Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean
Abstract
1. Introduction
2. Results
2.1. Phenotypic Analysis
2.2. Assessing the Heritability of Traits
2.3. GWAS Analysis
2.4. SNP Effect Size and Allele Frequency Distribution
2.5. Consistency and Differences in the Detection of Important SNPs (p < 0.05) by Different GWAS Methods
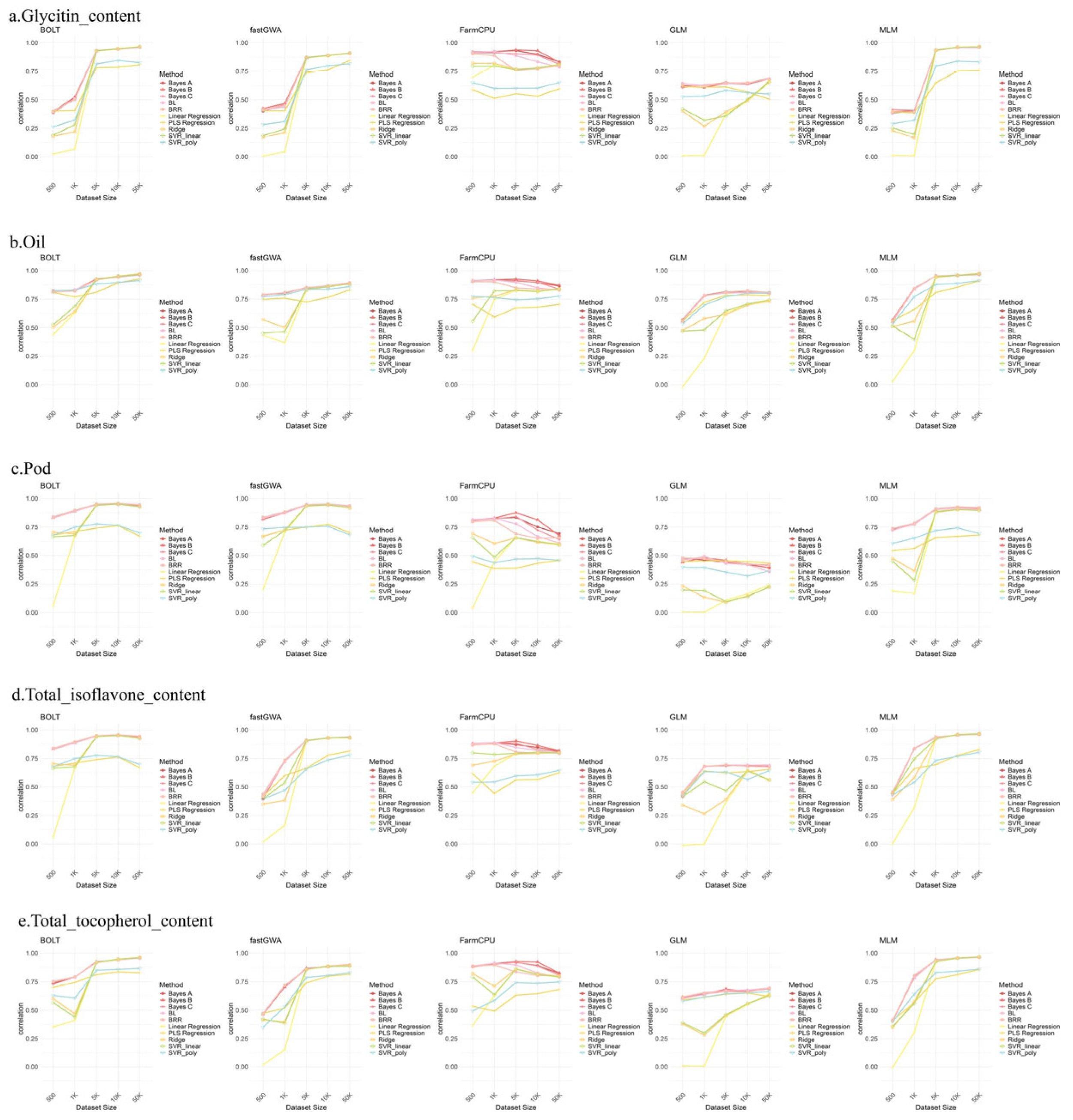
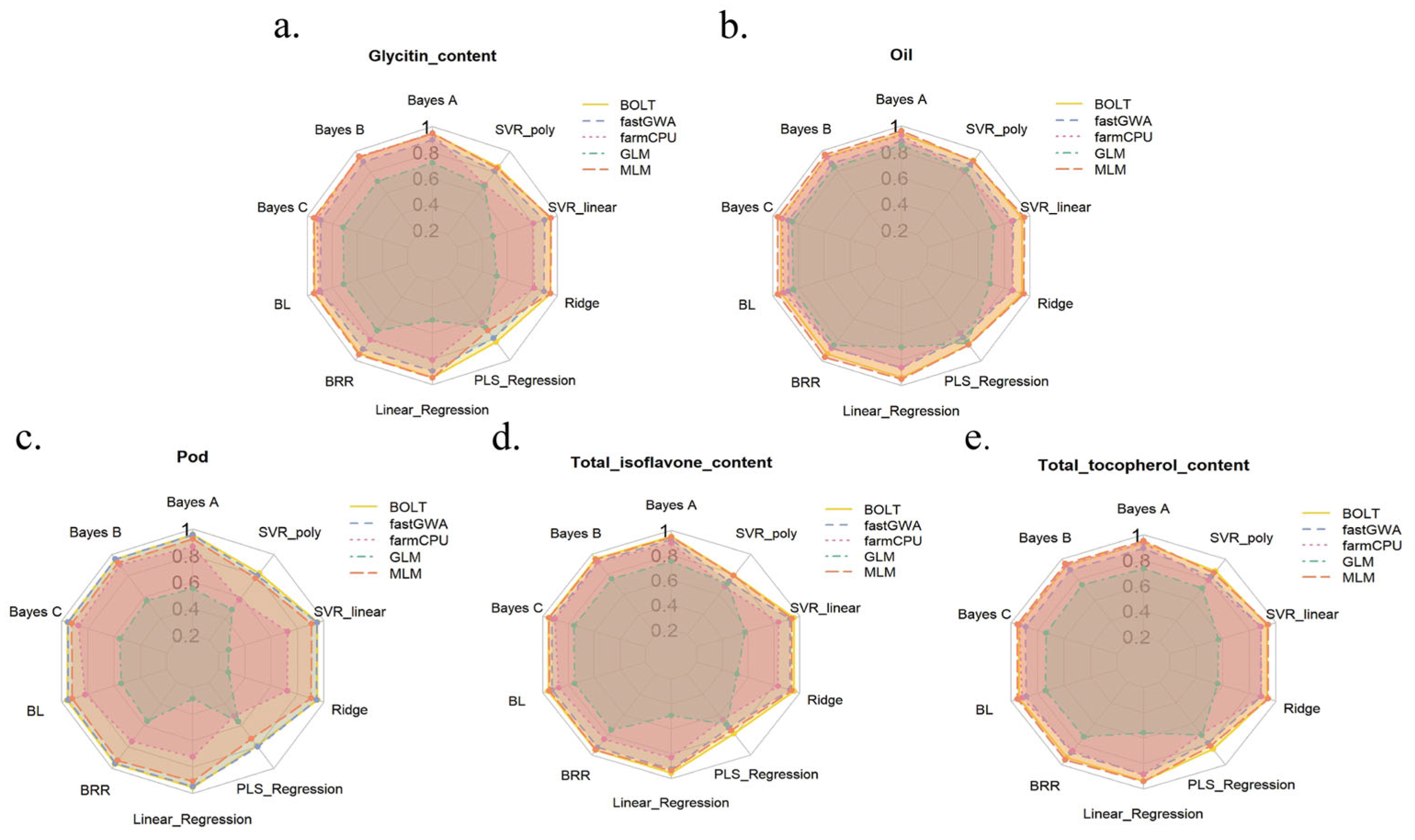
2.6. Genome Selection (GS) Analysis
3. Discussion
3.1. Comparison of GWAS Methods
3.2. Comparison of Genome Selection Models
3.3. Impact of SNP Number and Models
3.4. Role of Low-Frequency Alleles
3.5. Limitations of the Study
4. Materials and Methods
4.1. Data Sources
4.2. Estimation of Heritability
4.3. GWAS Methods
4.4. Division of Different Densities
4.5. GS Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mateos-Aparicio, I.; Redondo Cuenca, A.; Villanueva-Suárez, M.J.; Zapata-Revilla, M.A. Soybean, a promising health source. Nutr. Hosp. 2008, 23, 305–312. [Google Scholar]
- Yuan, X.; Jiang, X.; Zhang, M.; Wang, L.; Jiao, W.; Chen, H.; Mao, J.; Ye, W.; Song, Q. Integrative omics analysis elucidates the genetic basis underlying seed weight and oil content in soybean. Plant Cell 2024, 36, 2160–2175. [Google Scholar] [CrossRef] [PubMed]
- Setiadi, D.H.; Chass, G.A.; Torday, L.L.; Varro, A.; Papp, J.G. Vitamin E models. Shortened sidechain models of α, β, γ and δ tocopherol and tocotrienol—A density functional study. J. Mol. Struct. Theochem. 2003, 637, 11–26. [Google Scholar] [CrossRef]
