Vincristine Beyond Mitosis: Uncovering a First Link to G-Quadruplex DNA in Cancer Cells
Abstract
1. Introduction
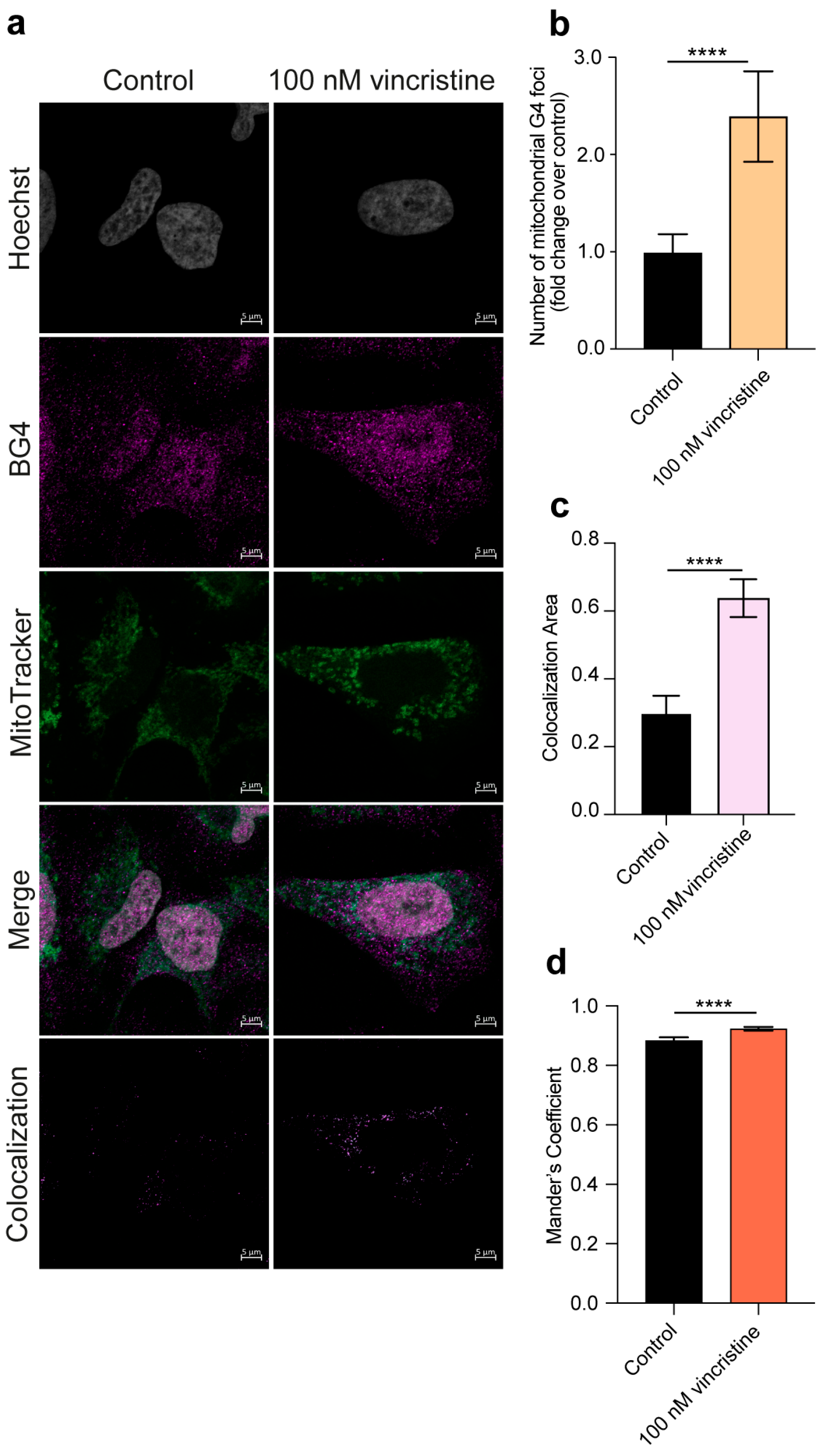
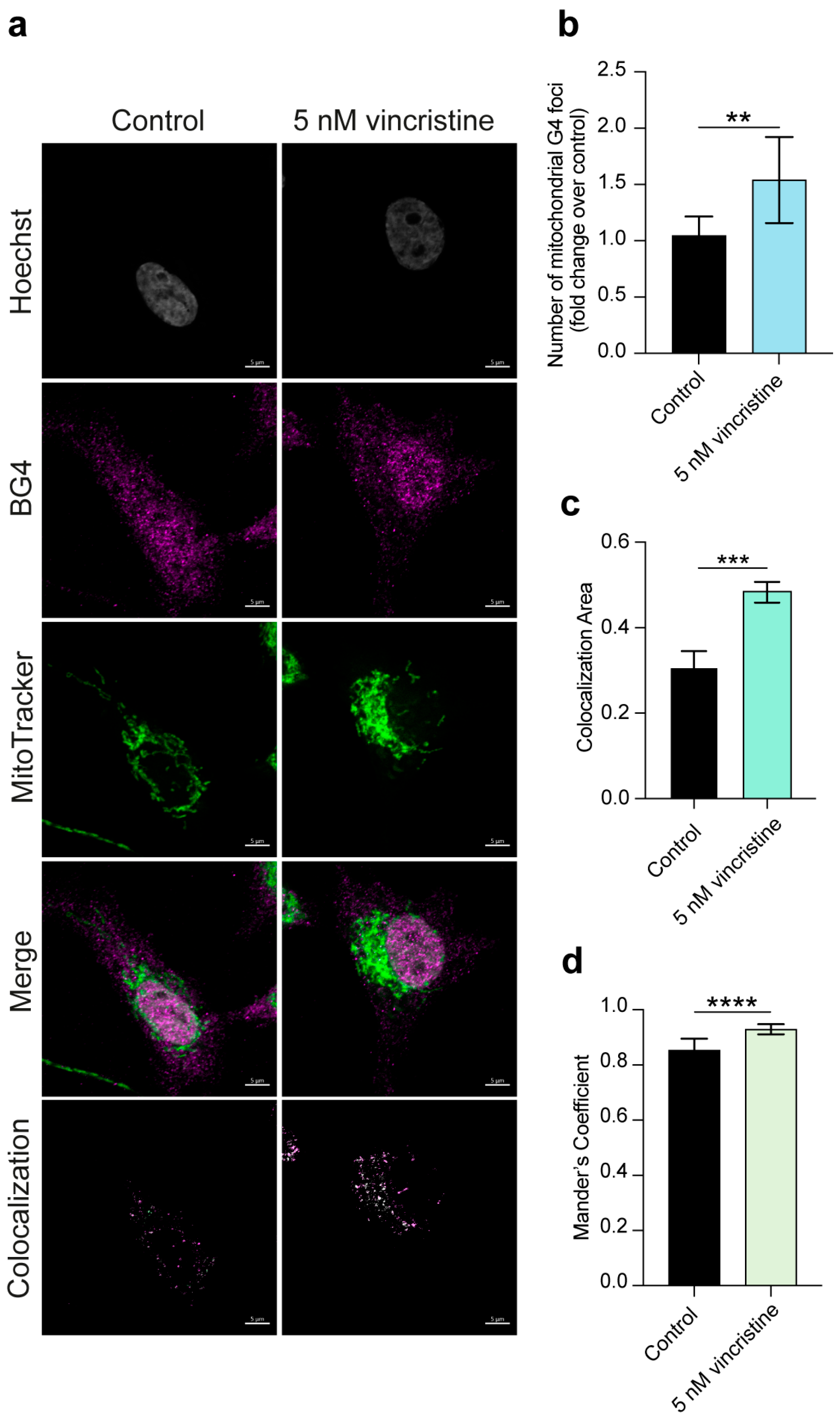
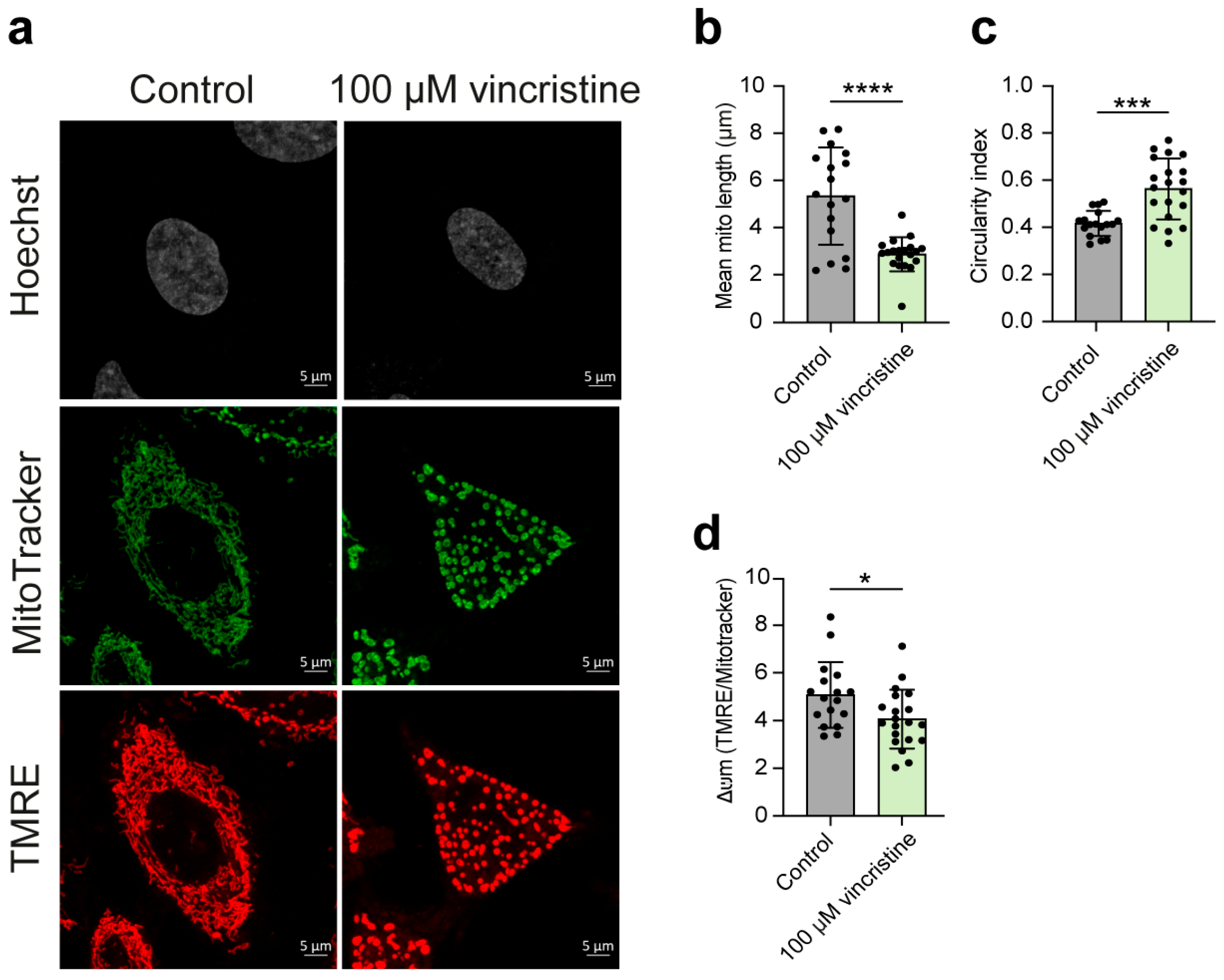
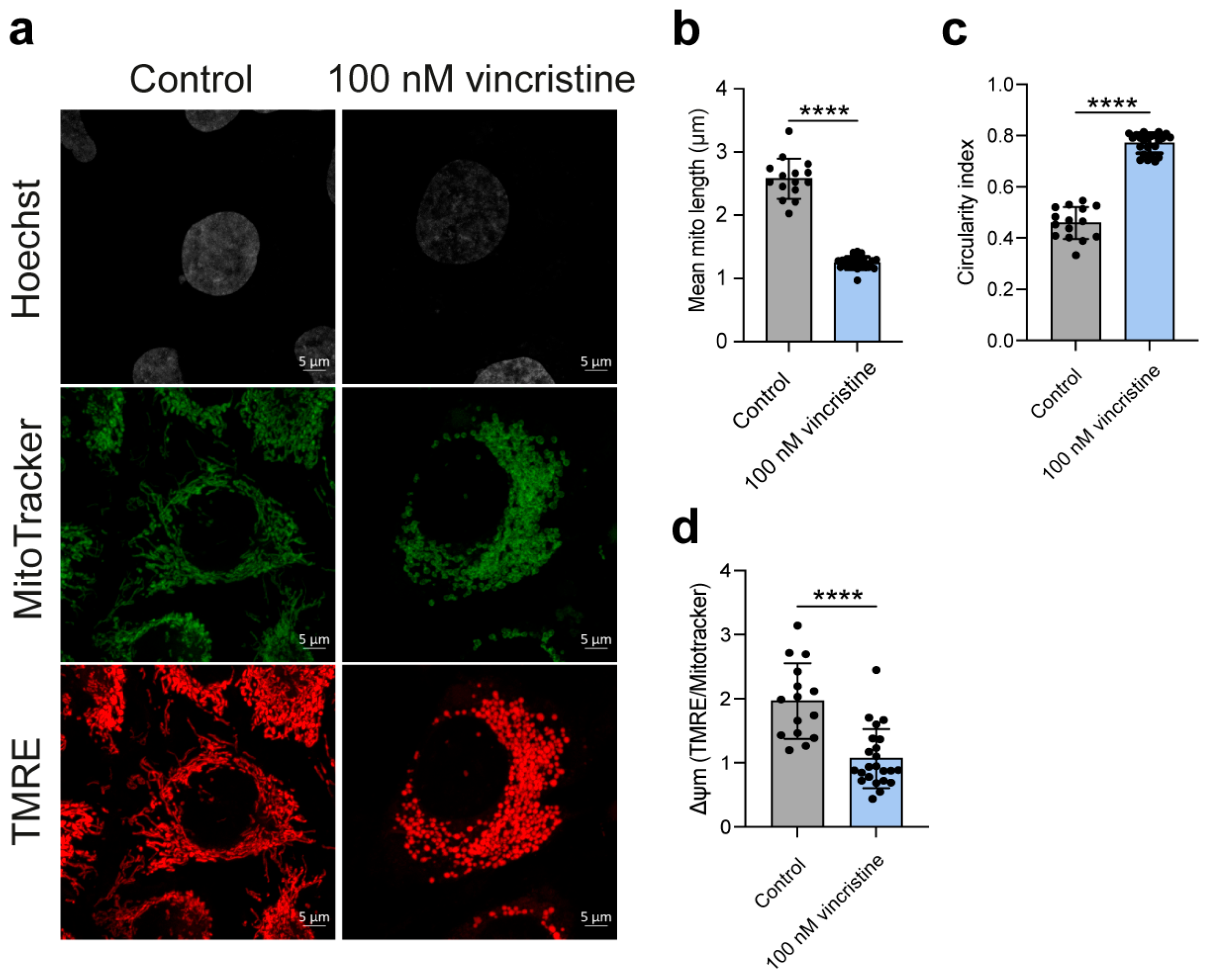
2. Results and Discussion
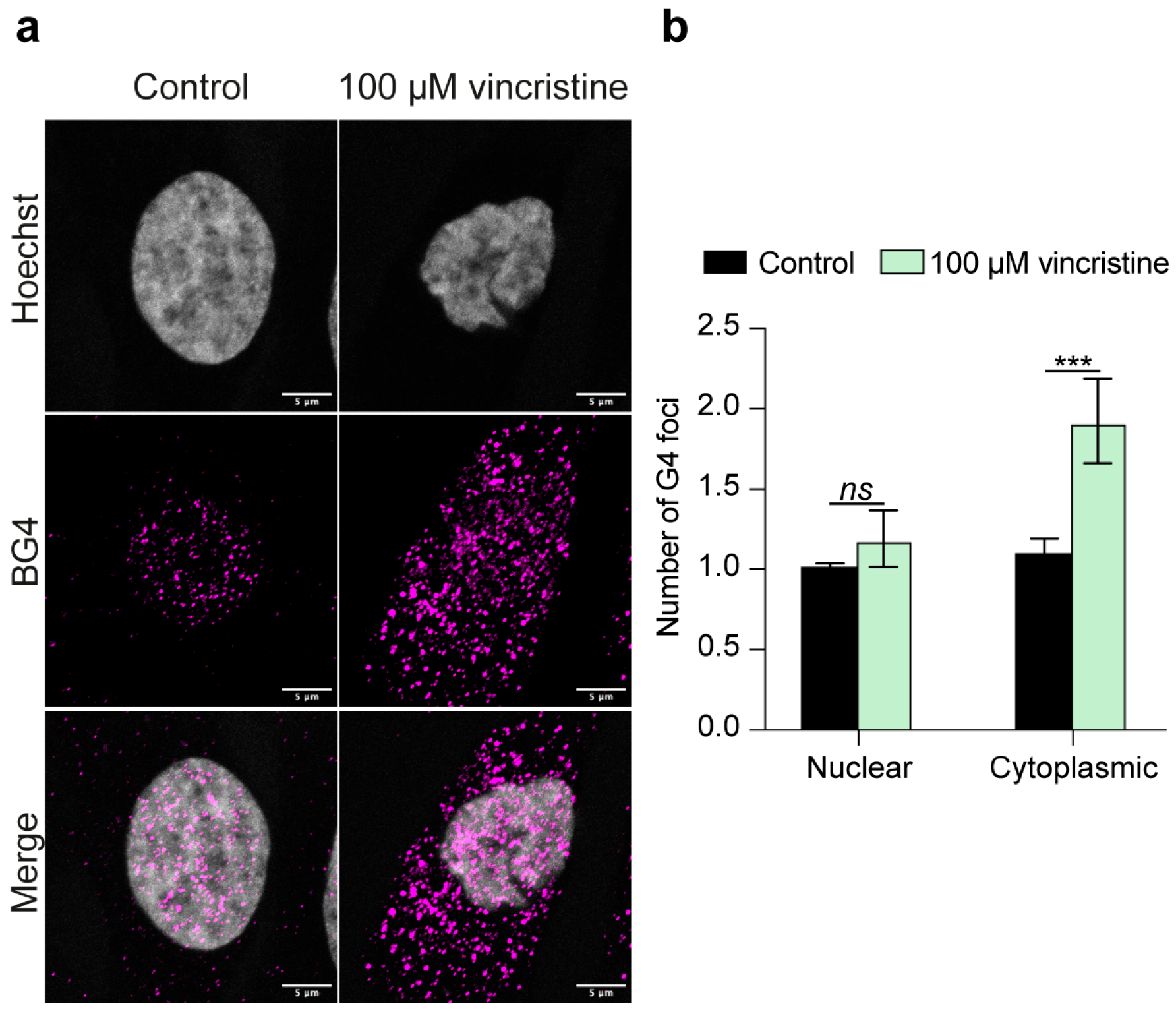
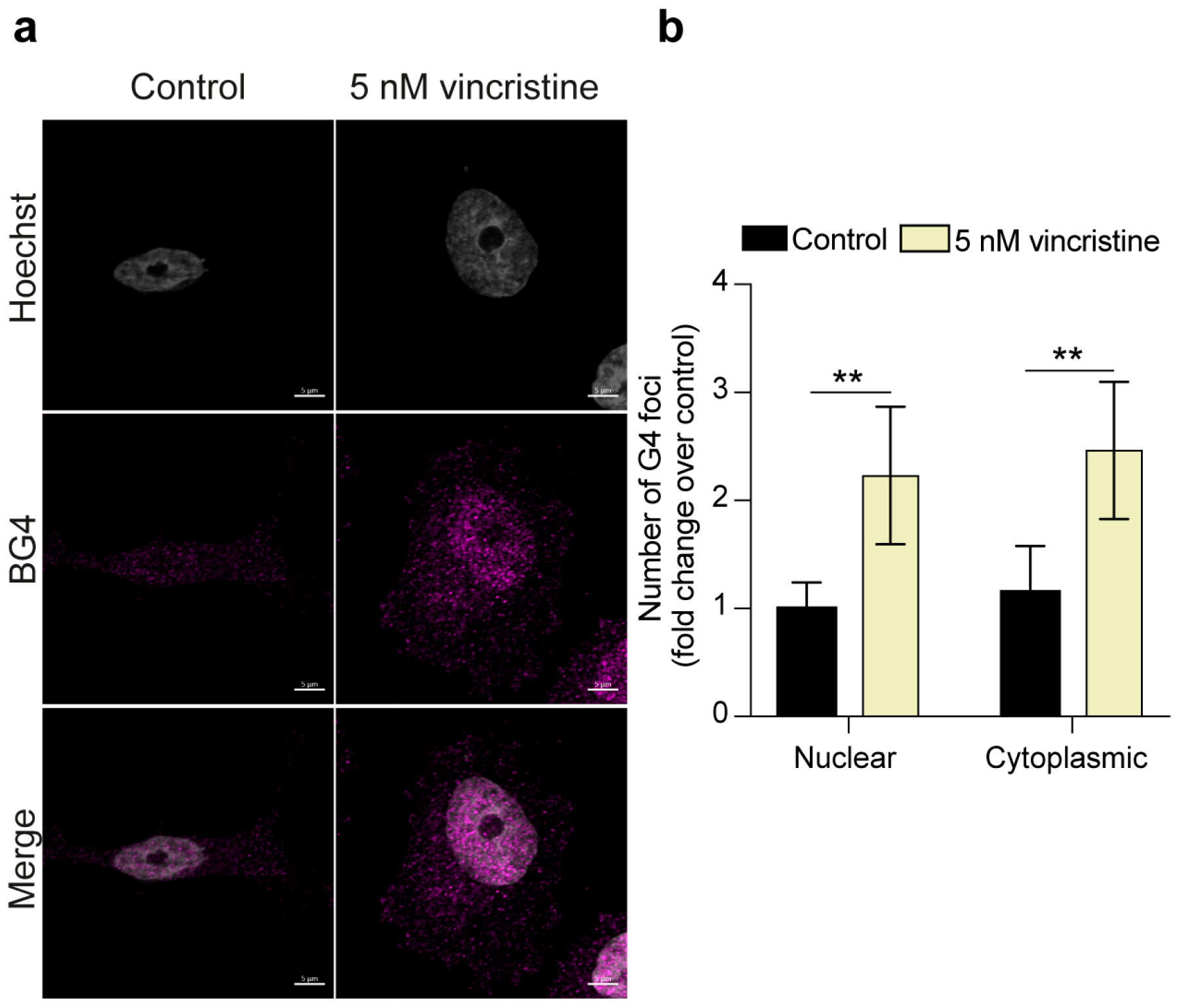
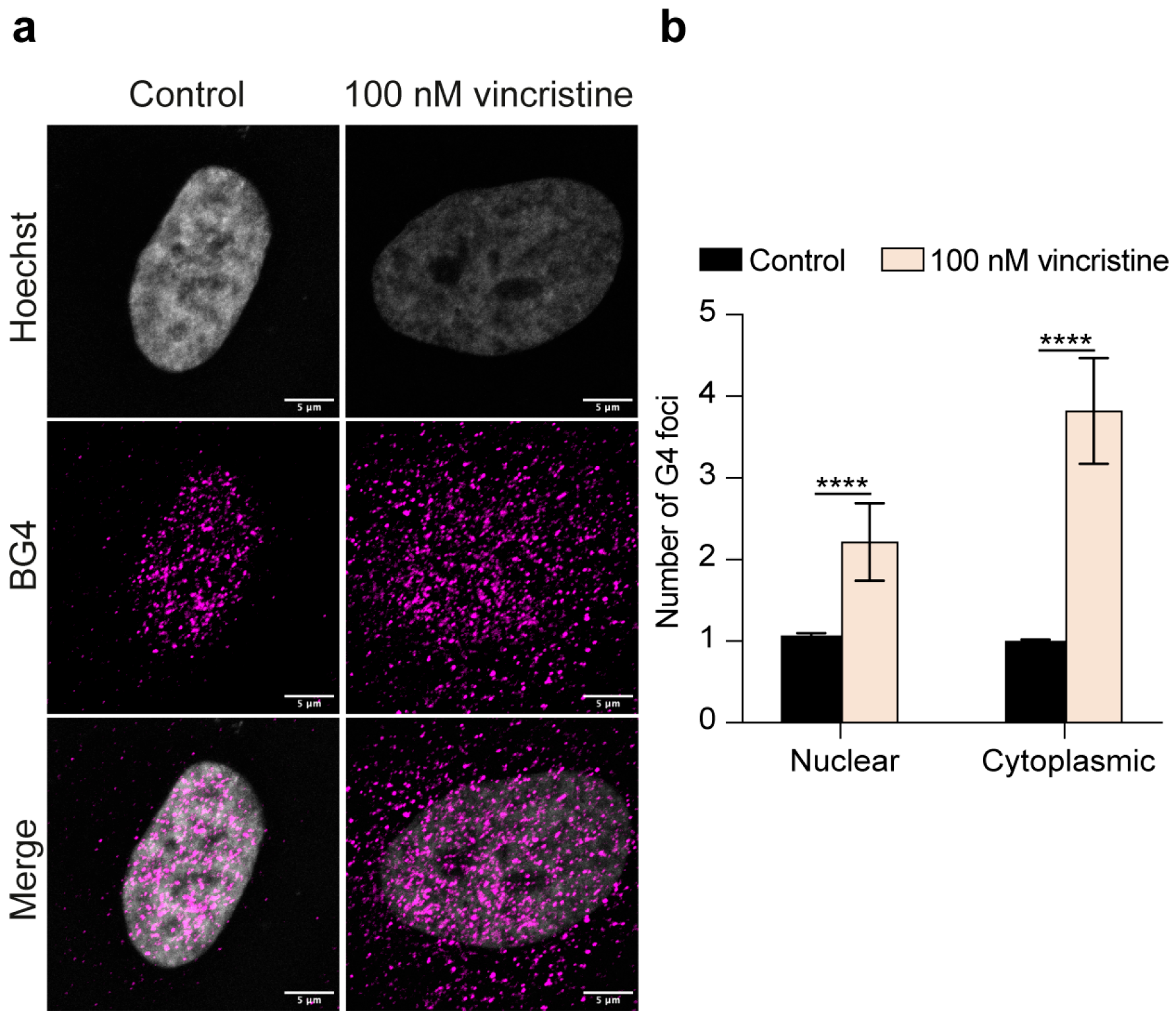
2.1. Immunofluorescence
2.2. Analysis of Mitochondrial Structure and Function
2.3. Nuclear Magnetic Resonance
2.4. Circular Dichroism Experiments
2.5. Isothermal Titration Calorimetry
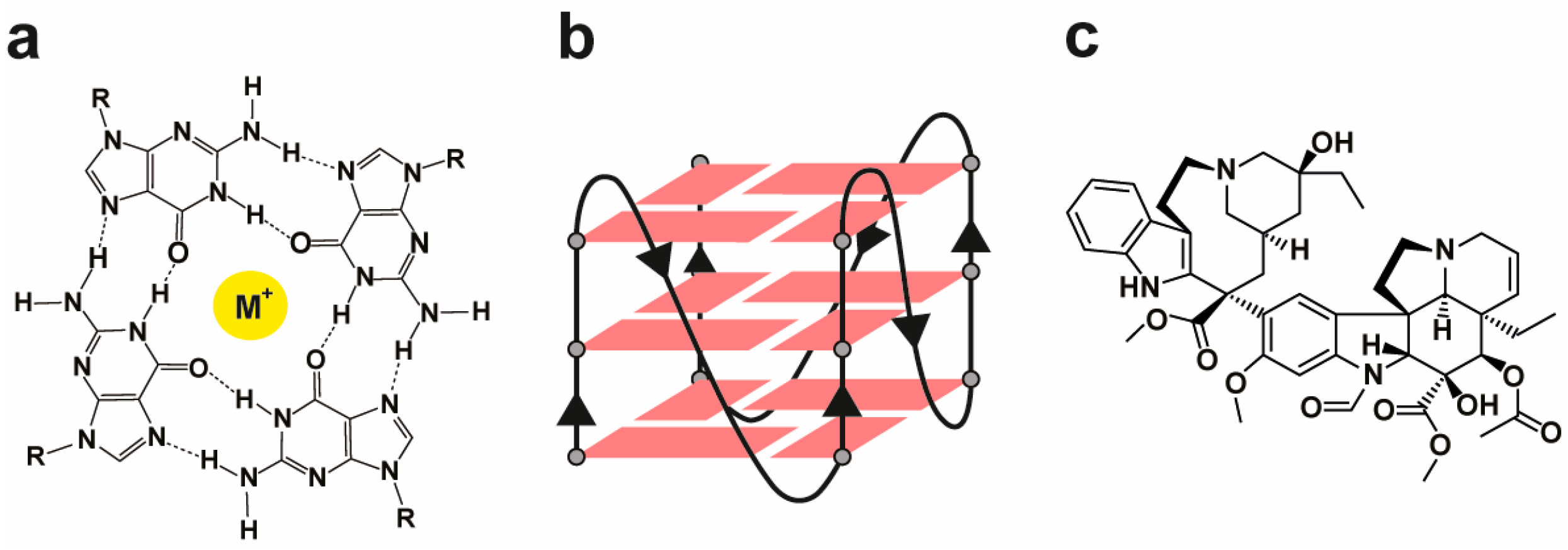
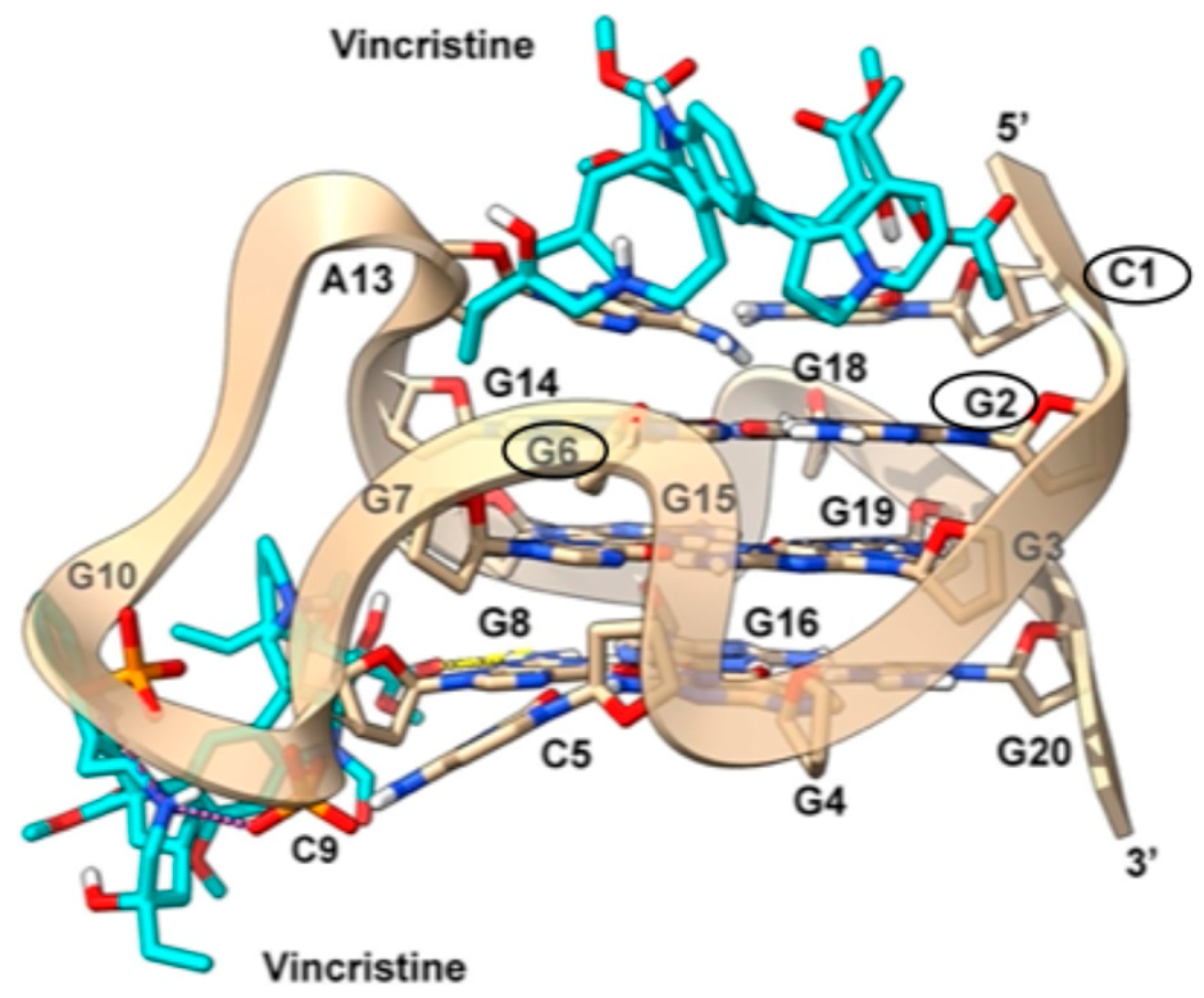
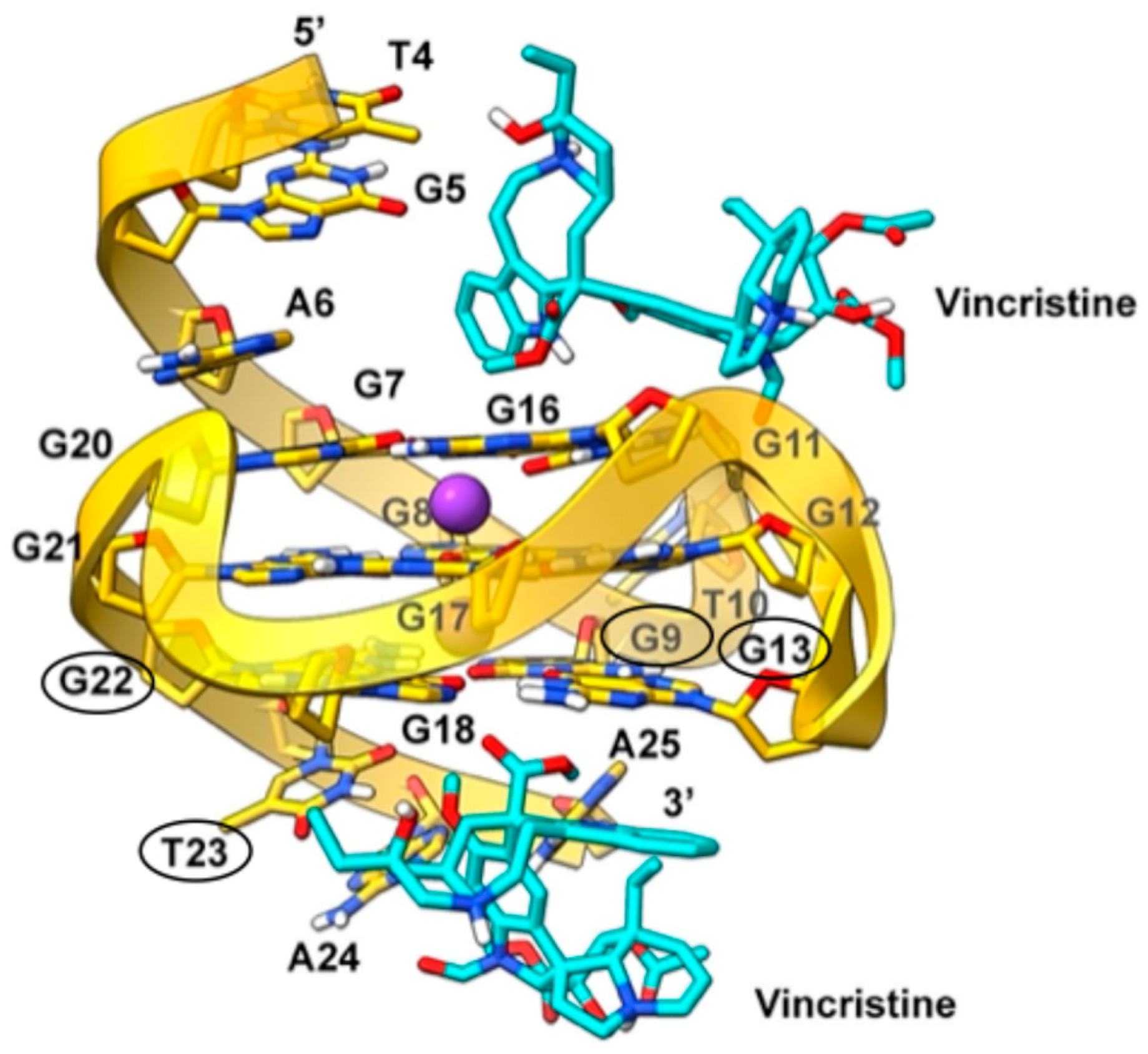
2.6. Molecular Modeling
2.6.1. c-KIT2 Gene Promoter
2.6.2. c-MYC Gene Promoter
3. Materials and Methods
3.1. Materials
3.2. Cell Culture
3.3. Immunofluorescence Assays
3.3.1. BG4 Immunofluorescence
3.3.2. MitoTracker Staining
3.4. Analysis of Mitochondrial Structure and Function
3.5. Oligonucleotide Preparation
3.6. Nuclear Magnetic Resonance Experiments
3.7. Circular Dichroism Spectroscopy
3.8. Isothermal Titration Calorimetry
3.9. Molecular Modeling
3.9.1. DNA Structure Selection and Preparation
3.9.2. Ligand Preparation
3.9.3. Molecular Docking Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BG4 | G-quadruplex-specific antibody |
| BSA | Bovine serum albumin |
| CD | Circular dichroism |
| c-KIT2 | Gene encoding the receptor tyrosine kinase protein |
| c-MYC | Cellular MYC proto-oncogene |
| DMEM | Dulbecco’s modified Eagle medium |
| DMSO | Dimethyl sulfoxide |
| dsDNA | Double-stranded DNA |
| FBS | Fetal bovine serum |
