Invasions 2020, 22, 983–995. [Google Scholar] [CrossRef]
- Hargrove, J.S.; Weyl, O.L.; Austin, J.D. Reconstructing the introduction history of an invasive fish predator in South Africa. Biol. Invasions 2017, 19, 2261–2276. [Google Scholar] [CrossRef]
- Ni, Y.; Zhu, C.D. Fishes of Taihu Lake; Shanghai Scientific and Technical Publishers: Beijing, China, 2005; p. 255. [Google Scholar]
- Yuan, C.M.; Xie, H.G. Freshwater Fishes in Jiangsu Province; Jiangsu Scientific and Technical Publishers: Nanjing, China, 1987; p. 315. [Google Scholar]
- Poulet, N.; Balaresque, P.; Aho, T.; Björklund, M. Genetic structure and dynamics of a small introduced population: The pikeperch, Sander lucioperca, in the Rhône delta. Genetica 2009, 135, 77–86. [Google Scholar] [CrossRef]
- Zhang, Z.F.; Zhang, X.; Xu, J.; Zhang, N.; Liu, Z.H.; Zhang, Y.H. Characteristics and influencing factors of planktonic crustacean community in Dongping Lake under the operation of the South-to-North water diversion project. Oceanol. Limnol. Sin. 2025. in print. Available online: https://link.cnki.net/urlid/37.1149.P.20250922.0938.002 (accessed on 3 August 2025).
- Liang, Y.Y.; Fang, T.; Li, J.; Yang, K.; Zhao, X.X.; Cui, K. Age, growth and reproductive traits of invasive goby Taenioides cirratus in the Chaohu Lake, China. J. Appl. Ichthyol. 2020, 36, 219–226. [Google Scholar] [CrossRef]
- Raas, M.W.D.; Dutheil, J.Y. The rate of adaptive molecular evolution in wild and domesticated Saccharomyces cerevisiae populations. Mol. Ecol. 2024, 33, e16980. [Google Scholar] [CrossRef]