- Defo, J.; Awany, D.; Ramesar, R. From SNP to pathway-based GWAS meta-analysis: Do current meta-analysis approaches resolve power and replication in genetic association studies? Brief. Bioinform. 2023, 24, bbac600. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Zhang, M.; Zhang, Z.; Liang, S.; Fan, L.; Yang, X.; Yuan, Y.; Pan, Y.; Zhou, G.; Liu, S.; et al. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol. J. 2022, 20, 1807–1818. [Google Scholar] [CrossRef]
- Qin, C.; Li, Y.H.; Li, D.; Zhang, X.; Kong, L.; Zhou, Y.; Lyu, X.; Ji, R.; Wei, X.; Cheng, Q.; et al. PH13 improves soybean shade traits and enhances yield for high-density planting at high latitudes. Nat. Commun. 2023, 14, 6813. [Google Scholar] [CrossRef]
- Shook, J.M.; Zhang, J.; Jones, S.E.; Singh, A.; Diers, B.W.; Singh, A.K. Meta-GWAS for quantitative trait loci identification in soybean. G3 (Bethesda) 2021, 11, jkab117. [Google Scholar] [CrossRef]
- Singh, V.K.; Chaturvedi, D.; Pundir, S.; Kumar, D.; Sharma, R.; Kumar, S.; Sharma, S.; Sharma, S. GWAS scans of cereal cyst nematode (Heterodera avenae) resistance in Indian wheat germplasm. Mol. Genet. Genom. 2023, 298, 579–601. [Google Scholar] [CrossRef]
- Wang, C.; Zeng, Y.; Wang, J.; Wang, T.; Li, X.; Shen, Z.; Meng, J.; Yao, X. A genome-wide association study of the racing performance traits in Yili horses based on Blink and FarmCPU models. Sci. Rep. 2024, 14, 27648. [Google Scholar] [CrossRef]