| G4 | G-quadruplex |
| HeLa | Human cervical cancer cell Line |
| ITC | Isothermal titration calorimetry |
| mtDNA | Mitochondrial DNA |
| NMR | Nuclear magnetic resonance |
| PBS | Phosphate-buffered saline |
| PDB | Protein data bank |
| rG4 | RNA G-quadruplex |
| RNase A | Ribonuclease A |
| TMRE | Tetramethylrhodamine Ethyl Ester |
| UTR | Untranslated region |
| U2OS | Human osteosarcoma cell line |
References
- MacDonald, W.J.; Purcell, C.; Pinho-Schwermann, M.; Stubbs, N.M.; Srinivasan, P.R.; El-Deiry, W.S. Heterogeneity in Cancer. Cancers 2025, 17, 441. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 19 May 2025).
- Kiri, S.; Ryba, T. Cancer, Metastasis, and the Epigenome. Mol. Cancer 2024, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Arciuolo, V.; Marzano, S.; Buono, R.; Grasso, N.; Di Porzio, A.; Randazzo, A.; Pagano, B.; Amato, J. Screening of G-Quadruplex DNA Ligands by Fluorescence Detection of Peptide Displacement. Eur. J. Med. Chem. Rep. 2025, 15, 100293. [Google Scholar] [CrossRef]
- Romano, F.; Di Porzio, A.; Iaccarino, N.; Riccardi, G.; Di Lorenzo, R.; Laneri, S.; Pagano, B.; Amato, J.; Randazzo, A. G-Quadruplexes in Cancer-Related Gene Promoters: From Identification to Therapeutic Targeting. Expert. Opin. Ther. Pat. 2023, 33, 745–773. [Google Scholar] [CrossRef]
- Moraca, F.; Marzano, S.; D’Amico, F.; Lupia, A.; Di Fonzo, S.; Vertecchi, E.; Salvati, E.; Di Porzio, A.; Catalanotti, B.; Randazzo, A.; et al. Ligand-Based Drug Repurposing Strategy Identified SARS-CoV-2 RNA G-Quadruplex Binders. Chem. Commun. 2022, 58, 11913–11916. [Google Scholar] [CrossRef]
