Integrating Structural Bioinformatics and Functional Mechanisms of Sesquiterpene Synthases CARS and CADS in Lavandula angustifolia (Lavender)
Abstract
1. Introduction
2. Results
2.1. Bioinformatics Analysis
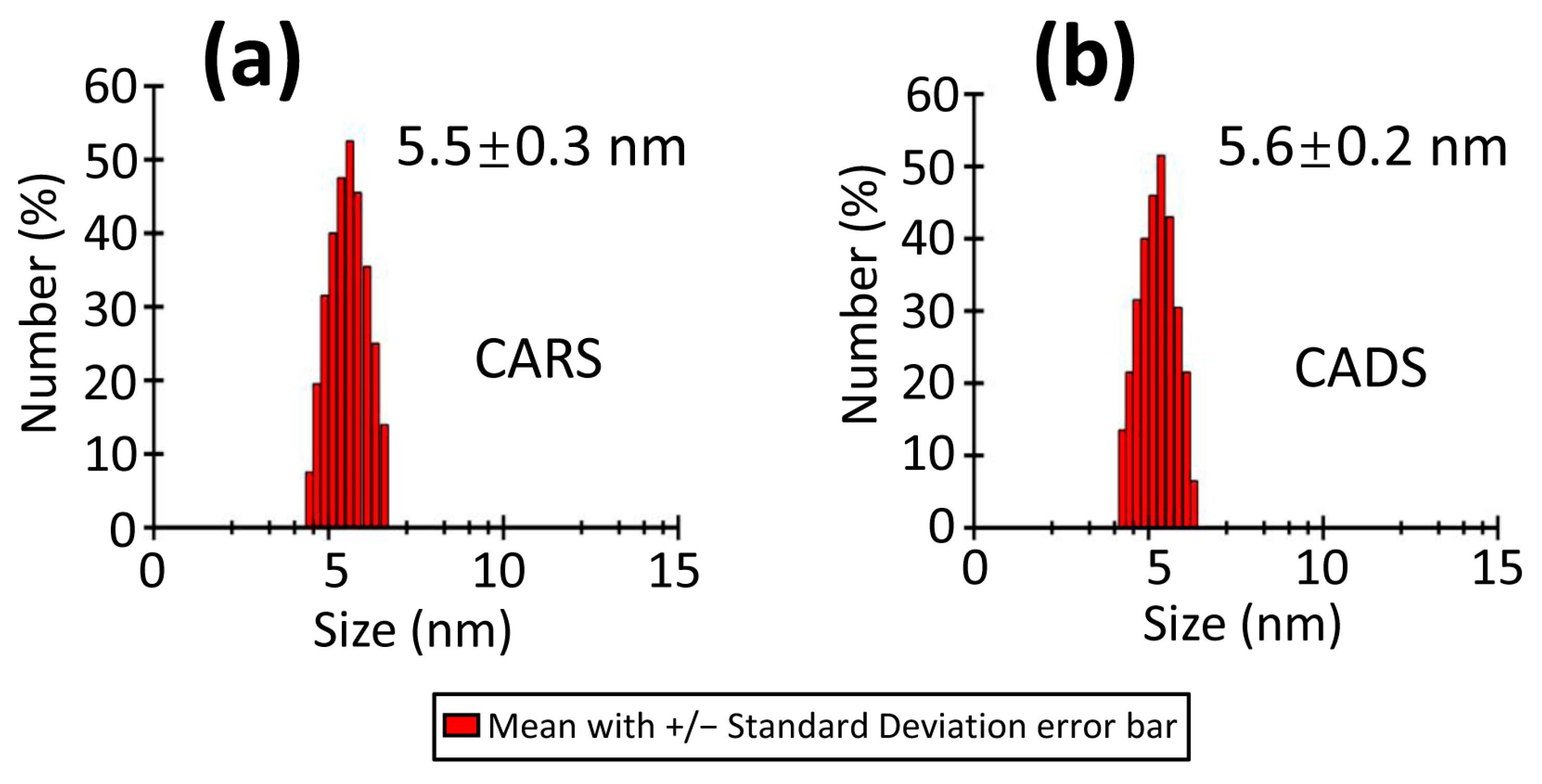
2.2. Characterization of CARS and CADS by Dynamic Light Scattering
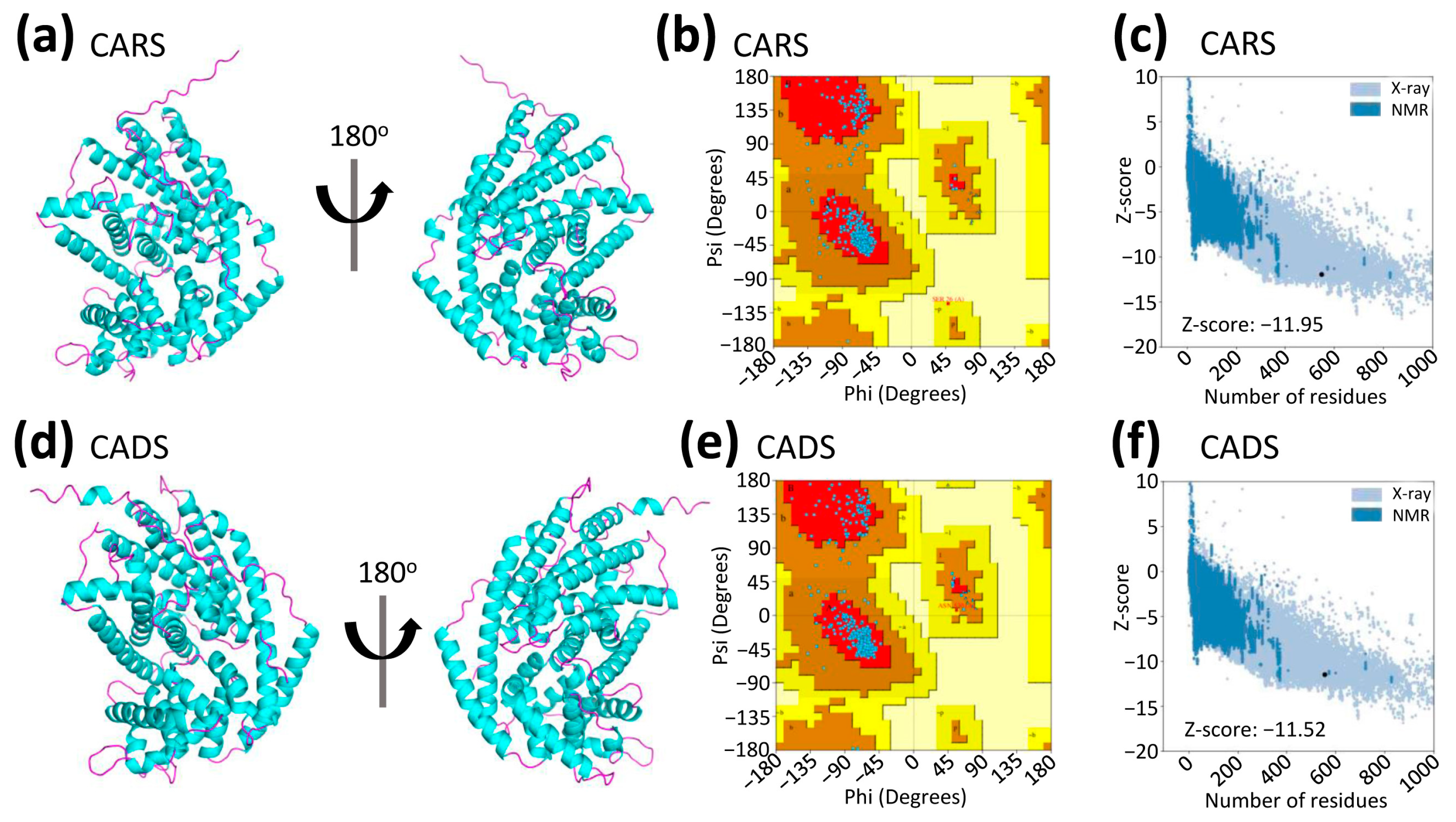
2.3. Prediction and Quality Assessment of Structural Models of CARS and CADS
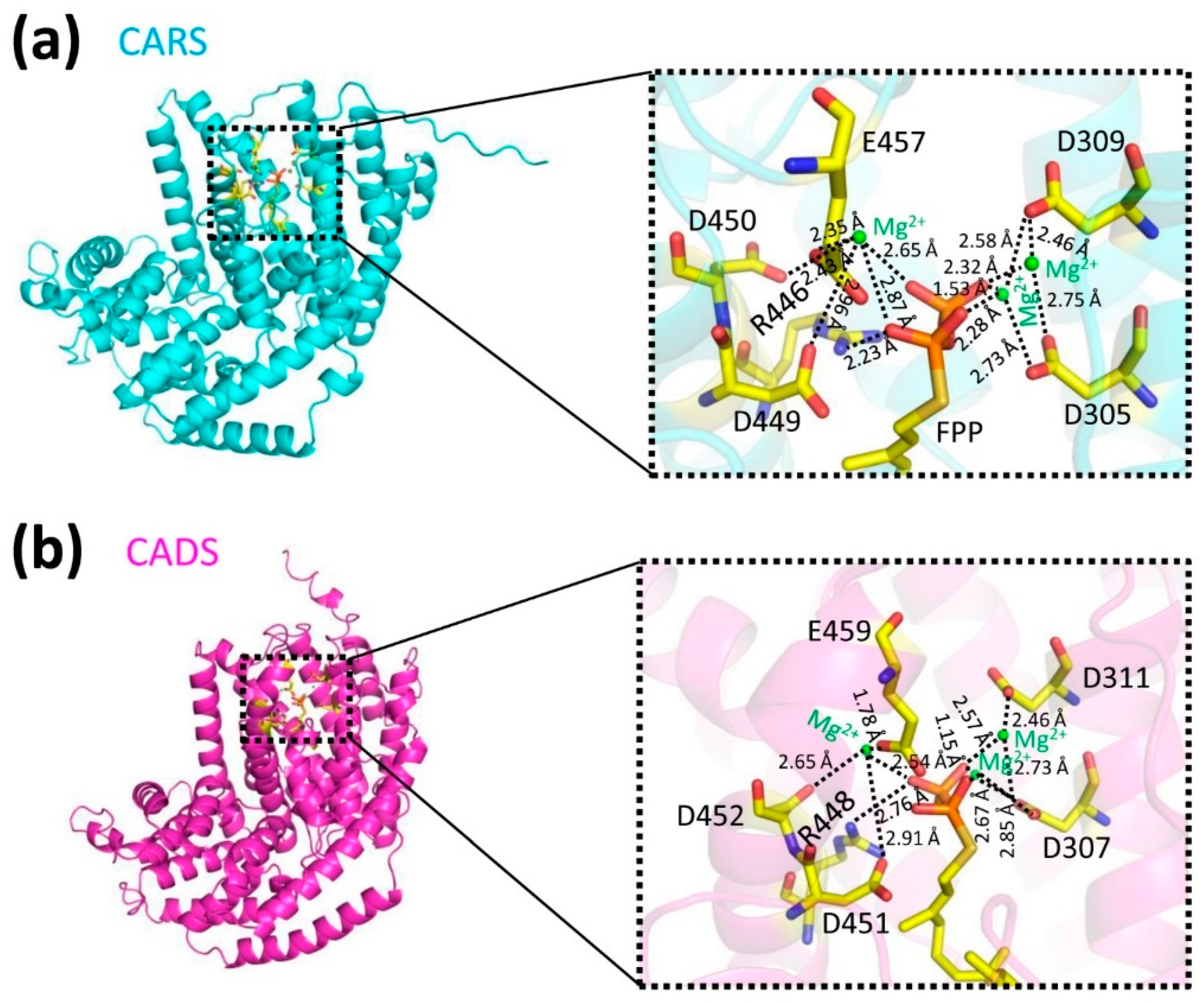
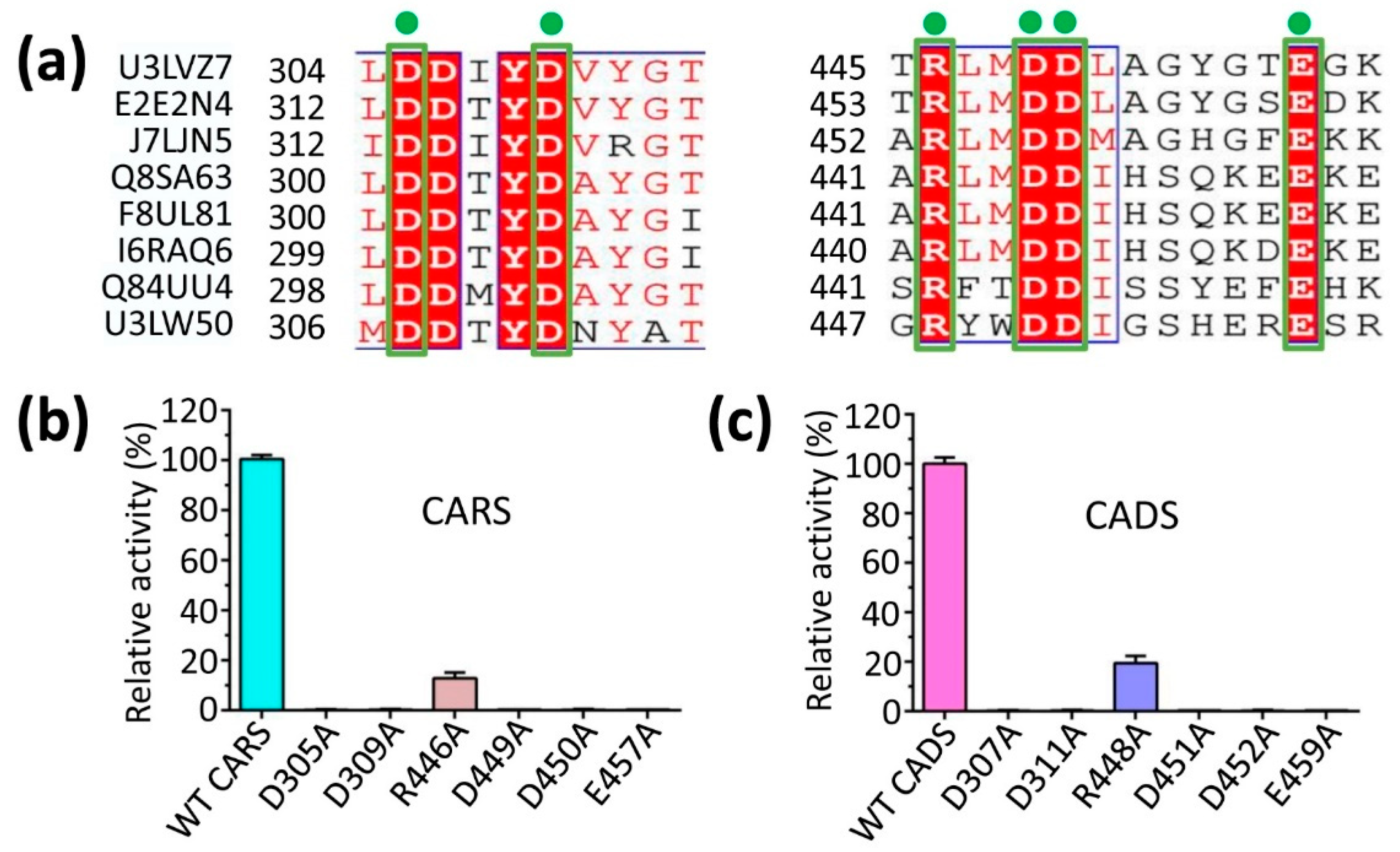
2.4. The Predicted Ligand Binding Sites of Protein-Substrate Complexes
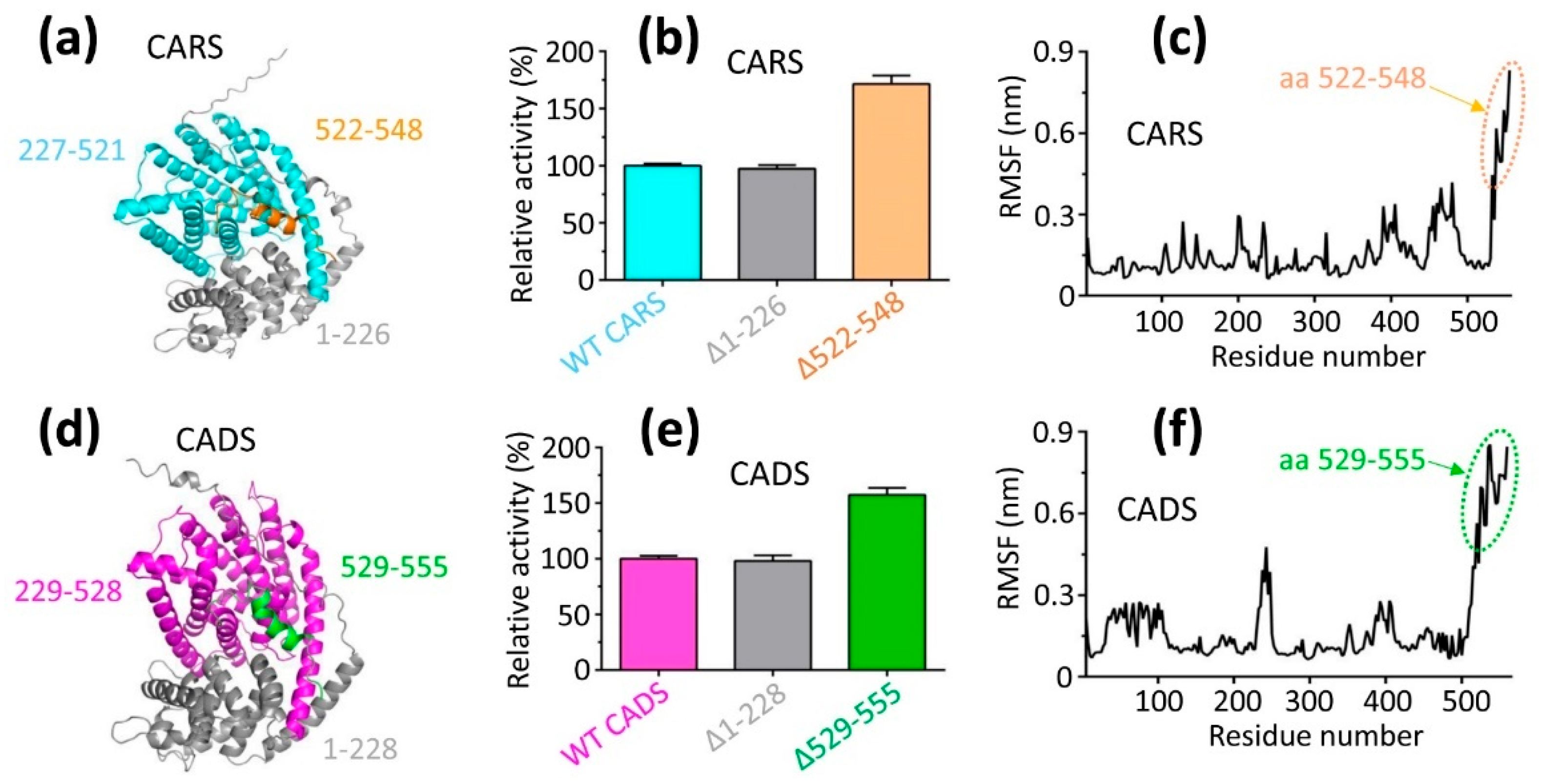
2.5. The Activities of Δ1–226 and Δ1–228 Match Those of Their Respective Wild-Type Proteins
2.6. Deleting 522–548 and 529–555 Resulted in a Dramatic Increase in the Activity Compared to Wild-Type CARS and CADS, Respectively
2.7. Kinetic Profiling for Different CARS and CADS Constructs
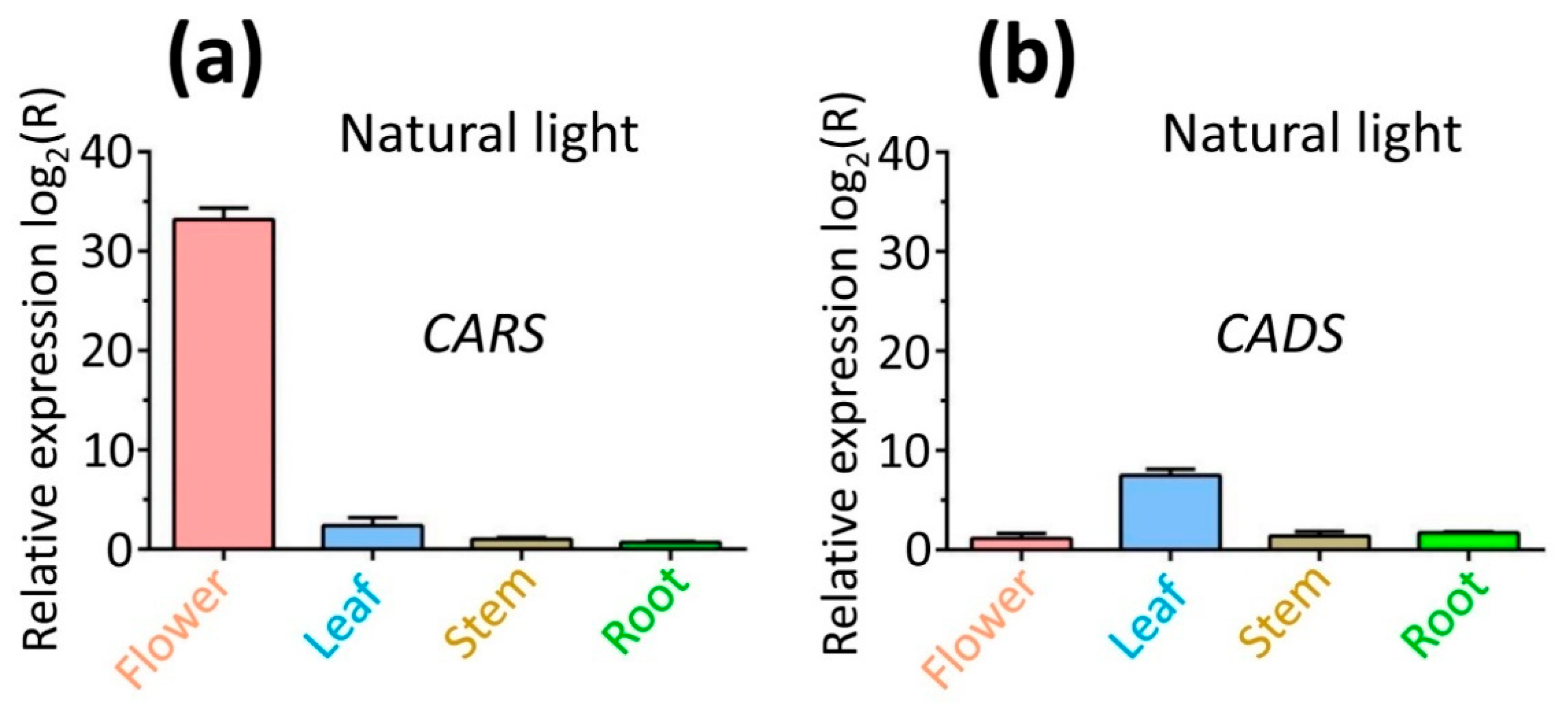
2.8. Expression Profiles of Genes CARS and CADS in Different Tissues Under Natural Light
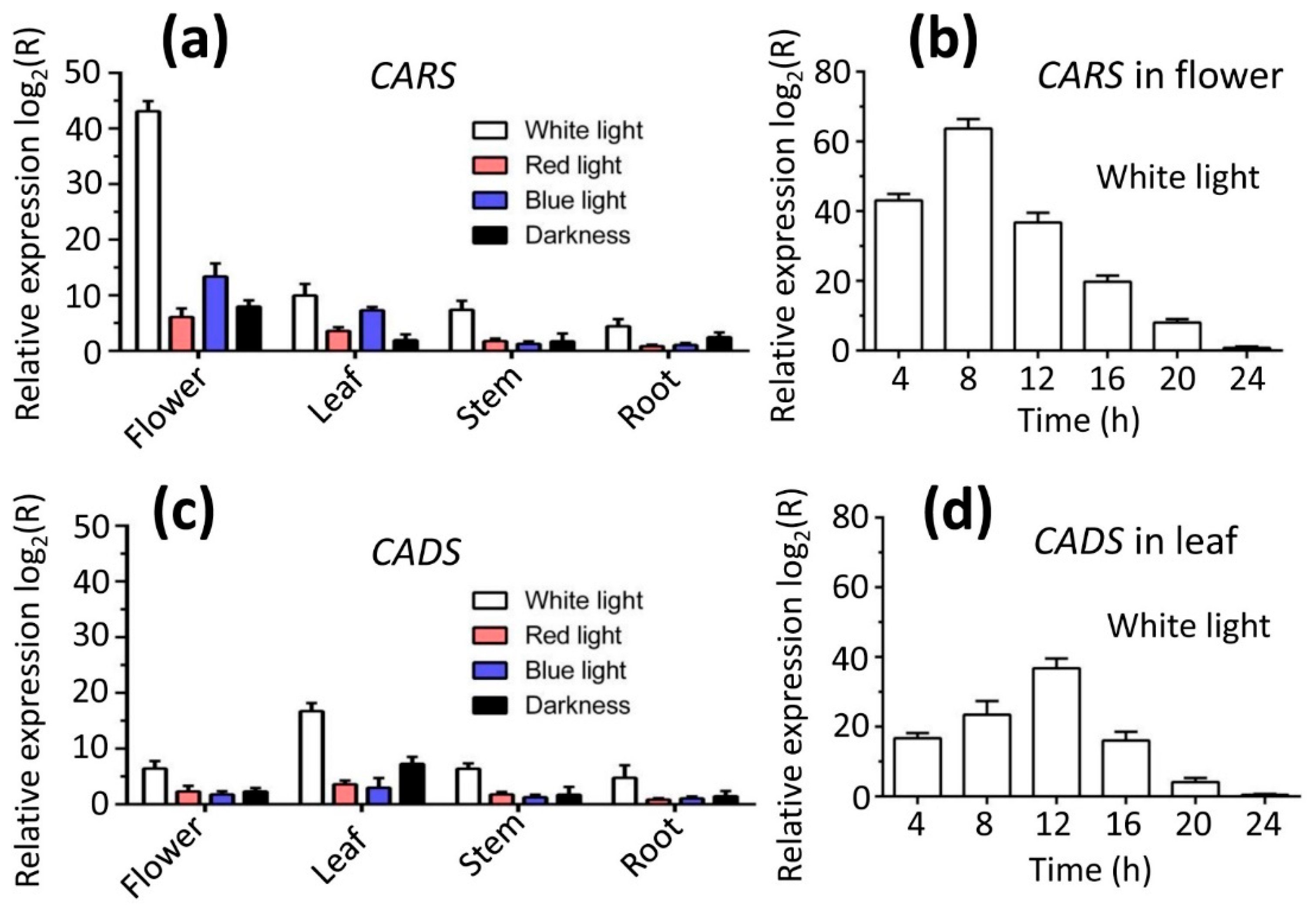
2.9. Effects of Various Light Qualities on Metabolites Resulting from CARS and CADS
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Accession codes
References
- Crișan, I.; Ona, A.; Vârban, D.; Muntean, L.; Vârban, R.; Stoie, A.; Mihăiescu, T.; Morea, A. Current Trends for Lavender (Lavandula angustifolia Mill.) Crops and Products with Emphasis on Essential Oil Quality. Plants 2023, 12, 357. [Google Scholar] [CrossRef]
- de Melo Alves Silva, L.C.; de Oliveira Mendes, F.d.C.; de Castro Teixeira, F.; de Lima Fernandes, T.E.; Barros Ribeiro, K.R.; da Silva Leal, K.C.; Dantas, D.V.; Neves Dantas, R.A. Use of Lavandula angustifolia essential oil as a complementary therapy in adult health care: A scoping review. Heliyon 2023, 9, e15446. [Google Scholar] [CrossRef]
- Landmann, C.; Fink, B.; Festner, M.; Dregus, M.; Engel, K.-H.; Schwab, W. Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Arch. Biochem. Biophys. 2007, 465, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, N.; Deng, H.; Song, D.; Maimaiti, M.; Nuerbieke, A.; Yekepeng, M.; Aili, K. Structural and functional insights into NAD(P)H-quinone oxidoreductases in lavender: Implications for abiotic stress tolerance and essential oil production. Front. Plant Sci. 2025, 16, 1661227. [Google Scholar] [CrossRef]