- Dai, K.; Wang, X.; Liu, H.; Qiao, P.; Wang, J.; Shi, W.; Guo, J.; Diao, X. Efficient identification of QTL for agronomic traits in foxtail millet (Setaria italica) using RTM- and MLM-GWAS. Theor. Appl. Genet. 2024, 137, 18. [Google Scholar] [CrossRef]
- Loh, P.R.; Kichaev, G.; Gazal, S.; Schoech, A.P.; Price, A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018, 50, 906–908. [Google Scholar] [CrossRef]
- Loya, H.; Kalantzis, G.; Cooper, F.; Palamara, P.F. A scalable variational inference approach for increased mixed-model association power. Nat. Genet. 2025, 57, 461–468. [Google Scholar] [CrossRef]
- Singer, W.M.; Shea, Z.; Yu, D.; Huang, H.; Mian, M.A.R.; Shang, C.; Rosso, M.L.; Song, Q.J.; Zhang, B. Genome-Wide Association Study and Genomic Selection for Proteinogenic Methionine in Soybean Seeds. Front. Plant Sci. 2022, 13, 859109. [Google Scholar] [CrossRef]
- Vianna, G.R.; Cunha, N.B.; Rech, E.L. Soybean seed protein storage vacuoles for expression of recombinant molecules. Curr. Opin. Plant Biol. 2023, 71, 102331. [Google Scholar] [CrossRef] [PubMed]
- Yoosefzadeh-Najafabadi, M.; Rajcan, I.; Eskandari, M. Optimizing genomic selection in soybean: An important improvement in agricultural genomics. Heliyon 2022, 8, e11873. [Google Scholar] [CrossRef] [PubMed]
- Bandillo, N.B.; Jarquin, D.; Posadas, L.G.; Lorenz, A.J.; Graef, G.L. Genomic selection performs as effectively as phenotypic selection for increasing seed yield in soybean. Plant Genome 2023, 16, e20285. [Google Scholar] [CrossRef] [PubMed]