- Gossmann, T.I.; Keightley, P.D.; Eyre-Walker, A. The effect of variation in the effective population size on the rate of adaptive molecular evolution in eukaryotes. Genome Biol. Evol. 2012, 4, 658–667. [Google Scholar] [CrossRef]
- Angst, P.; Ebert, D.; Fields, P.D. Demographic history shapes genomic variation in an intracellular parasite with a wide geographical distribution. Mol. Ecol. 2022, 31, 2528–2544. [Google Scholar] [CrossRef]
- Ruan, S.; Lu, Z.; Huang, W.; Zhang, Y.; Shan, X.; Song, W.; Ji, C. Renal metabolomic profiling of large yellow croaker Larimichthys crocea acclimated in low salinity waters. Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 46, 101083. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, L.; Zhao, J.; Liu, M.; Wang, K.; Zhou, Q.; Cao, Y.; Hu, R.; Wang, W.; Liu, Q. Comprehensive multi-omics and biochemical analysis to elucidate the molecular response mechanisms of gill and kidney tissues under acute salinity stress in Pseudobagras ussuriensis. BMC Genom. 2025, 26, 590. [Google Scholar] [CrossRef] [PubMed]
- Džugasová, V.; Obernauerová, M.; Horváthová, K.; Vachová, M.; Žáková, M.; Šubík, J. Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Curr. Genet. 1998, 34, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Gorgoglione, R.; Porcelli, V.; Santoro, A.; Daddabbo, L.; Vozza, A.; Monné, M.; Di Noia, M.A.; Palmieri, L.; Fiermonte, G.; Palmieri, F. The human uncoupling proteins 5 and 6 (UCP5/SLC25A14 and UCP6/SLC25A30) transport sulfur oxyanions, phosphate and dicarboxylates. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1860, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, X.; Yu, M.; Yang, P.; Wang, W. G6PD, bond by miR-24, regulates mitochondrial dysfunction and oxidative stress in phenylephrine-induced hypertrophic cardiomyocytes. Life Sci. 2020, 260, 118378. [Google Scholar] [CrossRef]
- Fjeld, K.; Beer, S.; Johnstone, M.; Zimmer, C.; Mössner, J.; Ruffert, C.; Krehan, M.; Zapf, C.; Njølstad, P.R.; Johansson, S. Length of variable numbers of tandem repeats in the carboxyl ester lipase (CEL) gene may confer susceptibility to alcoholic liver cirrhosis but not alcoholic chronic pancreatitis. PLoS ONE 2016, 11, e0165567. [Google Scholar] [CrossRef]
- Sohn, J.H.; Ji, Y.; Cho, C.-Y.; Nahmgoong, H.; Lim, S.; Jeon, Y.G.; Han, S.M.; Han, J.S.; Park, I.; Rhee, H.-W. Spatial regulation of reactive oxygen species via G6PD in brown adipocytes supports thermogenic function. Diabetes 2021, 70, 2756–2770. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Wang, Z.; Peng, Y. A comparative transcriptomics approach to analyzing the differences in cold resistance in Pomacea canaliculata between Guangdong and Hunan. J. Immunol. Res. 2020, 2020, 8025140. [Google Scholar] [CrossRef]
- Ciardiello, M.A.; Camardella, L.; di Prisco, G. Glucose-6-phosphate dehydrogenase from the blood cells of two Antarctic teleosts: Correlation with cold adaptation. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1995, 1250, 76–82. [Google Scholar] [CrossRef]