- Hedayati, S.; Tarahi, M.; Iraji, A.; Hashempur, M.H. Recent developments in the encapsulation of lavender essential oil. Adv. Colloid Interface Sci. 2024, 331, 103229. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Hamza, B.; Mir, R.H.; Fatima, K.; Malik, F. Lavender Plant: Farming and Health Benefits. Curr. Mol. Med. 2024, 24, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, X.; Luan, F.; Duan, J.; Zou, J.; Sun, J.; Shi, Y.; Guo, D.; Wang, C.; Wang, X. Therapeutic Potential of Essential Oils Against Ulcerative Colitis: A Review. J. Inflamm. Res. 2024, 17, 3527–3549. [Google Scholar] [CrossRef] [PubMed]
- Oseni, O.M.; Sajaditabar, R.; Mahmoud, S.S. Metabolic engineering of terpene metabolism in lavender. Beni-Suef Univ. J. Basic Appl. Sci. 2024, 13, 67. [Google Scholar] [CrossRef] [PubMed]
- Matera, R.; Lucchi, E.; Valgimigli, L. Plant Essential Oils as Healthy Functional Ingredients of Nutraceuticals and Diet Supplements: A Review. Molecules 2023, 28, 901. [Google Scholar] [CrossRef]
- Adal, A.M.; Sarker, L.S.; Malli, R.P.N.; Liang, P.; Mahmoud, S.S. RNA-Seq in the discovery of a sparsely expressed scent-determining monoterpene synthase in lavender (Lavandula). Planta 2018, 249, 271–290. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kuzuyama, T. Structural and Mechanistic Insight into Terpene Synthases that Catalyze the Irregular Non-Head-to-Tail Coupling of Prenyl Substrates. ChemBioChem 2018, 20, 29–33. [Google Scholar] [PubMed]
- Liu, D.; Deng, H.; Song, H. Insights into the functional mechanisms of the sesquiterpene synthase GEAS and GERDS in lavender. Int. J. Biol. Macromol. 2025, 299, 140195. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Zhang, L.; Yan, J.; Fu, Z.; Yang, T.; Zeng, H. Biochemistry of Terpenes and Recent Advances in Plant Protection. Int. J. Mol. Sci. 2021, 22, 5710. [Google Scholar] [CrossRef]
- Habán, M.; Korczyk-Szabó, J.; Čerteková, S.; Ražná, K. Lavandula Species, Their Bioactive Phytochemicals, and Their Biosynthetic Regulation. Int. J. Mol. Sci. 2023, 24, 8831. [Google Scholar] [CrossRef]
- Benabdelkader, T.; Guitton, Y.; Pasquier, B.; Magnard, J.L.; Jullien, F.; Kameli, A.; Legendre, L. Functional characterization of terpene synthases and chemotypic variation in three lavender species of section Stoechas. Physiol. Plant. 2014, 153, 43–57. [Google Scholar] [CrossRef]