- Mir, Z.A.; Chandra, T.; Saharan, A.; Budhlakoti, N.; Mishra, D.C.; Saharan, M.S.; Mir, R.R.; Singh, A.K.; Sharma, S.; Vikas, V.K.; et al. Recent advances on genome-wide association studies (GWAS) and genomic selection (GS); prospects for Fusarium head blight research in Durum wheat. Mol. Biol. Rep. 2023, 50, 3885–3901. [Google Scholar] [CrossRef]
- Zhou, X.; Xiang, X.; Zhang, M.; Cao, D.; Du, C.; Zhang, L.; Hu, J. Combining GS-assisted GWAS and transcriptome analysis to mine candidate genes for nitrogen utilization efficiency in Populus cathayana. BMC Plant Biol. 2023, 23, 182. [Google Scholar] [CrossRef]
- Zhang, X.; Su, J.; Jia, F.; He, Y.; Liao, Y.; Wang, Z.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; et al. Genetic architecture and genomic prediction of plant height-related traits in chrysanthemum. Hortic. Res. 2024, 11, uhad236. [Google Scholar] [CrossRef]
- Jiang, X.; Zeng, X.; Xu, M.; Li, M.; Zhang, F.; He, F.; Yang, T.; Wang, C.; Gao, T.; Long, R.; et al. The whole-genome dissection of root system architecture provides new insights for the genetic improvement of alfalfa (Medicago sativa L.). Hortic. Res. 2025, 12, uhae271. [Google Scholar] [CrossRef]
- Weissbrod, O.; Kanai, M.; Shi, H.; Gazal, S.; Peyrot, W.J.; Khera, A.V.; Okada, Y.; Martin, A.R.; Finucane, H.K.; Price, A.L. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022, 54, 450–458. [Google Scholar] [CrossRef]