- Ciardiello, M.A.; Camardella, L.; Carratore, V.; di Prisco, G. Enzymes in Antarctic fish: Glucose-6-phosphate dehydrogenase and glutamate dehydrogenase. Comp. Biochem. Physiol. Part A Physiol. 1997, 118, 1031–1036. [Google Scholar] [CrossRef]
- Liang, L.; Chang, Y.; He, X.; Tang, R. Transcriptome analysis to identify cold-responsive genes in amur carp (Cyprinus carpio haematopterus). PLoS ONE 2015, 10, e0130526. [Google Scholar] [CrossRef]
- Udayantha, H.; Lee, S.; Liyanage, D.; Lim, C.; Jeong, T.; Omeka, W.; Yang, H.; Kim, G.; Kim, J.; Lee, J. Identification of candidate variants and genes associated with temperature tolerance in olive flounders by Genome-Wide Association Study (GWAS). Aquaculture 2023, 576, 739858. [Google Scholar] [CrossRef]
- Lü, Z.; Liu, T.; Liu, Y.; Wang, Y.; Liu, J.; Liu, B.; Gong, L.; Liu, L. Climate adaptation and drift shape the genomes of two eel-goby sister species endemic to contrasting latitude. Animals 2023, 13, 3240. [Google Scholar] [CrossRef]
- Cho, S.-W.; Choi, K.Y.; Park, C.-S. A new putative cyclic nucleotide-gated channel gene, cng-3, is critical for thermotolerance in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2004, 325, 525–531. [Google Scholar] [CrossRef]
- Okahata, M.; Wei, A.D.; Ohta, A.; Kuhara, A. Cold acclimation via the KQT-2 potassium channel is modulated by oxygen in Caenorhabditis elegans. Sci. Adv. 2019, 5, eaav3631. [Google Scholar] [CrossRef]
- Himmel, N.J.; Letcher, J.M.; Sakurai, A.; Gray, T.R.; Benson, M.N.; Donaldson, K.J.; Cox, D.N. Identification of a neural basis for cold acclimation in Drosophila larvae. Iscience 2021, 24, 102657. [Google Scholar] [CrossRef]