- Malli, R.P.N.; Adal, A.M.; Sarker, L.S.; Liang, P.; Mahmoud, S.S. De novo sequencing of the Lavandula angustifolia genome reveals highly duplicated and optimized features for essential oil production. Planta 2018, 249, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Adal, A.M.; Najafianashrafi, E.; Sarker, L.S.; Mahmoud, S.S. Cloning, functional characterization and evaluating potential in metabolic engineering for lavender (+)-bornyl diphosphate synthase. Plant Mol. Biol. 2022, 111, 117–130. [Google Scholar] [CrossRef]
- Jullien, F.; Moja, S.; Bony, A.; Legrand, S.; Petit, C.; Benabdelkader, T.; Poirot, K.; Fiorucci, S.; Guitton, Y.; Nicolè, F.; et al. Isolation and functional characterization of a s-cadinol synthase, a new sesquiterpene synthase from Lavandula angustifolia. Plant Mol. Biol. 2014, 84, 227–241. [Google Scholar]
- Wayment-Steele, H.K.; Ojoawo, A.; Otten, R.; Apitz, J.M.; Pitsawong, W.; Hömberger, M.; Ovchinnikov, S.; Colwell, L.; Kern, D. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature 2023, 625, 832–839. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 66, 12–21. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Olson, A.J.; Goodsell, D.S. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007, 28, 1145–1152. [Google Scholar] [CrossRef]
- Su, M.; Yang, Q.; Du, Y.; Feng, G.; Liu, Z.; Li, Y.; Wang, R. Comparative Assessment of Scoring Functions: The CASF-2016 Update. J. Chem. Inf. Model. 2018, 59, 895–913. [Google Scholar] [CrossRef]
- Lohning, A.E.; Levonis, S.M.; Williams-Noonan, B.; Schweiker, S.S. A Practical Guide to Molecular Docking and Homology Modelling for Medicinal Chemists. Curr. Top. Med. Chem. 2017, 17, 2023–2040. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-C.; Lee, M.-C.; Chang, Y.-L.; Chen, W.-H.; Chen, H.-H. Diurnal regulation of the floral scent emission by light and circadian rhythm in the Phalaenopsis orchids. Bot. Stud. 2017, 58, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Chai, F.; Zhang, H.; Li, S.; Liang, Z.; Fan, P. Effects of sunlight exclusion on the profiles of monoterpene biosynthesis and accumulation in grape exocarp and mesocarp. Food Chem. 2017, 237, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhong, S.; Kong, L.; Fan, R.; Xu, Y.; Chen, Y.; Zhong, H. Emission and Transcriptional Regulation of Aroma Variation in Oncidium Twinkle ‘Red Fantasy’ Under Diel Rhythm. Plants 2024, 13, 3232. [Google Scholar] [CrossRef]