- Marzano, S.; Pagano, B.; Iaccarino, N.; Di Porzio, A.; De Tito, S.; Vertecchi, E.; Salvati, E.; Randazzo, A.; Amato, J. Targeting of Telomeric Repeat-Containing RNA g-Quadruplexes: From Screening to Biophysical and Biological Characterization of a New Hit Compound. Int. J. Mol. Sci. 2021, 22, 10315. [Google Scholar] [CrossRef]
- Maiocchi, A.; Pedrini, M.; Ferrari, V.; Carreira, A.S.A.; D’Amore, V.M.; Santoro, F.; Di Porzio, A.; Bosetti, M.; Cristofani, R.; Silvani, A.; et al. Design, Synthesis and Characterization of Aryl Bis-Guanyl Hydrazones as RNA Binders of C9orf72 G4C2 Extended Repeats. Eur. J. Med. Chem. 2025, 293, 117736. [Google Scholar] [CrossRef]
- Falabella, M.; Fernandez, R.J.; Johnson, F.B.; Kaufman, B.A. Potential Roles for G-Quadruplexes in Mitochondria. Curr. Med. Chem. 2019, 26, 2918–2932. [Google Scholar] [CrossRef]
- Romano, F.; Persico, C.; Barra, A.; Pinto, G.; Illiano, A.; Amoresano, A.; Aiello, I.; Abate, S.; D’Auria, L.; Martello, V.; et al. Unveiling the Biological Effects of DNA G-Quadruplex Ligands Through Multi-Omics Data Integration. Int. J. Biol. Macromol. 2025, 313, 144325. [Google Scholar] [CrossRef]
- Piccolo, M.; Russo, C.; Arciuolo, V.; Ferraro, M.G.; Abbate, V.; Di Porzio, A.; Cinquegrana, E.; Di Leva, F.S.; Pagano, B.; Randazzo, A.; et al. Design, Synthesis, and Anticancer Activity of Drug-like Iron Chelators/G-Quadruplex Binders as Synergic Dual Targeting Agents. J. Med. Chem. 2025, 68, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Persico, C.; Iaccarino, N.; Romano, F.; Giustiniano, M.; Russo, C.; Laneri, S.; Di Lorenzo, R.; Aiello, I.; Abate, S.; Izzo, L.; et al. Sensitization of Melanoma Cells to Standard Chemotherapy: G-Quadruplex Binders as Synergistic Agents. NAR Cancer 2024, 6, zcae042. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.; Mergny, J.L.; Cruz, C. G-Quadruplex Ligands in Cancer Therapy: Progress, Challenges, and Clinical Perspectives. Life Sci. 2024, 340, 122481. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Wilson, L. Microtubules as a Target for Anticancer Drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Sertel, S.; Fu, Y.; Zu, Y.; Rebacz, B.; Konkimalla, B.; Plinkert, P.K.; Krämer, A.; Gertsch, J.; Efferth, T. Molecular Docking and Pharmacogenomics of Vinca Alkaloids and Their Monomeric Precursors, Vindoline and Catharanthine. Biochem. Pharmacol. 2011, 81, 723–735. [Google Scholar] [CrossRef]
- Gidding, C. Vincristine Revisited. Crit. Rev. Oncol. Hematol. 1999, 29, 267–287. [Google Scholar] [CrossRef]
- Mohammadgholi, A.; Rabbani-Chadegani, A.; Fallah, S. Mechanism of the Interaction of Plant Alkaloid Vincristine with DNA and Chromatin: Spectroscopic Study. DNA Cell Biol. 2013, 32, 228–235. [Google Scholar] [CrossRef]
- Hui, W.W.I.; Simeone, A.; Zyner, K.G.; Tannahill, D.; Balasubramanian, S. Single-Cell Mapping of DNA G-Quadruplex Structures in Human Cancer Cells. Sci. Rep. 2021, 11, 23641, Correction in: Sci. Rep. 2022, 12, 908. [Google Scholar] [CrossRef]
- Wang, J.-X.; Wang, X.-D.; Hu, M.-H. Novel Quinoxaline Analogs as Telomeric G-Quadruplex Ligands Exert Antitumor Effects Related to Enhanced Immunomodulation. Eur. J. Med. Chem. 2024, 274, 116536. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative Visualization of DNA G-Quadruplex Structures in Human Cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Doimo, M.; Chaudhari, N.; Abrahamsson, S.; L’Hôte, V.; Nguyen, T.V.H.; Berner, A.; Ndi, M.; Abrahamsson, A.; Das, R.N.; Aasumets, K.; et al. Enhanced Mitochondrial G-Quadruplex Formation Impedes Replication Fork Progression Leading to MtDNA Loss in Human Cells. Nucleic Acids Res. 2023, 51, 7392–7408. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, G.; Parisi, C.; Sodano, F.; Catanzano, O.; Di Porzio, A.; Randazzo, A.; Sortino, S.; Quaglia, F. Light-Activatable Hyaluronic Acid-Derivatives Releasing Nitric Oxide and Their Delivery in the Skin. Adv. Heal. Mater. 2025, 14, e2500589. [Google Scholar] [CrossRef] [PubMed]
- Falabella, M.; Kolesar, J.E.; Wallace, C.; de Jesus, D.; Sun, L.; Taguchi, Y.V.; Wang, C.; Wang, T.; Xiang, I.M.; Alder, J.K.; et al. G-Quadruplex Dynamics Contribute to Regulation of Mitochondrial Gene Expression. Sci. Rep. 2019, 9, 5605. [Google Scholar] [CrossRef] [PubMed]
- Menale, C.; Trinchese, G.; Aiello, I.; Scalia, G.; Dentice, M.; Mollica, M.P.; Yoon, N.A.; Diano, S. Nutrient-Dependent Mitochondrial Fission Enhances Osteoblast Function. Nutrients 2023, 15, 2222. [Google Scholar] [CrossRef]
- Mitra, K.; Lippincott-Schwartz, J. Analysis of Mitochondrial Dynamics and Functions Using Imaging Approaches. Curr. Protoc. Cell Biol. 2010, 46, 4251–42521. [Google Scholar] [CrossRef]
- Hockenbery, D.; Nuñez, G.; Milliman, C.; Schreiber, R.D.; Korsmeyer, S.J. Bcl-2 Is an Inner Mitochondrial Membrane Protein That Blocks Programmed Cell Death. Nature 1990, 348, 334–336. [Google Scholar] [CrossRef]
- Hobbs, B.; Drant, J.; Williamson, M.P. The Measurement of Binding Affinities by NMR Chemical Shift Perturbation. J. Biomol. NMR 2022, 76, 153–163. [Google Scholar] [CrossRef]