- Ling, J.; Erol, F.; Gu, J.G.G. Role of KCNQ2 channels in orofacial cold sensitivity: KCNQ2 upregulation in trigeminal ganglion neurons after infraorbital nerve chronic constrictive injury. Neurosci. Lett. 2018, 664, 84–90. [Google Scholar] [CrossRef]
- Buffone, M.G.; Zhuang, T.; Ord, T.S.; Hui, L.; Moss, S.B.; Gerton, G.L. Recombinant mouse sperm ZP3-binding protein (ZP3R/sp56) forms a high order oligomer that binds eggs and inhibits mouse fertilization in vitro. J. Biol. Chem. 2008, 283, 12438–12445. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Huang, Q.; Wu, Z.; Cao, D.-d.; Ma, Z.; Xu, Q.; Hu, P.; Fu, Y.; Shen, Y.; Chan, J. Neofunctionalization of zona pellucida proteins enhances freeze-prevention in the eggs of Antarctic notothenioids. Nat. Commun. 2016, 7, 12987. [Google Scholar] [CrossRef] [PubMed]
- Spies, I.; Drinan, D.P.; Petrou, E.L.; Spurr, R.; Tarpey, C.; Hartinger, T.; Larson, W.; Hauser, L. Evidence for selection and spatially distinct patterns found in a putative zona pellucida gene in Pacific cod, and implications for management. Ecol. Evol. 2021, 11, 16661–16679. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Wilson, G.A.; Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef]
- Murga-Moreno, J.; Coronado-Zamora, M.; Hervas, S.; Casillas, S.; Barbadilla, A. iMKT: The integrative McDonald and Kreitman test. Nucleic Acids Res. 2019, 47, W283–W288. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Li, M.; Yang, Y.; Li, Z.; Xu, Y.; Wang, H.; Wang, D.; Zhang, Y.; Wang, H. Molecular mapping for fruit-related traits, and joint identification of candidate genes and selective sweeps for seed size in melon. Genomics 2022, 114, 110306. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Faske, T.M.; Agneray, A.C.; Jahner, J.P.; Sheta, L.M.; Leger, E.A.; Parchman, T.L. Genomic and common garden approaches yield complementary results for quantifying environmental drivers of local adaptation in rubber rabbitbrush, a foundational Great Basin shrub. Evol. Appl. 2021, 14, 2881–2900. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
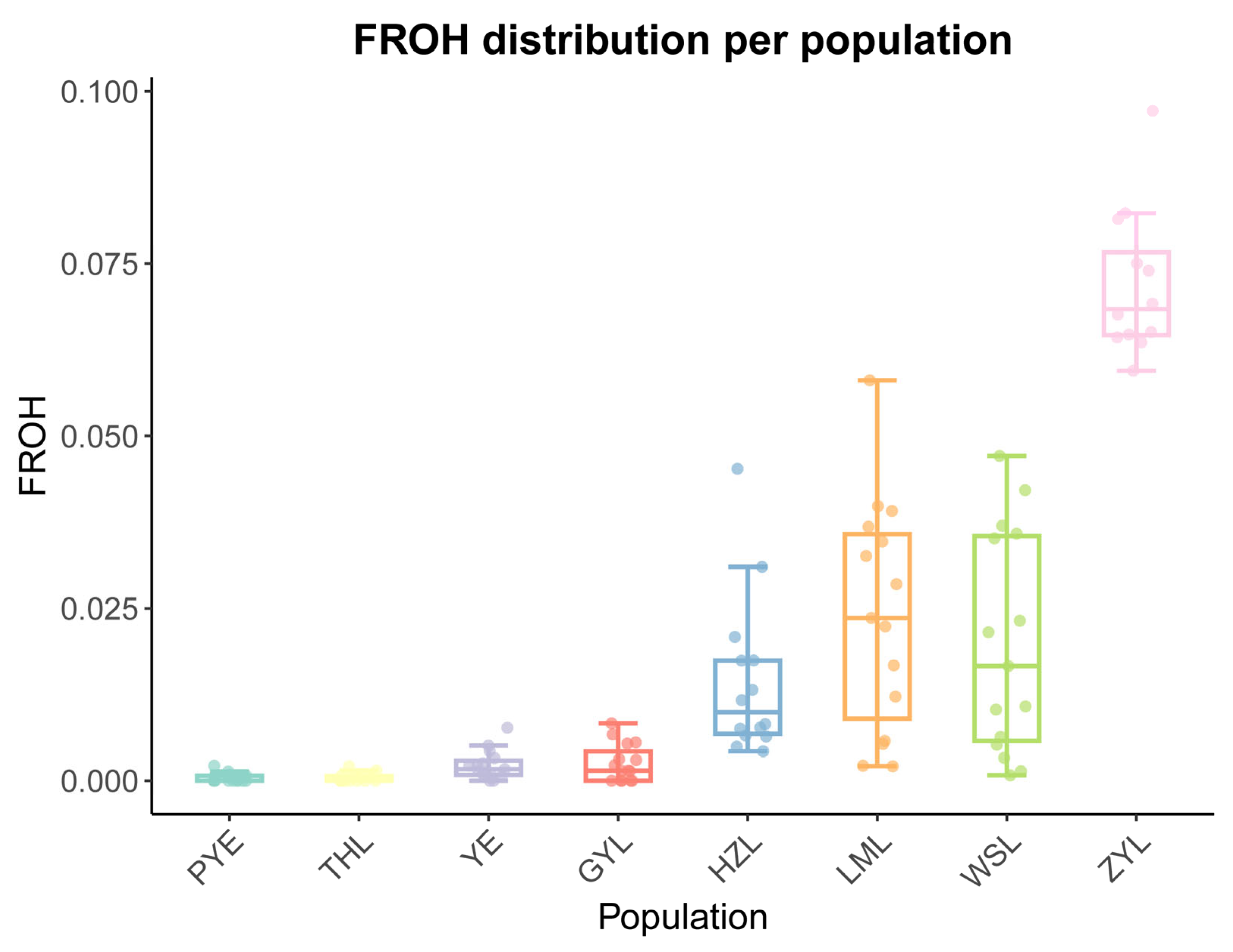
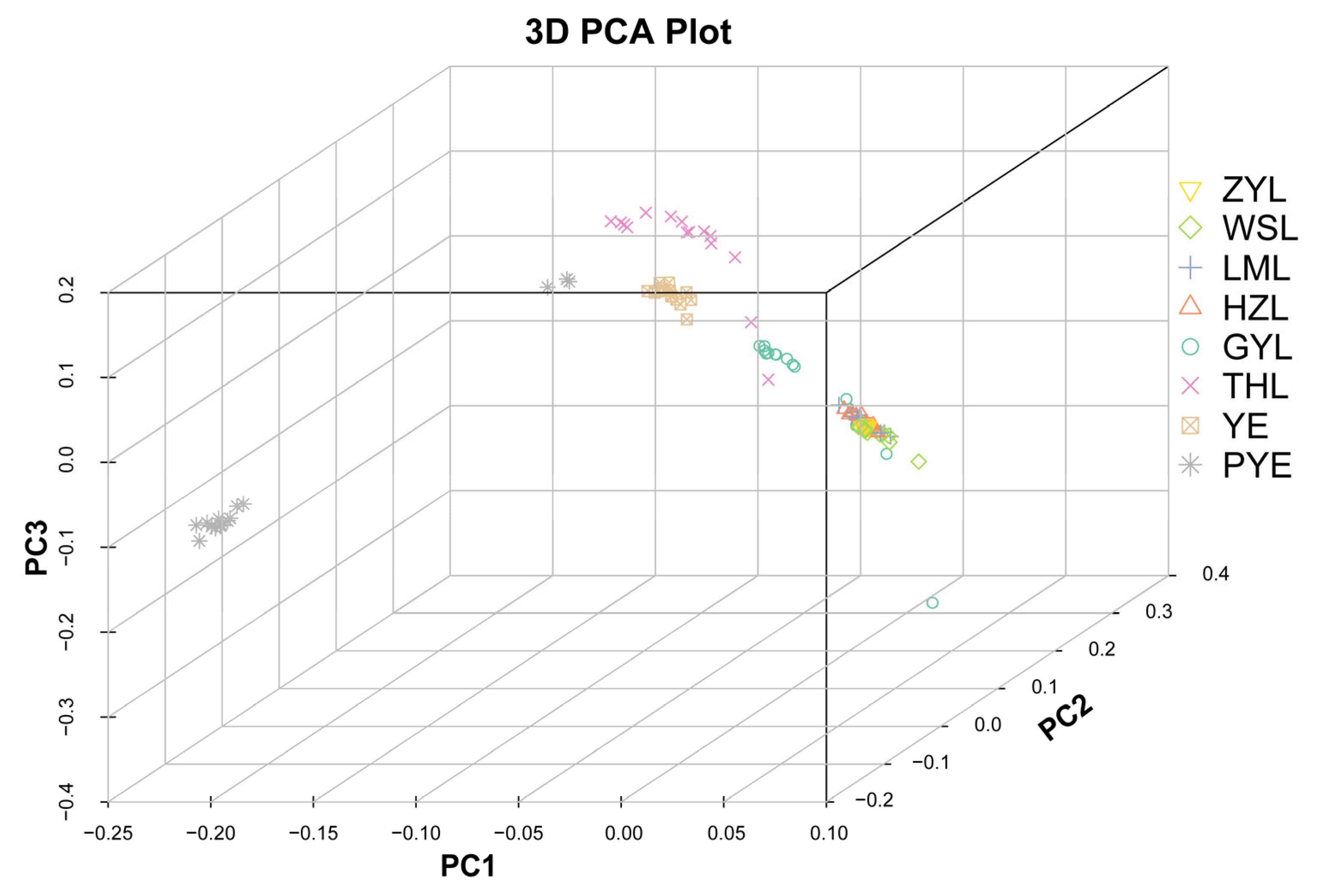
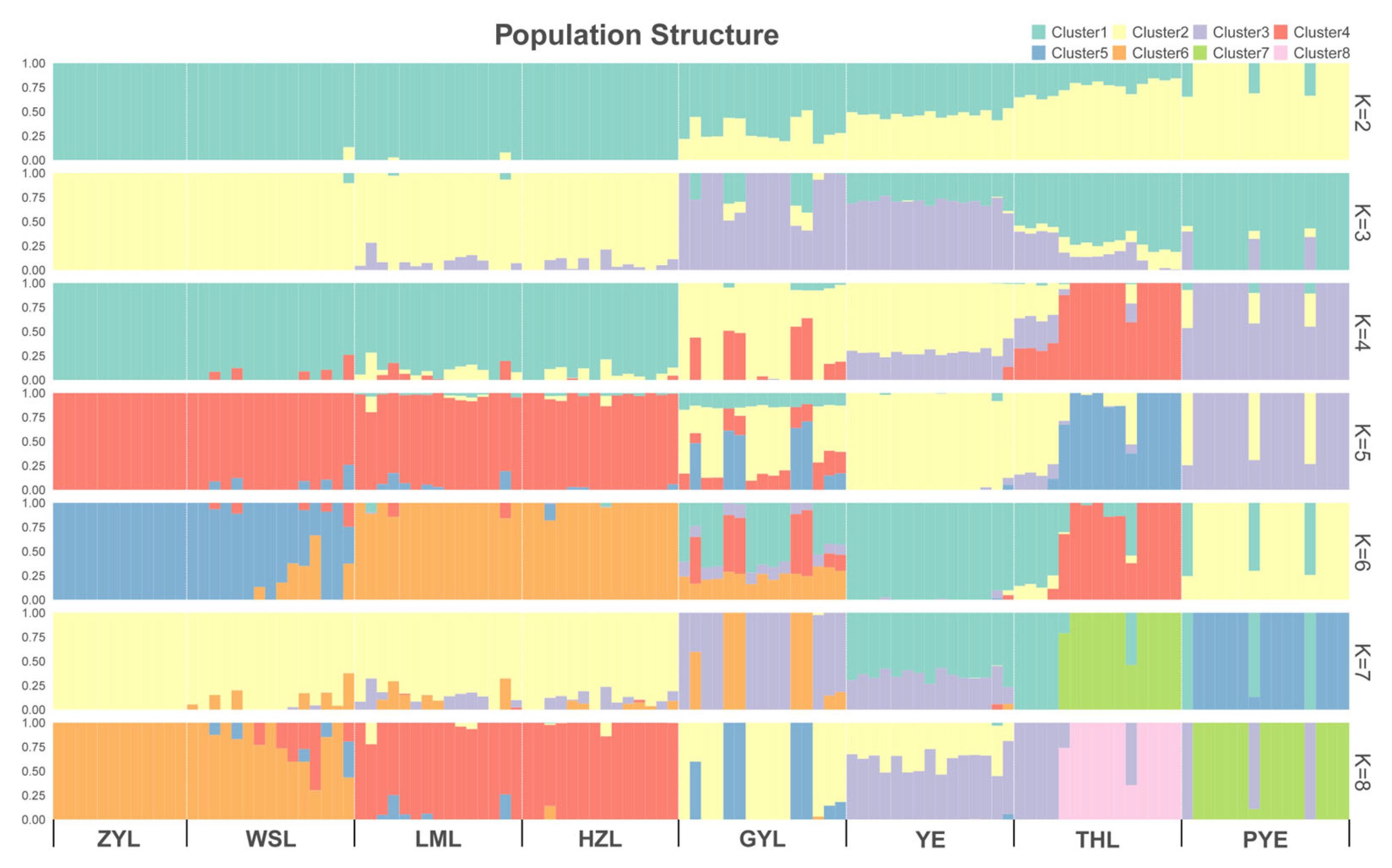
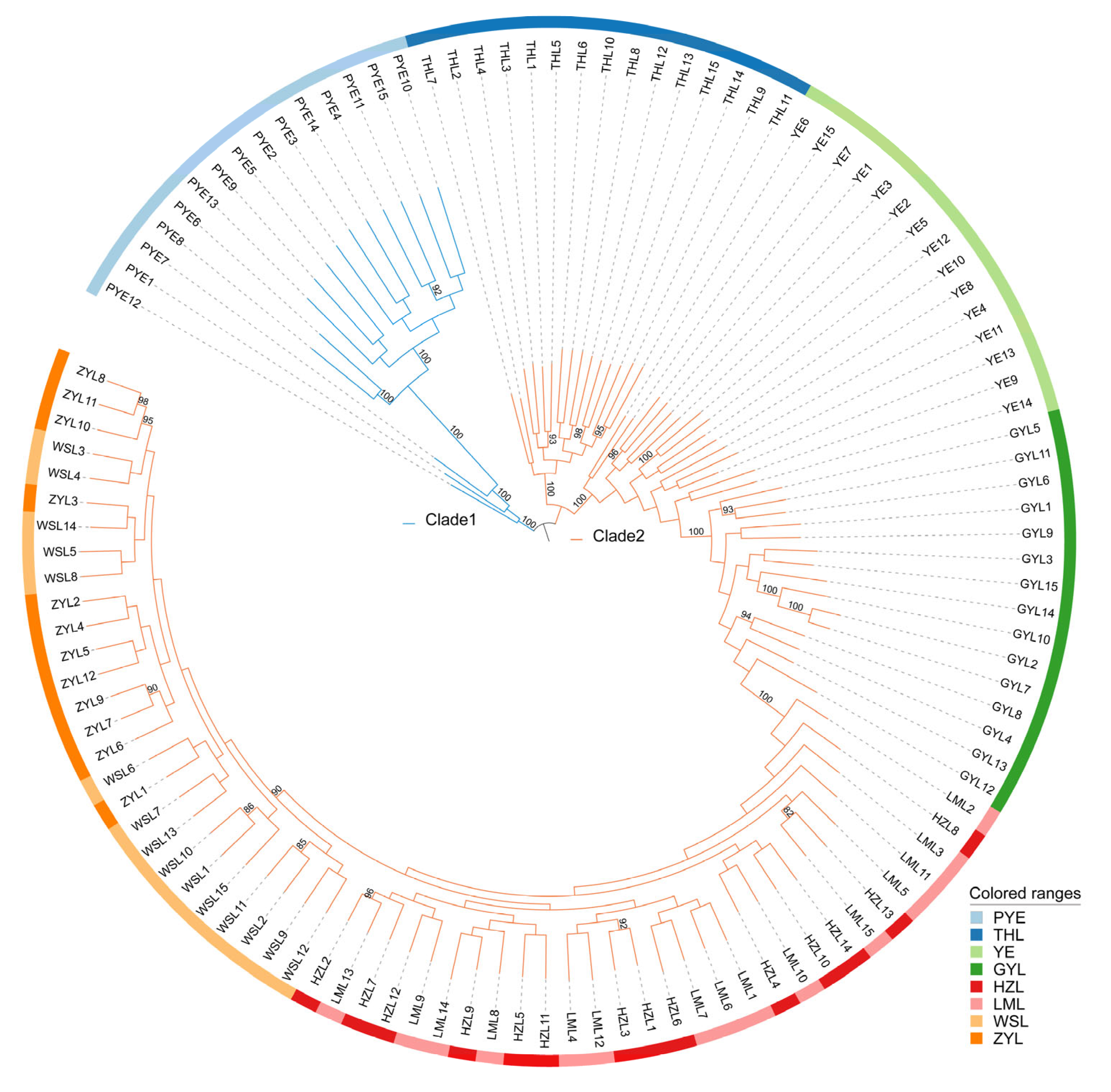

| Population | Sample Size | Ho | He | Pi | Polymorphic Loci (%) |
|---|---|---|---|---|---|
| ZYL | 12 | 0.129 | 0.122 | 0.128 | 44.493 |
| WSL | 15 | 0.132 | 0.128 | 0.133 | 50.406 |
| LML | 15 | 0.133 | 0.130 | 0.134 | 54.124 |
| HZL | 14 | 0.131 | 0.126 | 0.131 | 51.655 |
| GYL | 15 | 0.181 | 0.174 | 0.180 | 64.380 |
| THL | 15 | 0.217 | 0.212 | 0.220 | 79.435 |
| YE | 15 | 0.214 | 0.204 | 0.212 | 77.162 |
| PYE | 15 | 0.277 | 0.278 | 0.288 | 89.498 |
| Population | ZYL | WSL | LML | HZL | GYL | THL | YE | PYE |
|---|---|---|---|---|---|---|---|---|
| ZYL | ||||||||
| WSL | 0.021 | |||||||
| LML | 0.029 | 0.024 | ||||||
| HZL | 0.030 | 0.025 | 0.018 | |||||
| GYL | 0.056 | 0.053 | 0.047 | 0.049 | ||||
| THL | 0.075 | 0.074 | 0.070 | 0.072 | 0.054 | |||
| YE | 0.057 | 0.056 | 0.052 | 0.053 | 0.031 | 0.035 | ||