- Jan, M.; Liu, Z.; Rochaix, J.-D.; Sun, X. Retrograde and anterograde signaling in the crosstalk between chloroplast and nucleus. Front. Plant Sci. 2022, 13, 980237. [Google Scholar] [CrossRef]
- Dennis, G.; Posewitz, M.C. Advances in light system engineering across the phototrophic spectrum. Front. Plant Sci. 2024, 15, 1332456. [Google Scholar] [CrossRef]
- Schmitt, F.-J.; Friedrich, T. Adaptation processes in Halomicronema hongdechloris, an example of the light-induced optimization of the photosynthetic apparatus on hierarchical time scales. Front. Plant Sci. 2024, 15, 1359195. [Google Scholar] [CrossRef]
- Cavanagh, H.M.A.; Wilkinson, J.M. Biological activities of Lavender essential oil. Phytother. Res. 2002, 16, 301–308. [Google Scholar]
- Prosche, S.; Stappen, I. Flower Power: An Overview on Chemistry and Biological Impact of Selected Essential Oils from Blossoms. Planta Medica 2024, 90, 595–626. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Song, H.; Deng, H.; Abdiriyim, A.; Zhang, L.; Jiao, Z.; Li, X.; Liu, L.; Bai, S. Insights into the functional mechanisms of three terpene synthases from Lavandula angustifolia (Lavender). Front. Plant Sci. 2024, 15, 1497345. [Google Scholar] [CrossRef]
- Liu, D.; Du, Y.; Abdiriyim, A.; Zhang, L.; Song, D.; Deng, H.; Wen, X.; Zhang, Y.; Sun, B. Molecular functional mechanisms of two alcohol acetyltransferases in Lavandula x intermedia (lavandin). Front. Chem. 2025, 13, 1627286. [Google Scholar] [CrossRef]
- Vairinhos, J.; Miguel, M.G. Essential oils of spontaneous species of the genus Lavandula from Portugal: A brief review. Z. Für Naturforsch. C 2020, 75, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Li, J.; Dong, Y.; Zhang, W.; Bai, H.; Li, S.; Wang, S.; Li, H.; Shi, L. Terpene produced by coexpression of the TPS and P450 genes from Lavandula angustifolia protects plants from herbivore attacks during budding stages. BMC Plant Biol. 2023, 23, 477. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.W.-C.; Ghosal, G.; Wang, W.; Shen, X.; Wang, J.; Li, L.; Chen, J. Alpha Thalassemia/Mental Retardation Syndrome X-linked Gene Product ATRX Is Required for Proper Replication Restart and Cellular Resistance to Replication Stress. J. Biol. Chem. 2013, 288, 6342–6350. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, J.; Köllner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 10480. [Google Scholar] [CrossRef]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository—New features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T.; Elofsson, A. QMEANDisCo—Distance constraints applied on model quality estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar]
