Proposal of Bacillus altaicus sp. nov. Isolated from Soil in the Altai Region, Russia
Abstract
1. Introduction
2. Results
2.1. General Description and Morphological Characteristics of the Bacillus sp. al37.1 Strain
2.1.1. Initial 16S rRNA Sequence Analysis Suggests That the Strain Is Close to Bacillus mycoides
2.1.2. Assessing the Optimal Growth Conditions of the Strain
2.1.3. Morphological and Physiological Properties of the Isolate Distinguish It from Known Species
2.2. Genomic Analysis Suggests That al37.1T Represents a Novel Species, Bacillus altaicus
2.3. Bacillus altaicus sp. nov. Exhibits a Unique Composition of Fatty Acids and Biochemical Capabilities
2.3.1. Biochemical Characteristics
2.3.2. Chemotaxonomic Profile
2.4. Comparative Genomic Analysis Reveals That B. altaicus and Its Closest Relatives Within the B. mycoides Group Are Enriched with Functional Loci
2.4.1. B. altaicus and B. mycoides Group Species Hold Insecticidal Potential

2.4.2. Certain BGCs Represent the Metabolic Core of the B. mycoides Group
2.5. Genomic Inferences and Experimental Assays Demonstrate the Cytotoxic Properties of B. altaicus
2.5.1. Representatives of the B. mycoides Group Are Enriched with Enterotoxins and Other Virulence Factors
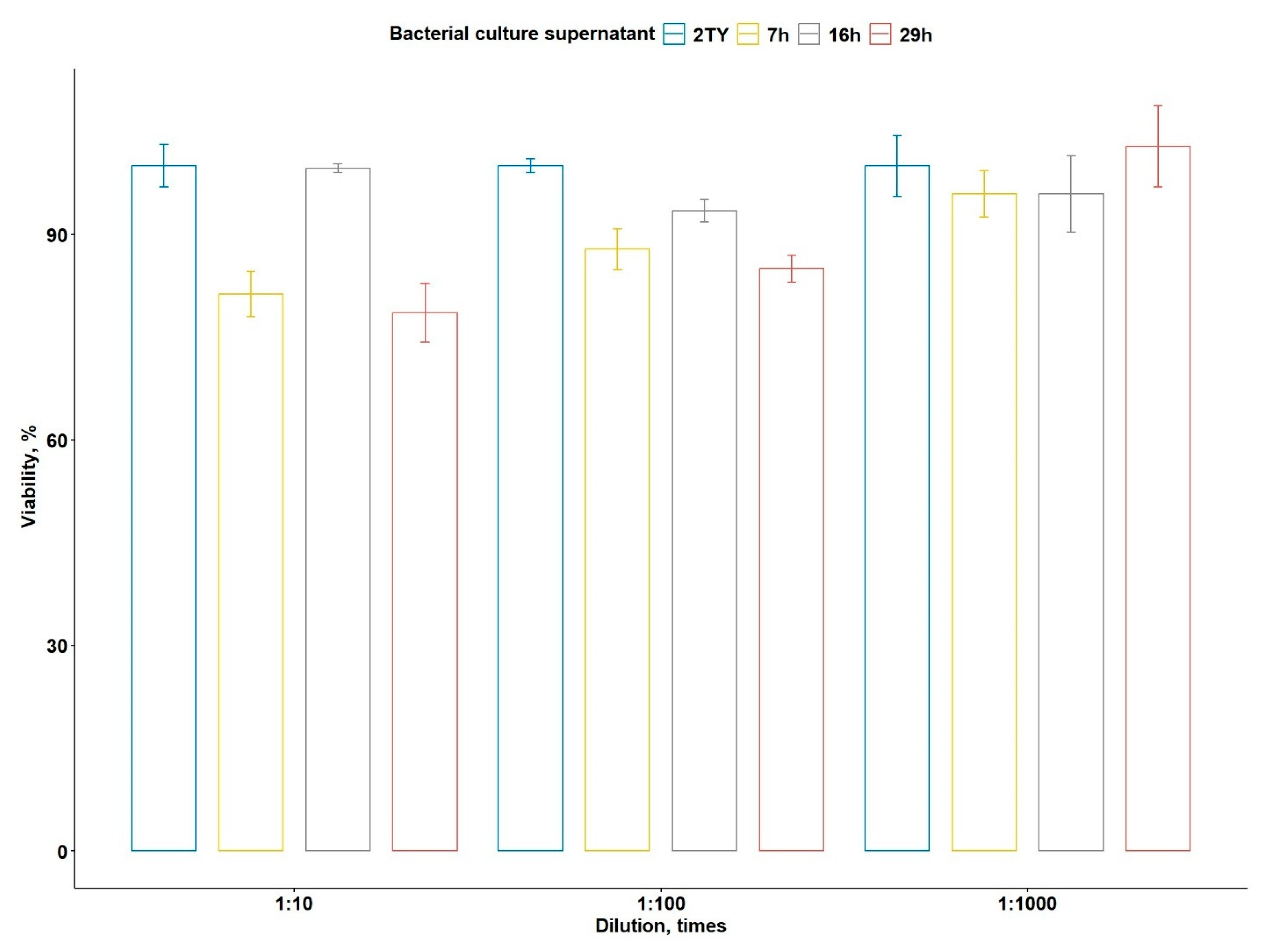
2.5.2. Cytotoxic Activity of B. altaicus al37.1T Against PANC-1 Cell Line
3. Discussion
4. Materials and Methods
4.1. Sampling Protocol
4.2. Isolation of Bacilli Strains from Soil Samples
4.3. Assessment of Optimal Growth Conditions
4.4. Morphological Description of the Strain
4.5. Sequencing of 16S rRNA Locus
4.6. Physiological and Biochemical Characterization
4.7. Chemotaxonomic Analysis
4.8. Genomic DNA Extraction, Quality Control, and Library Preparation
4.9. Genome Assembly and Annotation
4.10. Taxonomic Classification
4.11. Analysis of Functional Loci
4.12. Cytotoxicity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Timofeev, V.; Bakhteeva, I.; Khlopova, K.; Mironova, R.; Titareva, G.; Goncharova, Y.; Solomentsev, V.; Kravchenko, T.; Dyatlov, I.; Vergnaud, G. New Research on the Bacillus anthracis Genetic Diversity in Siberia. Pathogens 2023, 12, 1257. [Google Scholar] [CrossRef]
- Pisarenko, S.V.; Eremenko, E.I.; Ryazanova, A.G.; Kovalev, D.A.; Buravtseva, N.P.; Aksenova, L.Y.; Dugarzhapova, Z.F.; Evchenko, A.Y.; Kravets, E.V.; Semenova, O.V.; et al. Phylogenetic Analysis of Bacillus anthracis Strains from Western Siberia Reveals a New Genetic Cluster in the Global Population of the Species. BMC Genom. 2019, 20, 692. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, D.Y.; Berben, T.; Melton, E.D.; Overmars, L.; Vavourakis, C.D.; Muyzer, G. Microbial Diversity and Biogeochemical Cycling in Soda Lakes. Extremophiles 2014, 18, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Malkova, A.; Evdokimov, I.; Shirmanov, M.; Irkitova, A.; Dudnik, D. Development of a Microbiological Preparation for Crops Based on Bacillus pumilus Strains. BIO Web Conf. 2021, 36, 07012. [Google Scholar] [CrossRef]
- Lebedeva, E.; Panichev, A.; Kiselev, K.; Ryseva, Y.; Zaitseva, E. Taxonomic Composition and Physiological and Biochemical Properties of Cultivated Microorganisms Isolated from Kudurite Rocks of the Primorsky Krai and the Republic of Altai (Russia). Microbe 2024, 5, 100214. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, W.; Ning, W.; Song, B.; Osman, G.; Zhu, J.; Wang, W. Radiobacillus kanasensis sp. nov., a Halotolerant Bacterium Isolated from Woodland Soil. Int. J. Syst. Evol. Microbiol. 2023, 73, 005718. [Google Scholar] [CrossRef]