- Clifford, R.E.; Maihofer, A.X.; Stein, M.B.; Ryan, A.F.; Nievergelt, C.M. Novel Risk Loci in Tinnitus and Causal Inference With Neuropsychiatric Disorders Among Adults of European Ancestry. JAMA Otolaryngol. Head Neck Surg. 2020, 146, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Privé, F.; Vilhjálmsson, B.; Speed, D. Improved genetic prediction of complex traits from individual-level data or summary statistics. Nat. Commun. 2021, 12, 4192. [Google Scholar] [CrossRef] [PubMed]
- Arkwazee, H.A.; Wallace, L.T.; Hart, J.P.; Griffiths, P.D.; Myers, J.R. Genome-Wide Association Study (GWAS) of White Mold Resistance in Snap Bean. Genes 2022, 13, 2297. [Google Scholar] [CrossRef] [PubMed]
- Rennberger, G.; Branham, S.E.; Wechter, W.P. Genome-Wide Association Study of Resistance to Pseudomonas syringae in the USDA Collection of Citrullus amarus. Plant Dis. 2023, 107, 3464–3474. [Google Scholar] [CrossRef]
- Tan, H.; Meng, J.; Crozier, K.B. Multianalyte Detection with Metasurface-Based Midinfrared Microspectrometer. ACS Sens. 2024, 9, 5839–5847. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, L.; Kang, C.; Quan, B.; Zheng, Y.; Zhang, X.; Song, J.; Xia, T.; Wang, M. Near-infrared spectroscopy and machine learning-based technique to predict quality-related parameters in instant tea. Sci. Rep. 2022, 12, 3833. [Google Scholar] [CrossRef]
- Dehghan, A. Genome-Wide Association Studies. Methods Mol. Biol. 2018, 1793, 37–49. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Li, Z.; Yang, X.; Li, K.; Xie, A.; Dong, F.; Wang, S.; Yan, J.; Liu, J. An overview of detecting gene-trait associations by integrating GWAS summary statistics and eQTLs. Sci. China Life Sci. 2024, 67, 1133–1154. [Google Scholar] [CrossRef]