- Liu, D.; Yuan, C.; Guo, C.; Huang, M.; Lin, D. Structural and Functional Insights into the Stealth Protein CpsY of Mycobacterium tuberculosis. Biomolecules 2023, 13, 1611. [Google Scholar] [CrossRef]
- Api, A.M.; Bartlett, A.; Belsito, D.; Botelho, D.; Bruze, M.; Bryant-Freidrich, A.; Burton, G.A.; Cancellieri, M.A.; Chon, H.; Dagli, M.L.; et al. RIFM natural complex substance (NCS) fragrance ingredient safety assessment, lavandin abrialis, CAS registry number 8022-15-9, RIFM ID 1048554. Food Chem. Toxicol. 2024, 192, 114903. [Google Scholar] [CrossRef] [PubMed]
- Hedayati, S.; Tarahi, M.; Madani, A.; Mazloomi, S.M.; Hashempur, M.H. Towards a Greener Future: Sustainable Innovations in the Extraction of Lavender (Lavandula spp.) Essential Oil. Foods 2025, 14, 100. [Google Scholar] [CrossRef]
- Rashed, M.M.A.; Han, F.; Ghaleb, A.D.S.; Bao, N.; Dong, Z.; Zhai, K.-F.; Al Hashedi, S.A.; Lin, L.; Jafari, S.M. Traceability, authentication, and quality control of food-grade lavender essential oil: A comprehensive review. Adv. Colloid Interface Sci. 2025, 340, 103466. [Google Scholar] [CrossRef]
- Whitehead, J.N.; Leferink, N.G.H.; Hay, S.; Scrutton, N.S. Determinants of Product Outcome in Two Sesquiterpene Synthases from the Thermotolerant Bacterium Rubrobacter radiotolerans. ChemBioChem 2024, 26, e202400672. [Google Scholar] [CrossRef]
- Vujica, L.; Lončar, J.; Mišić, L.; Lučić, B.; Radman, K.; Mihaljević, I.; Bertoša, B.; Mesarić, J.; Horvat, M.; Smital, T. Environmental contaminants modulate transport activity of zebrafish (Danio rerio) multidrug and toxin extrusion protein 3 (Mate3/Slc47a2.1). Sci. Total Environ. 2023, 901, 165956. [Google Scholar] [PubMed]
- Scardino, V.; Di Filippo, J.I.; Cavasotto, C.N. How good are AlphaFold models for docking-based virtual screening? iScience 2023, 26, 105920. [Google Scholar] [CrossRef]
- Zhong, J.; Chen, Y.; Shi, H.; Zhou, T.; Wang, C.; Guo, Z.; Liang, Y.; Zhang, Q.; Sun, M. Identification and functional analysis of terpene synthases revealing the secrets of aroma formation in Chrysanthemum aromaticum. Int. J. Biol. Macromol. 2024, 279, 135377. [Google Scholar] [CrossRef]