- Oyunbileg, N.; Iizaka, Y.; Hamada, M.; Davaapurev, B.-O.; Fukumoto, A.; Tsetseg, B.; Kato, F.; Tamura, T.; Batkhuu, J.; Anzai, Y. Actinocatenispora comari sp. nov., an Endophytic Actinomycete Isolated from Aerial Parts of Comarum salesowianum. Int. J. Syst. Evol. Microbiol. 2021, 71, 004861. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Kublanov, I.V.; Khijniak, T.V. Natronospira proteinivora gen. nov., sp. nov, an Extremely Salt-Tolerant, Alkaliphilic Gammaproteobacterium from Hypersaline Soda Lakes. Int. J. Syst. Evol. Microbiol. 2017, 67, 2604–2608. [Google Scholar] [CrossRef]
- Yi, Y.; de Jong, A.; Frenzel, E.; Kuipers, O.P. Comparative Transcriptomics of Bacillus mycoides Strains in Response to Potato-Root Exudates Reveals Different Genetic Adaptation of Endophytic and Soil Isolates. Front. Microbiol. 2017, 8, 1487. [Google Scholar] [CrossRef]
- Ambrosini, A.; Stefanski, T.; Lisboa, B.B.; Beneduzi, A.; Vargas, L.K.; Passaglia, L.M.P. Diazotrophic Bacilli Isolated from the Sunflower Rhizosphere and the Potential of Bacillus mycoides B38V as Biofertiliser. Ann. Appl. Biol. 2016, 168, 93–110. [Google Scholar] [CrossRef]
- Neher, O.T.; Johnston, M.R.; Zidack, N.K.; Jacobsen, B.J. Evaluation of Bacillus mycoides Isolate BmJ and B. mojavensis Isolate 203-7 for the Control of Anthracnose of Cucurbits Caused by Glomerella cingulata var. orbiculare. Biol. Control 2009, 48, 140–146. [Google Scholar] [CrossRef]
- Bargabus, R.L.; Zidack, N.K.; Sherwood, J.E.; Jacobsen, B.J. Characterisation of Systemic Resistance in Sugar Beet Elicited by a Non-Pathogenic, Phyllosphere-Colonizing Bacillus mycoides, Biological Control Agent. Physiol. Mol. Plant Pathol. 2002, 61, 289–298. [Google Scholar] [CrossRef]
- Reddy, G.K.; Leferink, N.G.H.; Umemura, M.; Ahmed, S.T.; Breitling, R.; Scrutton, N.S.; Takano, E. Exploring Novel Bacterial Terpene Synthases. PLoS ONE 2020, 15, e0232220. [Google Scholar] [CrossRef]
- Di Franco, C.; Beccari, E.; Santini, T.; Pisaneschi, G.; Tecce, G. Colony Shape as a Genetic Trait in the Pattern-Forming Bacillus mycoides. BMC Microbiol. 2002, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Carroll, L.M.; Cheng, R.A.; Kovac, J. No Assembly Required: Using BTyper3 to Assess the Congruency of a Proposed Taxonomic Framework for the Bacillus cereus Group with Historical Typing Methods. Front. Microbiol. 2020, 11, 580691. [Google Scholar] [CrossRef]
- Guinebretière, M.-H.; Auger, S.; Galleron, N.; Contzen, M.; De Sarrau, B.; De Buyser, M.-L.; Lamberet, G.; Fagerlund, A.; Granum, P.E.; Lereclus, D.; et al. Bacillus cytotoxicus sp. nov. Is a Novel Thermotolerant Species of the Bacillus cereus Group Occasionally Associated with Food Poisoning. Int. J. Syst. Evol. Microbiol. 2013, 63, 31–40. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Lai, Q.; Zeng, R.; Ye, D.; Xu, J.; Shao, Z. Proposal of Nine Novel Species of the Bacillus cereus Group. Int. J. Syst. Evol. Microbiol. 2017, 67, 2499–2508. [Google Scholar] [CrossRef]
- Jiménez, G.; Urdiain, M.; Cifuentes, A.; López-López, A.; Blanch, A.R.; Tamames, J.; Kämpfer, P.; Kolstø, A.-B.; Ramón, D.; Martínez, J.F.; et al. Description of Bacillus toyonensis sp. nov., a Novel Species of the Bacillus cereus Group, and Pairwise Genome Comparisons of the Species of the Group by Means of ANI Calculations. Syst. Appl. Microbiol. 2013, 36, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. A Structure-Based Nomenclature for Bacillus thuringiensis and Other Bacteria-Derived Pesticidal Proteins. J. Invertebr. Pathol. 2021, 186, 107438. [Google Scholar] [CrossRef]