- Uitterlinden, A.G. An Introduction to Genome-Wide Association Studies: GWAS for Dummies. Semin. Reprod. Med. 2016, 34, 196–204. [Google Scholar] [CrossRef]
- Fan, J.; Shen, Y.; Chen, C.; Chen, X.; Yang, X.; Liu, H.; Chen, R.; Liu, S.; Zhang, B.; Zhang, M.; et al. A large-scale integrated transcriptomic atlas for soybean organ development. Mol. Plant 2025, 18, 669–689. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Gai, J. Genome-Wide Association Studies (GWAS). Methods Mol. Biol. 2023, 2638, 123–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shao, Z.; Kong, Y.; Du, H.; Li, W.; Yang, Z.; Li, X.; Ke, H.; Sun, Z.; Shao, J.; et al. High-quality genome of a modern soybean cultivar and resequencing of 547 accessions provide insights into the role of structural variation. Nat. Genet. 2024, 56, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Wray, N.R.; Goddard, M.E.; Visscher, P.M. GCTA-GREML accounts for linkage disequilibrium when estimating genetic variance from genome-wide SNPs. Proc. Natl. Acad. Sci. USA 2016, 113, E4579–E4580. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Liu, X.; Yin, L.; Zhang, H.; Li, X.; Zhao, S. Performing Genome-Wide Association Studies Using rMVP. Methods Mol. Biol. 2022, 2481, 219–245. [Google Scholar] [CrossRef]