- Liu, D.; Li, N.; Yu, F.; Du, Y.; Song, H.; Yao, W. Mechanistic Insights into the Bornyl Diphosphate Synthase from Lavandula angustifolia. Curr. Issues Mol. Biol. 2025, 47, 517. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Rzewski, A.; Sun, H.; Robbins, P.D.; Gambotto, A. UpGene: Application of a Web-Based DNA Codon Optimization Algorithm. Biotechnol. Prog. 2008, 20, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Menzella, H.G. Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli. Microb. Cell Factories 2011, 10, 15. [Google Scholar] [CrossRef]
- Webster, G.R.; Teh, A.Y.; Ma, J.K. Synthetic gene design—The rationale for codon optimization and implications for molecular pharming in plants. Biotechnol. Bioeng. 2016, 114, 492–502. [Google Scholar] [CrossRef]
- Yu, K.; Ang, K.S.; Lee, D.-Y. Synthetic Gene Design Using Codon Optimization On-Line (COOL). Methods Mol. Biol. 2017, 1472, 13–34. [Google Scholar] [PubMed]
- Kaur, J.; Kumar, A.; Kaur, J. Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 2018, 106, 803–822. [Google Scholar] [CrossRef]
- Koblan, L.W.; Doman, J.L.; Wilson, C.; Levy, J.M.; Tay, T.; A Newby, G.; Maianti, J.P.; Raguram, A.; Liu, D.R. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 2018, 36, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Papamichail, D.; Liu, H.; Machado, V.; Gould, N.; Coleman, J.R.; Papamichail, G. Codon Context Optimization in Synthetic Gene Design. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 15, 452–459. [Google Scholar] [CrossRef]
- Ranaghan, M.J.; Li, J.J.; Laprise, D.M.; Garvie, C.W. Assessing optimal: Inequalities in codon optimization algorithms. BMC Biol. 2021, 19, 1–13. [Google Scholar] [CrossRef]
- Gouet, P.; Robert, X.; Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Chistyakov, V.V.; Thornton, J.M. PDBsum more: New summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2004, 33, D266–D268. [Google Scholar] [CrossRef]
- Laskowski, R.A. PDBsum new things. Nucleic Acids Res. 2009, 37, D355–D359. [Google Scholar] [CrossRef] [PubMed]
- de Beer, T.A.P.; Berka, K.; Thornton, J.M.; Laskowski, R.A. PDBsum additions. Nucleic Acids Res. 2013, 42, D292–D296. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J. PDBsum: Structural summaries of PDB entries. Protein Sci. 2017, 27, 129–134. [Google Scholar] [CrossRef]