- Nishiwaki, H.; Nakashima, K.; Ishida, C.; Kawamura, T.; Matsuda, K. Cloning, Functional Characterization, and Mode of Action of a Novel Insecticidal Pore-Forming Toxin, Sphaericolysin, Produced by Bacillus Sphaericus. Appl. Environ. Microbiol. 2007, 73, 3404–3411. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Prihoda, D.; Palicka, A.; Soukup, J.; Klempir, O.; Rampula, L.; Durcak, J.; Wurst, M.; Kotowski, J.; Chang, D.; et al. A Deep Learning Genome-Mining Strategy for Biosynthetic Gene Cluster Prediction. Nucleic Acids Res. 2019, 47, e110. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef]
- Vilas-Bôas, G.T.; Peruca, A.P.S.; Arantes, O.M.N. Biology and Taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can. J. Microbiol. 2007, 53, 673–687. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Lereclus, D.; Koehler, T.M. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7, 10-1128. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; Kovac, J. Proposal of a Taxonomic Nomenclature for the Bacillus cereus Group Which Reconciles Genomic Definitions of Bacterial Species with Clinical and Industrial Phenotypes. mBio 2020, 11, 10-1128. [Google Scholar] [CrossRef]
- Palmer, M.; Steenkamp, E.T.; Blom, J.; Hedlund, B.P.; Venter, S.N. All ANIs Are Not Created Equal: Implications for Prokaryotic Species Boundaries and Integration of ANIs into Polyphasic Taxonomy. Int. J. Syst. Evol. Microbiol. 2020, 70, 2937–2948. [Google Scholar] [CrossRef]
- Bavykin, S.G.; Lysov, Y.P.; Zakhariev, V.; Kelly, J.J.; Jackman, J.; Stahl, D.A.; Cherni, A. Use of 16S rRNA, 23S rRNA, and gyrB Gene Sequence Analysis to Determine Phylogenetic Relationships of Bacillus cereus Group Microorganisms. J. Clin. Microbiol. 2004, 42, 3711–3730, Erratum in J. Clin. Microbiol. 2006, 44, 2676. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Wintzingerode, F.; Rainey, F.A.; Kroppenstedt, R.M.; Stackebrandt, E. Identification of Environmental Strains of Bacillus mycoides by Fatty Acid Analysis and Species-Specific 16S rDNA Oligonucleotide Probe. FEMS Microbiol. Ecol. 2006, 24, 201–209. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, D.; Andreeva, M.; Li, Y.; Fan, L.; Tang, M. Temporal-Spatial Variability of Modern Climate in the Altai Mountains during 1970-2015. PLoS ONE 2020, 15, e0230196. [Google Scholar] [CrossRef]
- Diomande, S.E.; Nguyen-The, C.; Guinebretiere, M.-H.; Broussolle, V.; Brillard, J. Role of Fatty Acids in Bacillus Environmental Adaptation. Front. Microbiol. 2015, 6, 813. [Google Scholar] [CrossRef]
- Prakash, O.; Nimonkar, Y.; Shaligram, S.; Joseph, N.; Shouche, Y.S. Response of Cellular Fatty Acids to Environmental Stresses in Endophytic Micrococcus spp. Ann. Microbiol. 2015, 65, 2209–2218. [Google Scholar] [CrossRef]
- Bajerski, F.; Wagner, D.; Mangelsdorf, K. Cell Membrane Fatty Acid Composition of Chryseobacterium frigidisoli PB4T, Isolated from Antarctic Glacier Forefield Soils, in Response to Changing Temperature and pH Conditions. Front. Microbiol. 2017, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.; Caramujo, M. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed]
- Rozanov, A.S.; Myagkaya, I.N.; Korzhuk, A.V.; Ershov, N.I.; Kirichenko, I.S.; Gustaytis, M.A.; Saryg-ool, B.Y.; Malov, V.I.; Shipova, A.A.; Lazareva, E.V.; et al. Metagenomics Data of Microbial Communities in Bacterial Mats and Bottom Sediments in Water Bodies within the Kurai Mercury Province (Gorny Altai, Russia). Data Brief. 2021, 36, 107099. [Google Scholar] [CrossRef]