- Pagano, A.; Iaccarino, N.; Abdelhamid, M.A.S.; Brancaccio, D.; Garzarella, E.U.; Di Porzio, A.; Novellino, E.; Waller, Z.A.E.; Pagano, B.; Amato, J.; et al. Common G-Quadruplex Binding Agents Found to Interact With i-Motif-Forming DNA: Unexpected Multi-Target-Directed Compounds. Front. Chem. 2018, 6, 281. [Google Scholar] [CrossRef]
- Kuryavyi, V.; Phan, A.T.; Patel, D.J. Solution Structures of All Parallel-Stranded Monomeric and Dimeric G-Quadruplex Scaffolds of the Human c-Kit2 Promoter. Nucleic Acids Res. 2010, 38, 6757–6773. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Jones, R.A.; Yang, D. Solution Structure of the Biologically Relevant G-Quadruplex Element in the Human c-MYC Promoter. Implications for G-Quadruplex Stabilization. Biochemistry 2005, 44, 2048–2058. [Google Scholar] [CrossRef]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the Human Telomere in K+ Solution: An Intramolecular (3 + 1) G-Quadruplex Scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Iida, K.; Nagasawa, K. Topologies of G-Quadruplex: Biological Functions and Regulation by Ligands. Biochem. Biophys. Res. Commun. 2020, 531, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Adrian, M.; Heddi, B.; Phan, A.T. NMR Spectroscopy of G-Quadruplexes. Methods 2012, 57, 11–24. [Google Scholar] [CrossRef]
- Dai, J.; Chen, D.; Jones, R.A.; Hurley, L.H.; Yang, D. NMR Solution Structure of the Major G-Quadruplex Structure Formed in the Human BCL2 Promoter Region. Nucleic Acids Res. 2006, 34, 5133–5144. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.M.; Bollum, F.J. A Circular Dichroism Study of Poly DG, Poly DC, and Poly DG:DC. Biopolymers 1974, 13, 2087–2102. [Google Scholar] [CrossRef]