- Laskowski, R.A. PDBsum1: A standalone program for generating PDBsum analyses. Protein Sci. 2022, 31, e4473. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Deserno, M.; Holm, C. How to mesh up Ewald sums. I. A theoretical and numerical comparison of various particle mesh routines. J. Chem. Phys. 1998, 109, 7678–7693. [Google Scholar] [CrossRef]
- Deserno, M.; Holm, C. How to mesh up Ewald sums. II. An accurate error estimate for the particle–particle–particle-mesh algorithm. J. Chem. Phys. 1998, 109, 7694–7701. [Google Scholar] [CrossRef]
- Hoover, W.G.; Holian, B.L. Kinetic moments method for the canonical ensemble distribution. Phys. Lett. A 1996, 211, 253–257. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.-Y.; Li, S.; Xing, F.; Chen, L.-L. CRISPR-P: A Web Tool for Synthetic Single-Guide RNA Design of CRISPR-System in Plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Larson, M.H.; A Gilbert, L.; Wang, X.; A Lim, W.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yan, P.; Shen, W.; Li, X.; Zhou, P.; Li, Y. Modular construction of plasmids by parallel assembly of linear vector com-ponents. Anal. Biochem. 2013, 437, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]

| Target Genes | Codon Optimization | Codon Adaptation Index (CAI) Value | GC Content Value |
|---|---|---|---|
| CARS | Before codon optimization | 49.7% | 40.0% |
| After codon optimization | 82.4% | 49.7% | |
| CADS | Before codon optimization | 56.7% | 42.0% |
| After codon optimization | 83.1% | 51.4% |
| Constructs | Km (μM) | Kcat (min−1) |
|---|---|---|
| Wild-type CARS | 11.34 ± 0.13 | 6.27 ± 0.11 |
| ∆1–226 | 9.57 ± 0.24 | 14.39 ± 0.32 |
| ∆522–548 | 4.96 ± 0.17 | 23.47 ± 0.69 |
| Wild-type CADS | 15.86 ± 0.21 | 5.96 ± 0.23 |
| ∆1–228 | 11.13 ± 0.37 | 17.62 ± 0.14 |
| ∆529–555 | 7.13 ± 0.29 | 26.38 ± 0.53 |
| Sesquiterpene Synthases | Tissues | |||
|---|---|---|---|---|
| Flower | Leaf | Stem | Root | |
| CARS | 412.37 ± 9.78 | 19.23 ± 1.34 | 3.84 ± 0.56 | 2.41 ± 0.25 |
| CADS | 11.37 ± 1.16 | 123.45 ± 2.39 | 1.02 ± 0.52 | 0.96 ± 0.12 |
| Tissue | Light | Metabolites (Quantity μg/g Dry Tissue) |
|---|---|---|
| Flower | White light | 473.19 ± 8.35 |
| Red light | 9.38 ± 0.26 | |
| Blue light | 31.15 ± 1.86 | |
| Dark light | 10.92 ± 1.87 | |
| Leaf | White light | 26.38 ± 2.39 |
| Red light | 4.39 ± 0.36 | |
| Blue light | 13.49 ± 1.27 | |
| Dark light | 1.29 ± 0.35 | |