- Vavourakis, C.D.; Ghai, R.; Rodriguez-Valera, F.; Sorokin, D.Y.; Tringe, S.G.; Hugenholtz, P.; Muyzer, G. Metagenomic Insights into the Uncultured Diversity and Physiology of Microbes in Four Hypersaline Soda Lake Brines. Front. Microbiol. 2016, 7, 211. [Google Scholar] [CrossRef] [PubMed]
- Kadnikov, V.V.; Mardanov, A.V.; Ivasenko, D.A.; Antsiferov, D.V.; Beletsky, A.V.; Karnachuk, O.V.; Ravin, N.V. Lignite Coal Burning Seam in the Remote Altai Mountains Harbors a Hydrogen-Driven Thermophilic Microbial Community. Sci. Rep. 2018, 8, 6730. [Google Scholar] [CrossRef] [PubMed]
- Karnachuk, O.V.; Rusanov, I.I.; Panova, I.A.; Kadnikov, V.V.; Avakyan, M.R.; Ikkert, O.P.; Lukina, A.P.; Beletsky, A.V.; Mardanov, A.V.; Knyazev, Y.V.; et al. The Low-Temperature Germinating Spores of the Thermophilic Desulfofundulus Contribute to an Extremely High Sulfate Reduction in Burning Coal Seams. Front. Microbiol. 2023, 14, 1204102. [Google Scholar] [CrossRef]
- Fu, Q.; Qiu, Y.; Zhao, J.; Li, J.; Xie, S.; Liao, Q.; Fu, X.; Huang, Y.; Yao, Z.; Dai, Z.; et al. Monotonic Trends of Soil Microbiomes, Metagenomic and Metabolomic Functioning across Ecosystems along Water Gradients in the Altai Region, Northwestern China. Sci. Total Environ. 2024, 912, 169351. [Google Scholar] [CrossRef]
- Ma, X.; Fan, L.; Yang, M.; Li, J.; Yan, M.; Yang, Z.; Chen, X.; Zhang, B.; Li, Y.; Gao, Y. Allocation Strategy of Nonstructural Carbohydrates in Spiraea L. across Different Grassland Types in the Altai Mountains. Front. Plant Sci. 2025, 16, 1562363. [Google Scholar] [CrossRef] [PubMed]
- Boyarskikh, I.G.; Artemov, I.A.; Kuznetsov, A.A.; Kostikova, V.A. Changes in Profiles of Classes and of Individual Polyphenols in Leaves of Spiraea chamaedryfolia and Spiraea media along an Altitudinal Gradient. Plants 2023, 12, 2977. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Ye, M.; He, Q.; Zeng, G.; Li, M.; Chen, W.; Pan, X.; Qian, J.; Lv, Y. Elevation Gradient Effects on Grassland Species Diversity and Phylogenetic in the Two-River Source Forest Region of the Altai Mountains, Xinjiang, China. Front. Plant Sci. 2025, 16, 1487582. [Google Scholar] [CrossRef] [PubMed]
- Novikova, S.V.; Sharov, V.V.; Oreshkova, N.V.; Simonov, E.P.; Krutovsky, K.V. Genetic Adaptation of Siberian Larch (Larix sibirica Ledeb.) to High Altitudes. Int. J. Mol. Sci. 2023, 24, 4530. [Google Scholar] [CrossRef]
- Best, H.L.; Williamson, L.J.; Lipka-Lloyd, M.; Waller-Evans, H.; Lloyd-Evans, E.; Rizkallah, P.J.; Berry, C. The Crystal Structure of Bacillus thuringiensis Tpp80Aa1 and Its Interaction with Galactose-Containing Glycolipids. Toxins 2022, 14, 863. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Z.; Zhang, J.; Wan, Y.; Jin, W.; Li, Y.; Fang, X. Cry80Aa1, a Novel Bacillus thuringiensis Toxin with Mosquitocidal Activity to Culex pipiens pallens. J. Invertebr. Pathol. 2020, 173, 107386. [Google Scholar] [CrossRef]
- Dammak, I.; Dammak, M.; Tounsi, S. Histopathological and Combinatorial Effects of the Metalloprotease InhA1 and Cry Proteins of Bacillus thuringiensis against Spodoptera littoralis. Int. J. Biol. Macromol. 2015, 81, 759–762. [Google Scholar] [CrossRef]
- Luo, X.; Chen, L.; Huang, Q.; Zheng, J.; Zhou, W.; Peng, D.; Ruan, L.; Sun, M. Bacillus thuringiensis Metalloproteinase Bmp1 Functions as a Nematicidal Virulence Factor. Appl. Environ. Microbiol. 2013, 79, 460–468. [Google Scholar] [CrossRef]