- Grealey, J.; Lannelongue, L.; Saw, W.Y.; Marten, J.; Méric, G.; Ruiz-Carmona, S.; Inouye, M. The Carbon Footprint of Bioinformatics. Mol. Biol. Evol. 2022, 39, msac034. [Google Scholar] [CrossRef]
- Huang, Q.; Han, X.; Tong, Z.; Deng, Y.; Xie, L.; Liu, S.; Xie, B.; Zhang, W. A Comprehensive Assessment of Ultraviolet-Radiation-Induced Mutations in Flammulina filiformis Using Whole-Genome Resequencing. J. Fungi 2024, 10, 228. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; de Los Campos, G.; Wu, H.; Vazquez, A.I.; Jones, K. Fast analysis of biobank-size data and meta-analysis using the BGLR R-package. G3 (Bethesda) 2025, 15, jkae288. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; de Los Campos, G. Multitrait Bayesian shrinkage and variable selection models with the BGLR-R package. Genetics 2022, 222, iyac112. [Google Scholar] [CrossRef]
- Yang, F.; Wang, X.; Ma, H.; Li, J. Transformers-sklearn: A toolkit for medical language understanding with transformer-based models. BMC Med. Inf. Decis. Mak. 2021, 21, 90. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef]
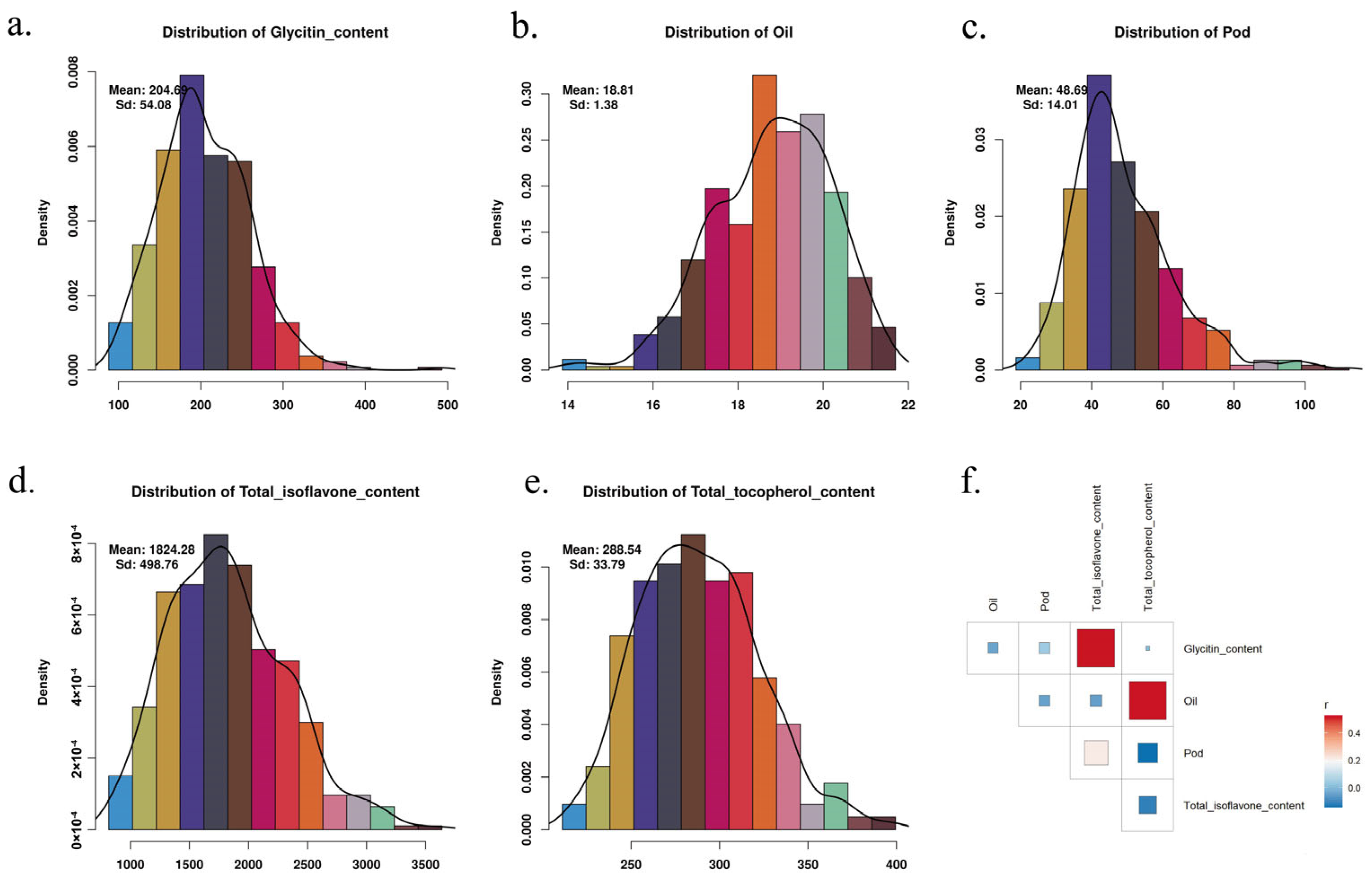
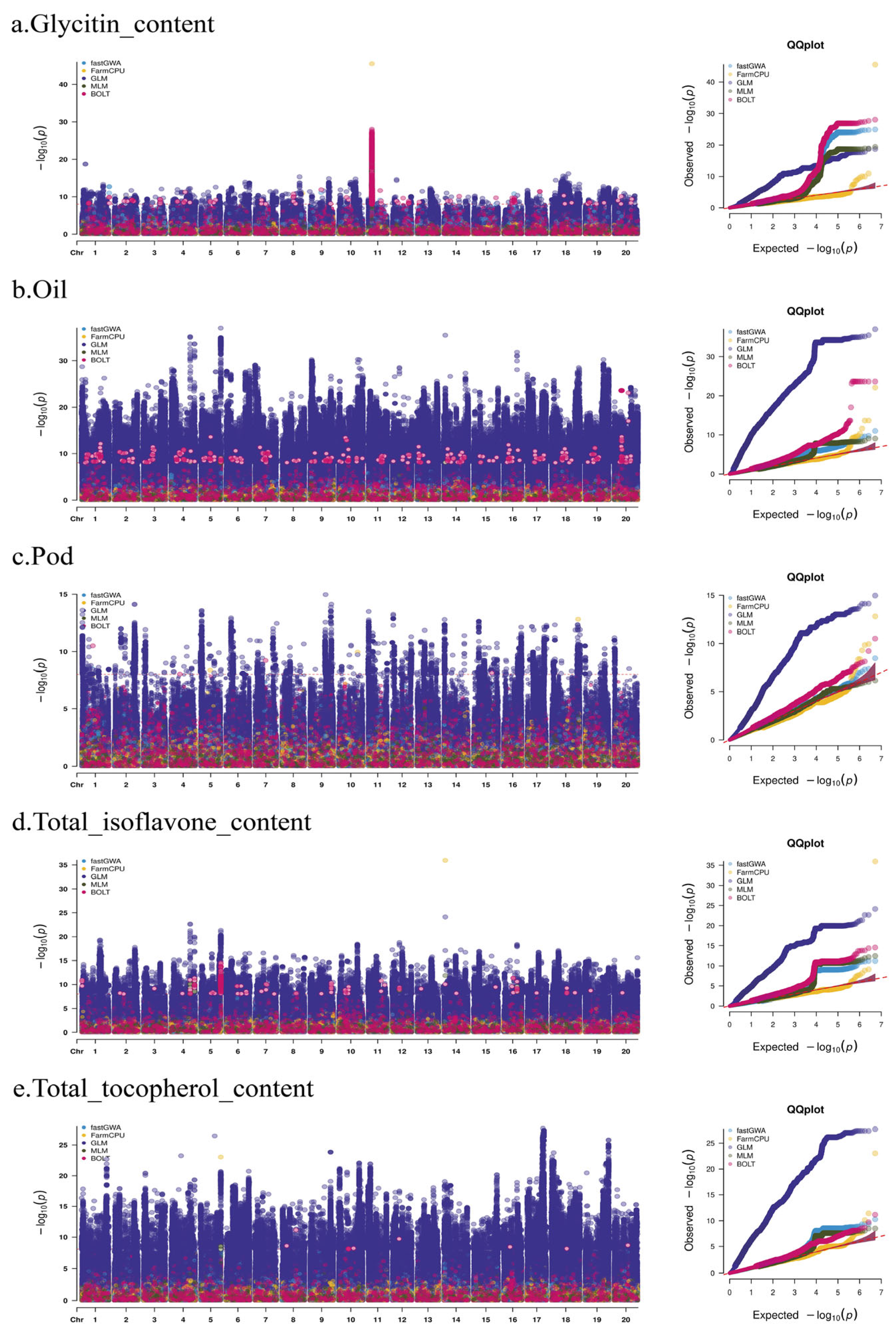
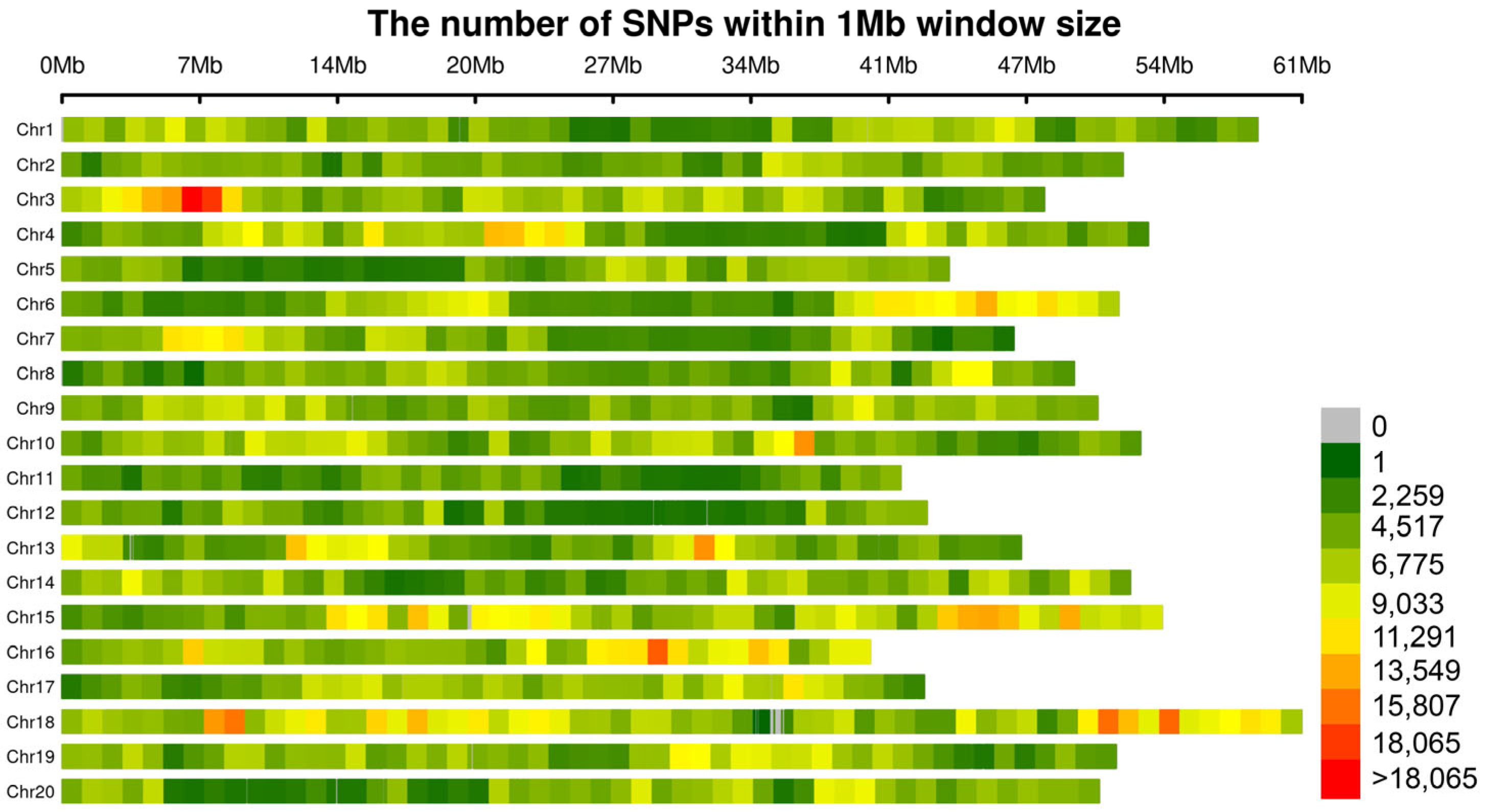

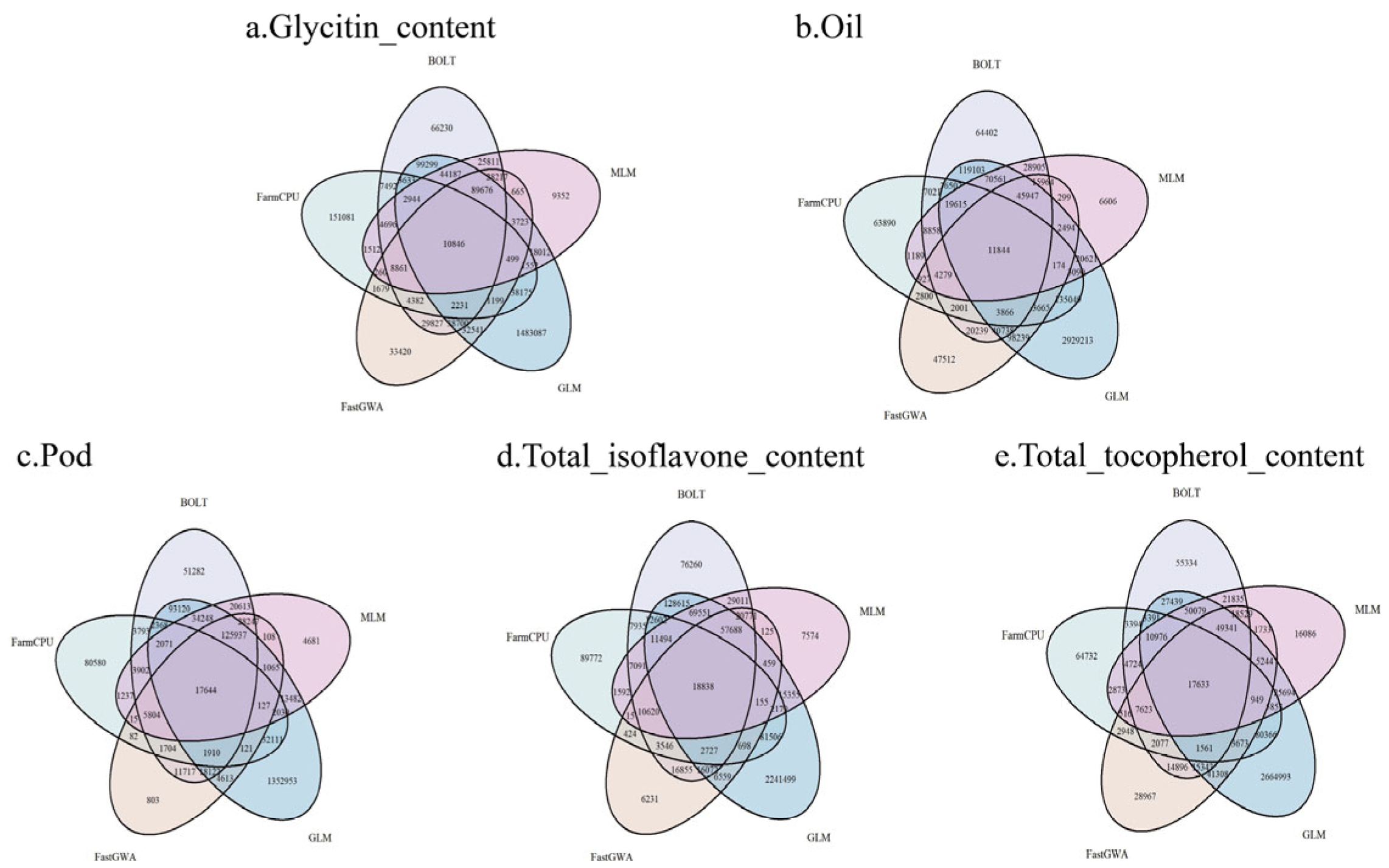
| Trait | Heritability | Pval | Vg | Ve |
|---|---|---|---|---|
| Glycitin_content | 0.58337 | 2.70049 × 10−12 | 1705.95 ± 243.969 | 1218.31 ± 125.792 |
| Oil | 0.62777 | 8.4279 × 10−20 | 1.18832 ± 0.13047 | 0.70460 ± 0.06681 |
| Pod | 0.17354 | 0.00136 | 34.0584 ± 10.6366 | 162.193 ± 15.3207 |