| Stem | White light | 7.15 ± 1.13 |
| Red light | 1.37 ± 0.19 | |
| Blue light | 2.81 ± 0.68 | |
| Dark light | 1.89 ± 0.37 | |
| Root | White light | 4.27 ± 0.73 |
| Red light | 1.02 ± 0.21 | |
| Blue light | 1.37 ± 0.56 | |
| Dark light | 1.86 ± 0.93 |
| Tissue | Light | Metabolites (Quantity μg/g Dry Tissue) |
|---|---|---|
| Flower | White light | 19.68 ± 1.45 |
| Red light | 5.62 ± 0.97 | |
| Blue light | 7.18 ± 1.03 | |
| Dark light | 6.37 ± 0.52 | |
| Leaf | White light | 176.39 ± 6.47 |
| Red light | 8.39 ± 1.25 | |
| Blue light | 7.84 ± 0.87 | |
| Dark light | 31.75 ± 1.84 | |
| Stem | White light | 12.89 ± 1.25 |
| Red light | 4.56 ± 0.37 | |
| Blue light | 6.12 ± 0.94 | |
| Dark light | 6.08 ± 0.79 | |
| Root | White light | 9.47 ± 1.05 |
| Red light | 3.12 ± 0.47 | |
| Blue light | 3.48 ± 0.92 | |
| Dark light | 4.39 ± 0.96 |
| Time (h) Under White Light | CARS (in Flower) | CADS (in Leaf) |
|---|---|---|
| 4 | 473.19 ± 8.35 | 176.39 ± 6.47 |
| 8 | 645.38 ± 13.47 | 235.69 ± 7.92 |
| 12 | 398.59 ± 11.27 | 318.79 ± 11.41 |
| 16 | 237.93 ± 9.37 | 164.94 ± 7.38 |
| 20 | 67.96 ± 5.71 | 53.79 ± 4.18 |
| 24 | 1.39 ± 0.47 | 2.49 ± 0.73 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Li, N.; Deng, H.; Song, D.; Song, H. Integrating Structural Bioinformatics and Functional Mechanisms of Sesquiterpene Synthases CARS and CADS in Lavandula angustifolia (Lavender). Int. J. Mol. Sci. 2025, 26, 9568. https://doi.org/10.3390/ijms26199568
Liu D, Li N, Deng H, Song D, Song H. Integrating Structural Bioinformatics and Functional Mechanisms of Sesquiterpene Synthases CARS and CADS in Lavandula angustifolia (Lavender). International Journal of Molecular Sciences. 2025; 26(19):9568. https://doi.org/10.3390/ijms26199568
Chicago/Turabian StyleLiu, Dafeng, Na Li, Huashui Deng, Daoqi Song, and Hongjun Song. 2025. "Integrating Structural Bioinformatics and Functional Mechanisms of Sesquiterpene Synthases CARS and CADS in Lavandula angustifolia (Lavender)" International Journal of Molecular Sciences 26, no. 19: 9568. https://doi.org/10.3390/ijms26199568
APA StyleLiu, D., Li, N., Deng, H., Song, D., & Song, H. (2025). Integrating Structural Bioinformatics and Functional Mechanisms of Sesquiterpene Synthases CARS and CADS in Lavandula angustifolia (Lavender). International Journal of Molecular Sciences, 26(19), 9568. https://doi.org/10.3390/ijms26199568