- Wan, L.; Lin, J.; Du, H.; Zhang, Y.; Bravo, A.; Soberón, M.; Sun, M.; Peng, D. Bacillus thuringiensis Targets the Host Intestinal Epithelial Junctions for Successful Infection of Caenorhabditis elegans. Environ. Microbiol. 2019, 21, 1086–1098. [Google Scholar] [CrossRef]
- Martínez-Zavala, S.A.; Barboza-Pérez, U.E.; Hernández-Guzmán, G.; Bideshi, D.K.; Barboza-Corona, J.E. Chitinases of Bacillus thuringiensis: Phylogeny, Modular Structure, and Applied Potentials. Front. Microbiol. 2020, 10, 3032. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Ru, J.; Zhang, Y.; Wang, Q.; Li, Y. Fengycin Produced by Bacillus subtilis 9407 Plays a Major Role in the Biocontrol of Apple Ring Rot Disease. Microbiol. Res. 2017, 199, 89–97. [Google Scholar] [CrossRef]
- Medeot, D.B.; Fernandez, M.; Morales, G.M.; Jofré, E. Fengycins From Bacillus amyloliquefaciens MEP218 Exhibit Antibacterial Activity by Producing Alterations on the Cell Surface of the Pathogens Xanthomonas axonopodis pv. vesicatoria and Pseudomonas aeruginosa PA01. Front. Microbiol. 2020, 10, 3107. [Google Scholar] [CrossRef]
- Nithyapriya, S.; Lalitha, S.; Sayyed, R.Z.; Reddy, M.S.; Dailin, D.J.; El Enshasy, H.A.; Luh Suriani, N.; Herlambang, S. Production, Purification, and Characterization of Bacillibactin Siderophore of Bacillus subtilis and Its Application for Improvement in Plant Growth and Oil Content in Sesame. Sustainability 2021, 13, 5394. [Google Scholar] [CrossRef]
- Chakraborty, K.; Kizhakkekalam, V.K.; Joy, M.; Chakraborty, R.D. Bacillibactin Class of Siderophore Antibiotics from a Marine Symbiotic Bacillus as Promising Antibacterial Agents. Appl. Microbiol. Biotechnol. 2022, 106, 329–340. [Google Scholar] [CrossRef]
- Li, B.; Li, Q.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Responses of Beneficial Bacillus amyloliquefaciens SQR9 to Different Soilborne Fungal Pathogens through the Alteration of Antifungal Compounds Production. Front. Microbiol. 2014, 5, 636. [Google Scholar] [CrossRef]
- Dimopoulou, A.; Theologidis, I.; Benaki, D.; Koukounia, M.; Zervakou, A.; Tzima, A.; Diallinas, G.; Hatzinikolaou, D.G.; Skandalis, N. Direct Antibiotic Activity of Bacillibactin Broadens the Biocontrol Range of Bacillus amyloliquefaciens MBI600. mSphere 2021, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Koppisch, A.T.; Browder, C.C.; Moe, A.L.; Shelley, J.T.; Kinkel, B.A.; Hersman, L.E.; Iyer, S.; Ruggiero, C.E. Petrobactin Is the Primary Siderophore Synthesized by Bacillus anthracis str. Sterne under Conditions of Iron Starvation. BioMetals 2005, 18, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Koppisch, A.T.; Dhungana, S.; Hill, K.K.; Boukhalfa, H.; Heine, H.S.; Colip, L.A.; Romero, R.B.; Shou, Y.; Ticknor, L.O.; Marrone, B.L.; et al. Petrobactin Is Produced by Both Pathogenic and Non-Pathogenic Isolates of the Bacillus cereus Group of Bacteria. BioMetals 2008, 21, 581–589. [Google Scholar] [CrossRef]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for Plant Growth Promotion and Stress Resilience: What Have We Learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef]
- Zhu, S.; Hegemann, J.D.; Fage, C.D.; Zimmermann, M.; Xie, X.; Linne, U.; Marahiel, M.A. Insights into the Unique Phosphorylation of the Lasso Peptide Paeninodin. J. Biol. Chem. 2016, 291, 13662–13678. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, M.N.; Shikov, A.E.; Savina, I.A.; Shmatov, F.M.; Nizhnikov, A.A.; Antonets, K.S. Genomic Insights into the Bactericidal and Fungicidal Potential of Bacillus mycoides b12.3 Isolated in the Soil of Olkhon Island in Lake Baikal, Russia. Microorganisms 2024, 12, 2450. [Google Scholar] [CrossRef]