- del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem.—Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef]
- Cantor, C.R.; Warshaw, M.M.; Shapiro, H. Oligonucleotide Interactions. III. Circular Dichroism Studies of the Conformation of Deoxyoligonucleolides. Biopolymers 1970, 9, 1059–1077. [Google Scholar] [CrossRef]
- Pagano, B.; Mattia, C.A.; Giancola, C. Applications of Isothermal Titration Calorimetry in Biophysical Studies of G-Quadruplexes. Int. J. Mol. Sci. 2009, 10, 2935–2957. [Google Scholar] [CrossRef]
- Hänsel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-Quadruplex Structures Mark Human Regulatory Chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Reed, C.R.; Kennedy, S.D.; Horowitz, R.H.; Keedakkatt Puthenpeedikakkal, A.M.; Stern, H.A.; Mathews, D.H. Modeling and NMR Data Elucidate the Structure of a G-Quadruplex-Ligand Interaction for a Pu22T-Cyclometalated Iridium(III) System. J. Phys. Chem. B 2024, 128, 11634–11643. [Google Scholar] [CrossRef]
- Peterková, K.; Durník, I.; Marek, R.; Plavec, J.; Podbevšek, P. C-Kit2 G-Quadruplex Stabilized via a Covalent Probe: Exploring G-Quartet Asymmetry. Nucleic Acids Res. 2021, 49, 8947–8960. [Google Scholar] [CrossRef]
- Calabrese, D.R.; Chen, X.; Leon, E.C.; Gaikwad, S.M.; Phyo, Z.; Hewitt, W.M.; Alden, S.; Hilimire, T.A.; He, F.; Michalowski, A.M.; et al. Chemical and Structural Studies Provide a Mechanistic Basis for Recognition of the MYC G-Quadruplex. Nat. Commun. 2018, 9, 4229. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2024-3: Protein Preparation Wizard 2024. Available online: https://www.schrodinger.com/life-science/download/release-notes/release-2024-3/ (accessed on 17 December 2024).