| Total_isoflavone_content | 0.46592 | 9.9697 × 10−9 | 11590 ± 20,222.9 | 132,855 ± 13,745.6 |
| Total_tocopherol_content | 0.58590 | 7.36219 × 10−14 | 668.987 ± 89.4219 | 472.818 ± 45.6027 |
| Model | Time |
| BOLT | 1 min 58 s |
| fastGWA | 1 min 12 s |
| FarmCPU | 5 min 46 s |
| GLM | 4 min 49 s |
| MLM | 5 min 40 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.; Tang, X.; Zhu, X.; Zhang, R.; Yao, Y.; Cao, D.; He, W.; Liu, Q.; Luan, X.; Shu, Y.; et al. Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean. Int. J. Mol. Sci. 2025, 26, 9586. https://doi.org/10.3390/ijms26199586
Xue Y, Tang X, Zhu X, Zhang R, Yao Y, Cao D, He W, Liu Q, Luan X, Shu Y, et al. Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean. International Journal of Molecular Sciences. 2025; 26(19):9586. https://doi.org/10.3390/ijms26199586
Chicago/Turabian StyleXue, Yongguo, Xiaofei Tang, Xiaoyue Zhu, Ruixin Zhang, Yubo Yao, Dan Cao, Wenjin He, Qi Liu, Xiaoyan Luan, Yongjun Shu, and et al. 2025. "Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean" International Journal of Molecular Sciences 26, no. 19: 9586. https://doi.org/10.3390/ijms26199586
APA StyleXue, Y., Tang, X., Zhu, X., Zhang, R., Yao, Y., Cao, D., He, W., Liu, Q., Luan, X., Shu, Y., & Liu, X. (2025). Leveraging GWAS-Identified Markers in Combination with Bayesian and Machine Learning Models to Improve Genomic Selection in Soybean. International Journal of Molecular Sciences, 26(19), 9586. https://doi.org/10.3390/ijms26199586