- Sivaramalingam, S.S.; Jothivel, D.; Govindarajan, D.K.; Kadirvelu, L.; Sivaramakrishnan, M.; Chithiraiselvan, D.D.; Kandaswamy, K. Structural and Functional Insights of Sortases and Their Interactions with Antivirulence Compounds. Curr. Res. Struct. Biol. 2024, 8, 100152. [Google Scholar] [CrossRef]
- Chen, F.; Liu, B.; Wang, D.; Wang, L.; Deng, X.; Bi, C.; Xiong, Y.; Wu, Q.; Cui, Y.; Zhang, Y.; et al. Role of Sortase A in the Pathogenesis of Staphylococcus aureus -Induced Mastitis in Mice. FEMS Microbiol. Lett. 2014, 351, 95–103. [Google Scholar] [CrossRef]
- Aljghami, M.E.; Barghash, M.M.; Majaesic, E.; Bhandari, V.; Houry, W.A. Cellular Functions of the ClpP Protease Impacting Bacterial Virulence. Front. Mol. Biosci. 2022, 9, 1054408. [Google Scholar] [CrossRef]
- Prüß, B.M.; Dietrich, R.; Nibler, B.; Märtlbauer, E.; Scherer, S. The Hemolytic Enterotoxin HBL Is Broadly Distributed among Species of the Bacillus cereus Group. Appl. Environ. Microbiol. 1999, 65, 5436–5442. [Google Scholar] [CrossRef] [PubMed]
- Senesi, S.; Ghelardi, E. Production, Secretion and Biological Activity of Bacillus cereus Enterotoxins. Toxins 2010, 2, 1690–1703. [Google Scholar] [CrossRef]
- Bağcıoğlu, M.; Fricker, M.; Johler, S.; Ehling-Schulz, M. Detection and Identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via Machine Learning Based FTIR Spectroscopy. Front. Microbiol. 2019, 10, 902. [Google Scholar] [CrossRef]
- Fiedoruk, K.; Drewnowska, J.M.; Mahillon, J.; Zambrzycka, M.; Swiecicka, I. Pan-Genome Portrait of Bacillus mycoides Provides Insights into the Species Ecology and Evolution. Microbiol. Spectr. 2021, 9, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Bulgari, D.; Filisetti, S.; Montagna, M.; Gobbi, E.; Faoro, F. Pathogenic Potential of Bacteria Isolated from Commercial Biostimulants. Arch. Microbiol. 2022, 204, 162. [Google Scholar] [CrossRef]
- Méndez Acevedo, M.; Carroll, L.M.; Mukherjee, M.; Mills, E.; Xiaoli, L.; Dudley, E.G.; Kovac, J. Novel Effective Bacillus cereus Group Species “Bacillus clarus” Is Represented by Antibiotic-Producing Strain ATCC 21929 Isolated from Soil. mSphere 2020, 5, e00882-20. [Google Scholar] [CrossRef]
- van Schaik, W.; Château, A.; Dillies, M.-A.; Coppée, J.-Y.; Sonenshein, A.L.; Fouet, A. The Global Regulator CodY Regulates Toxin Gene Expression in Bacillus anthracis and Is Required for Full Virulence. Infect. Immun. 2009, 77, 4437–4445. [Google Scholar] [CrossRef]
- Sastalla, I.; Fattah, R.; Coppage, N.; Nandy, P.; Crown, D.; Pomerantsev, A.P.; Leppla, S.H. The Bacillus cereus Hbl and Nhe Tripartite Enterotoxin Components Assemble Sequentially on the Surface of Target Cells and Are Not Interchangeable. PLoS ONE 2013, 8, e76955. [Google Scholar] [CrossRef]
- Prince, C.; Kovac, J. Regulation of Enterotoxins Associated with Bacillus cereus sensu lato Toxicoinfection. Appl. Environ. Microbiol. 2022, 88, e00405-22. [Google Scholar] [CrossRef]
- Tohya, M.; Hishinuma, T.; Watanabe, S.; Shimojima, M.; Ogawa, M.; Tada, T.; Kirikae, T. Three Novel Species of the Bacillus cereus Group Isolated from Clinical Samples in Japan. Int. J. Syst. Evol. Microbiol. 2021, 71, 004993. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.E.; Malovichko, Y.V.; Nizhnikov, A.A.; Antonets, K.S. Current Methods for Recombination Detection in Bacteria. Int. J. Mol. Sci. 2022, 23, 6257. [Google Scholar] [CrossRef]