| PYE | 0.130 | 0.097 | 0.094 | 0.095 | 0.074 | 0.055 | 0.052 |
| Population | α-Value | Fisher p-Value |
|---|---|---|
| ZYL | 0.135 | 3.17 × 10−21 |
| WSL | 0.126 | 2.14 × 10−20 |
| LML | 0.136 | 6.68 × 10−25 |
| HZL | 0.123 | 1.36 × 10−19 |
| GYL | 0.101 | 1.43 × 10−16 |
| THL | 0.099 | 4.56 × 10−19 |
| YE | 0.120 | 8.54 × 10−28 |
| PYE | 0.149 | 1.23 × 10−52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, K.; Liu, T.; Xu, A.; Yu, J.; Yang, Y.; Liu, J.; Li, F.; Zhu, D.; Gong, L.; Liu, L.; et al. Genomic Signatures of Adaptive Evolution in Taenioides sp. During Northward Invasion. Int. J. Mol. Sci. 2025, 26, 9613. https://doi.org/10.3390/ijms26199613
Huang K, Liu T, Xu A, Yu J, Yang Y, Liu J, Li F, Zhu D, Gong L, Liu L, et al. Genomic Signatures of Adaptive Evolution in Taenioides sp. During Northward Invasion. International Journal of Molecular Sciences. 2025; 26(19):9613. https://doi.org/10.3390/ijms26199613
Chicago/Turabian StyleHuang, Kun, Tianwei Liu, An Xu, Jing Yu, Yijing Yang, Jing Liu, Fenghui Li, Denghui Zhu, Li Gong, Liqin Liu, and et al. 2025. "Genomic Signatures of Adaptive Evolution in Taenioides sp. During Northward Invasion" International Journal of Molecular Sciences 26, no. 19: 9613. https://doi.org/10.3390/ijms26199613
APA StyleHuang, K., Liu, T., Xu, A., Yu, J., Yang, Y., Liu, J., Li, F., Zhu, D., Gong, L., Liu, L., & Lü, Z. (2025). Genomic Signatures of Adaptive Evolution in Taenioides sp. During Northward Invasion. International Journal of Molecular Sciences, 26(19), 9613. https://doi.org/10.3390/ijms26199613