- Schrödinger Release 2024-3: Maestro. Available online: https://www.schrodinger.com/platform/products/maestro/ (accessed on 17 December 2024).
- Olsson, M.H.M.; SØndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical p K a Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving Force Field Accuracy on Challenging Regimes of Chemical Space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.S.; Dalal, P.; Tebben, A.J.; Cheney, D.L.; Shelley, J.C. Macrocycle Conformational Sampling with Macromodel. J. Chem. Inf. Model. 2014, 54, 2680–2696. [Google Scholar] [CrossRef]
- Schrödinger Release 2024-3: MacroModel. Available online: https://www.schrodinger.com/platform/products/macromodel/ (accessed on 17 December 2024).
- Olanders, G.; Alogheli, H.; Brandt, P.; Karlén, A. Conformational Analysis of Macrocycles: Comparing General and Specialized Methods. J. Comput. Aided Mol. Des. 2020, 34, 231–252. [Google Scholar] [CrossRef]
- Schrödinger Release 2024-3: LigPrep. Available online: https://www.schrodinger.com/platform/products/ligprep/ (accessed on 17 December 2024).
- Santos-Martins, D.; Solis-Vasquez, L.; Tillack, A.F.; Sanner, M.F.; Koch, A.; Forli, S. Accelerating AutoDock4 with GPUs and Gradient-Based Local Search. J. Chem. Theory Comput. 2021, 17, 1060–1073. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel Procedure for Modeling Ligand/Receptor Induced Fit Effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Miller, E.B.; Murphy, R.B.; Sindhikara, D.; Borrelli, K.W.; Grisewood, M.J.; Ranalli, F.; Dixon, S.L.; Jerome, S.; Boyles, N.A.; Day, T.; et al. Reliable and Accurate Solution to the Induced Fit Docking Problem for Protein-Ligand Binding. J. Chem. Theory Comput. 2021, 17, 2630–2639. [Google Scholar] [CrossRef]


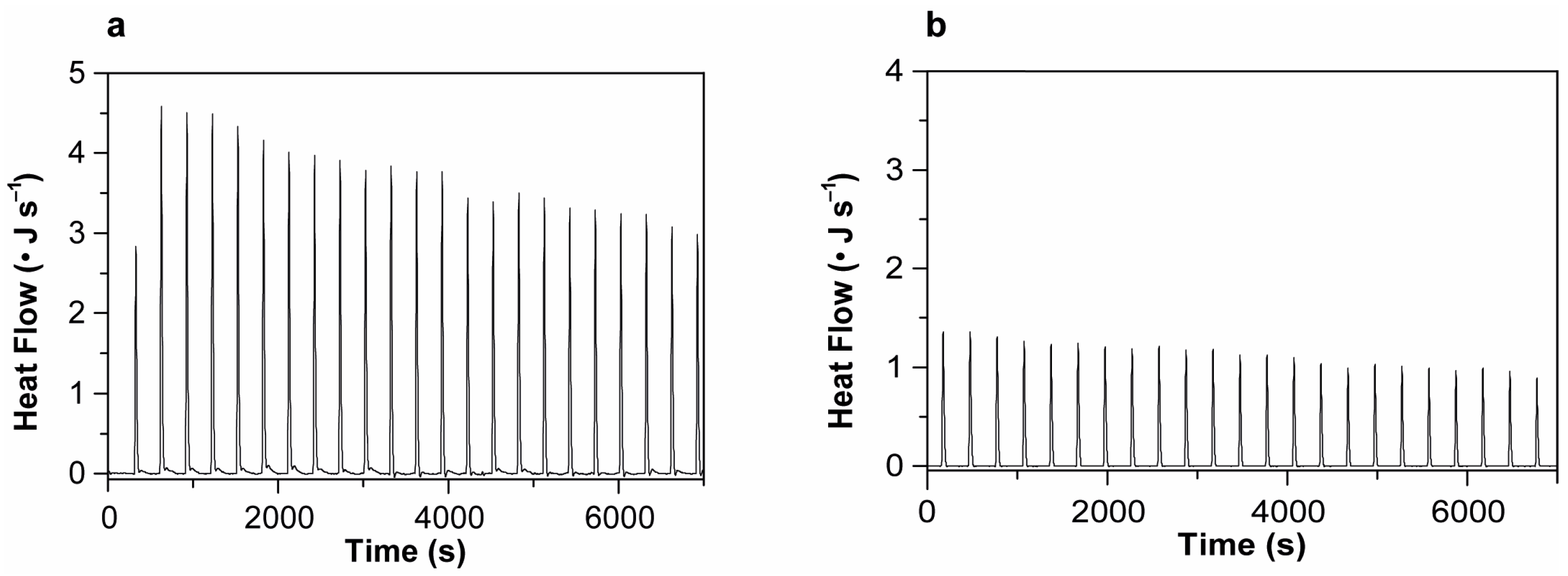
| + Vincristine | ΔH° (kJmol−1) * |
|---|---|
| c-KIT2 | −52.8 ± 5.8 |
| c-MYC | −11.7 ± 2.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Porzio, A.; Persico, C.; Romano, F.; Barra, A.; Aiello, I.; D’Auria, L.; Abate, S.; D’Aria, F.; Giancola, C.; Cinquegrana, E.; et al. Vincristine Beyond Mitosis: Uncovering a First Link to G-Quadruplex DNA in Cancer Cells. Int. J. Mol. Sci. 2025, 26, 9606. https://doi.org/10.3390/ijms26199606
Di Porzio A, Persico C, Romano F, Barra A, Aiello I, D’Auria L, Abate S, D’Aria F, Giancola C, Cinquegrana E, et al. Vincristine Beyond Mitosis: Uncovering a First Link to G-Quadruplex DNA in Cancer Cells. International Journal of Molecular Sciences. 2025; 26(19):9606. https://doi.org/10.3390/ijms26199606
Chicago/Turabian StyleDi Porzio, Anna, Carolina Persico, Francesca Romano, Alessandra Barra, Immacolata Aiello, Ludovica D’Auria, Sara Abate, Federica D’Aria, Concetta Giancola, Elpidio Cinquegrana, and et al. 2025. "Vincristine Beyond Mitosis: Uncovering a First Link to G-Quadruplex DNA in Cancer Cells" International Journal of Molecular Sciences 26, no. 19: 9606. https://doi.org/10.3390/ijms26199606
APA StyleDi Porzio, A., Persico, C., Romano, F., Barra, A., Aiello, I., D’Auria, L., Abate, S., D’Aria, F., Giancola, C., Cinquegrana, E., Di Leva, F. S., Amato, J., Marzano, S., Iaccarino, N., & Randazzo, A. (2025). Vincristine Beyond Mitosis: Uncovering a First Link to G-Quadruplex DNA in Cancer Cells. International Journal of Molecular Sciences, 26(19), 9606. https://doi.org/10.3390/ijms26199606