- Van Rossum, T.; Ferretti, P.; Maistrenko, O.M.; Bork, P. Diversity within Species: Interpreting Strains in Microbiomes. Nat. Rev. Microbiol. 2020, 18, 491–506. [Google Scholar] [CrossRef]
- Wright, E.S.; Baum, D.A. Exclusivity Offers a Sound yet Practical Species Criterion for Bacteria despite Abundant Gene Flow. BMC Genom. 2018, 19, 724. [Google Scholar] [CrossRef] [PubMed]
- Travers, R.S.; Martin, P.A.W.; Reichelderfer, C.F. Selective Process for Efficient Isolation of Soil Bacillus spp. Appl. Environ. Microbiol. 1987, 53, 1263–1266. [Google Scholar] [CrossRef]
- Antoniou, P.; Hamilton, J.; Koopman, B.; Jain, R.; Holloway, B.; Lyberatos, G.; Svoronos, S.A. Effect of Temperature and Ph on the Effective Maximum Specific Growth Rate of Nitrifying Bacteria. Water Res. 1990, 24, 97–101. [Google Scholar] [CrossRef]
- Reynolds, E.S. The Use of Lead Citrate at High PH as an Electron-Opaque Stain in Electron Microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Kosolapova, A.O.; Belousov, M.V.; Sulatsky, M.I.; Tsyganova, A.V.; Sulatskaya, A.I.; Bobylev, A.G.; Shtark, O.Y.; Tsyganov, V.E.; Volkov, K.V.; Zhukov, V.A.; et al. RopB Protein of Rhizobium leguminosarum bv. viciae Adopts Amyloid State during Symbiotic Interactions with Pea (Pisum sativum L.). Front. Plant Sci. 2022, 13, 1014699. [Google Scholar] [CrossRef]
- Ellis, E.A. Poststaining Grids for Transmission Electron Microscopy. In Electron Microscopy, Methods and Protocols; Humana Press: Totowa, NJ, USA, 2007; pp. 97–106. [Google Scholar]
- Stewart, G.S.; Johnstone, K.; Hagelberg, E.; Ellar, D.J. Commitment of Bacterial Spores to Germinate A Measure of the Trigger Reaction. Biochem. J. 1981, 198, 101–106. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Steward, G.F.; Martinez, J.; Azam, F. A Simple, Rapid Method for Demonstrating Bacterial Flagella. Appl. Environ. Microbiol. 2000, 66, 3632–3636. [Google Scholar] [CrossRef]
- Aygan, A.; Arikan, B. An Overview on Bacterial Motility Detection. Int. J. Agric. Biol. 2007, 9, 193–196. [Google Scholar]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Bochner, B.R. Sleuthing out Bacterial Identities. Nature 1989, 339, 157–158. [Google Scholar] [CrossRef] [PubMed]
- Wragg, P.; Randall, L.; Whatmore, A.M. Comparison of Biolog GEN III MicroStation Semi-Automated Bacterial Identification System with Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and 16S Ribosomal RNA Gene Sequencing for the Identification of Bacteria of Veterinary Interest. J. Microbiol. Methods 2014, 105, 16–21. [Google Scholar] [CrossRef]
- Chojniak, J.; Jałowiecki, Ł.; Dorgeloh, E.; Hegedusova, B.; Ejhed, H.; Magnér, J.; Płaza, G. Application of the BIOLOG System for Characterization of Serratia marcescens ss marcescens Isolated from Onsite Wastewater Technology (OSWT). Acta Biochim. Pol. 2015, 62, 799–805. [Google Scholar] [CrossRef]
- Solovchenko, A.; Gorelova, O.; Selyakh, I.; Pogosyan, S.; Baulina, O.; Semenova, L.; Chivkunova, O.; Voronova, E.; Konyukhov, I.; Scherbakov, P.; et al. A Novel CO2-Tolerant Symbiotic Desmodesmus (Chlorophyceae, Desmodesmaceae): Acclimation to and Performance at a High Carbon Dioxide Level. Algal Res. 2015, 11, 399–410. [Google Scholar] [CrossRef]
- Kates, M. Techniques of Lipidology: Analysis and Identification of Lipids, 2nd Revised ed.; Elsevier: Amsterdam, The Netherlands, 1986; ISBN 0444807322. [Google Scholar]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, M.N.; Nesterenko, M.A.; Shikov, A.E.; Nizhnikov, A.A.; Antonets, K.S. Draft Genome Sequence Data of Lysinibacillus sphaericus Strain 1795 with Insecticidal Properties. Data 2023, 8, 167. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 17 April 2024).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, H.; Liu, M.; Zhang, W.; Song, H.; Lan, H.; Wei, Y.; Niu, B.; Schmidt, B.; Liu, W. RabbitQC: High-Speed Scalable Quality Control for Sequencing Data. Bioinformatics 2021, 37, 573–574. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Barrett, T.; Clark, K.; Gevorgyan, R.; Gorelenkov, V.; Gribov, E.; Karsch-Mizrachi, I.; Kimelman, M.; Pruitt, K.D.; Resenchuk, S.; Tatusova, T.; et al. BioProject and BioSample Databases at NCBI: Facilitating Capture and Organization of Metadata. Nucleic Acids Res. 2012, 40, D57–D63. [Google Scholar] [CrossRef] [PubMed]
- Tonkin-Hill, G.; MacAlasdair, N.; Ruis, C.; Weimann, A.; Horesh, G.; Lees, J.A.; Gladstone, R.A.; Lo, S.; Beaudoin, C.; Floto, R.A.; et al. Producing Polished Prokaryotic Pangenomes with the Panaroo Pipeline. Genome Biol. 2020, 21, 180. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-Sites: Rapid Efficient Extraction of SNPs from Multi-FASTA Alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using Ggtree to Visualize Data on Tree-like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Sand, A.; Holt, M.K.; Johansen, J.; Brodal, G.S.; Mailund, T.; Pedersen, C.N.S. TqDist: A Library for Computing the Quartet and Triplet Distances between Binary or General Trees. Bioinformatics 2014, 30, 2079–2080. [Google Scholar] [CrossRef]
- Didelot, X.; Wilson, D.J. ClonalFrameML: Efficient Inference of Recombination in Whole Bacterial Genomes. PLoS Comput. Biol. 2015, 11, e1004041. [Google Scholar] [CrossRef]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard Operating Procedure for Calculating Genome-to-Genome Distances Based on High-Scoring Segment Pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable Analysis of Bacterial Genome Variation at the Population Level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, J.; Bo, D.; Yu, Y.; Ye, W.; Peng, D.; Sun, M. BtToxin_Digger: A Comprehensive and High-Throughput Pipeline for Mining Toxin Protein Genes from Bacillus thuringiensis. Bioinformatics 2021, 38, 250–251. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Valerio, S.; Lev Hacohen, A.; Schöppe, R.; Liesegang, H. IDOPS, a Profile HMM-Based Tool to Detect Pesticidal Sequences and Compare Their Genetic Context. Front. Microbiol. 2021, 12, 1471. [Google Scholar] [CrossRef] [PubMed]
- Shikov, A.E.; Malovichko, Y.V.; Skitchenko, R.K.; Nizhnikov, A.A.; Antonets, K.S. No More Tears: Mining Sequencing Data for Novel Bt Cry Toxins with CryProcessor. Toxins 2020, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Subramaniyam, S.; Kim, C.-B.; Manavalan, B. SortPred: The First Machine Learning Based Predictor to Identify Bacterial Sortases and Their Classes Using Sequence-Derived Information. Comput. Struct. Biotechnol. J. 2022, 20, 165–174. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb.prot095505. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
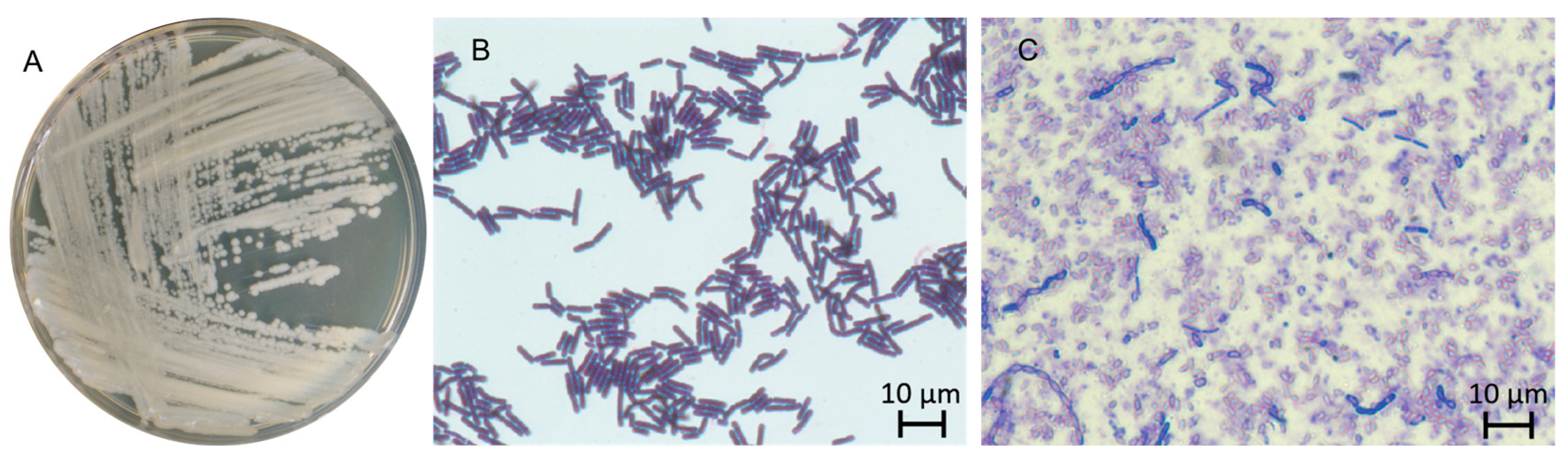

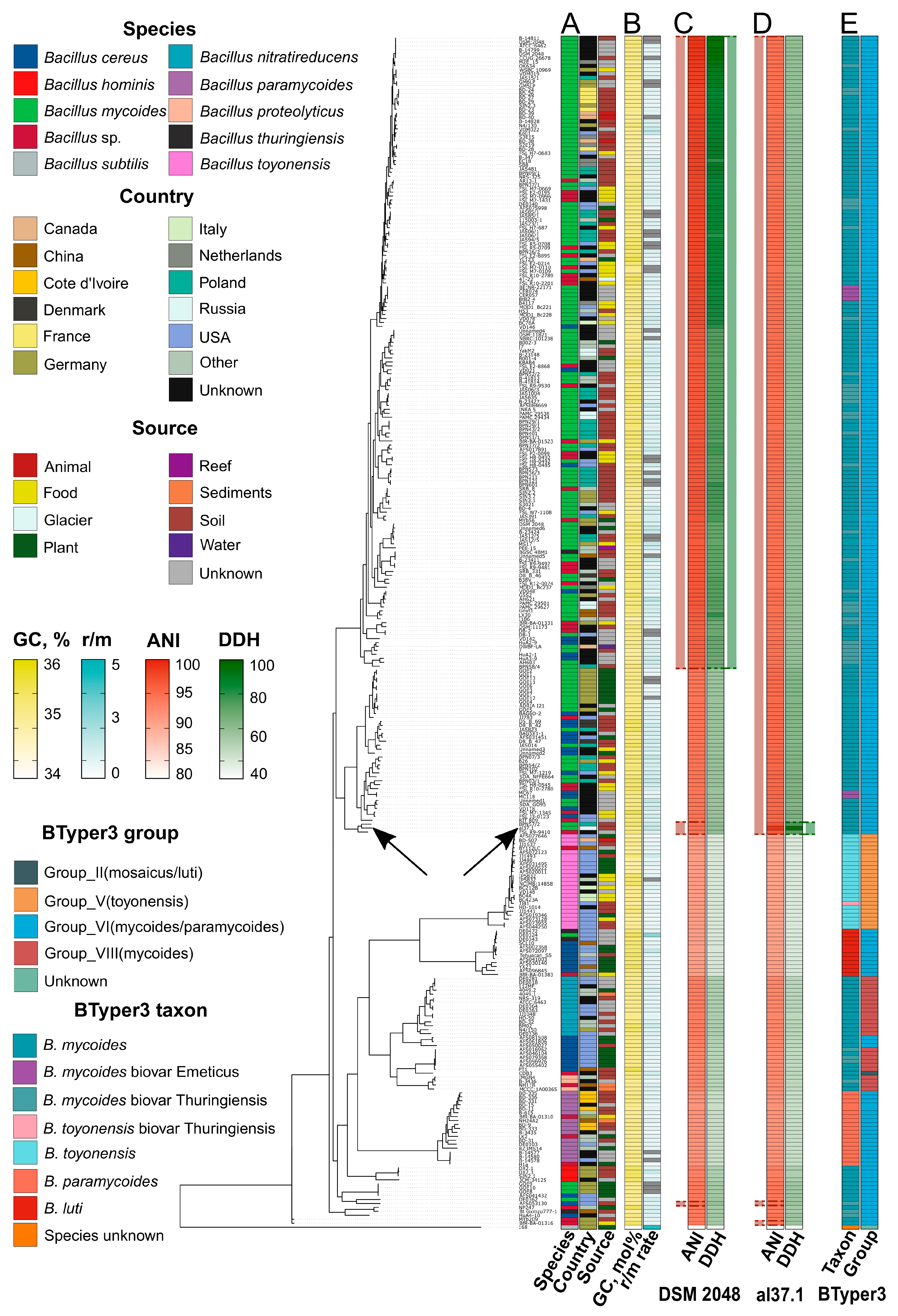
| Feature/Strain | DSM 2048T | al37.1T | NCIMB 14858 (BCT-7112T) | 4049T | MCCC 1A00365 (TD42T) | NH24A2T | JCM 34125 (BML-BC059T) | 168 |
|---|---|---|---|---|---|---|---|---|
| Assembly | GCF_029024805.1 | GCF_042136375.1 | GCF_000496285.1 | GCF_001884135.1 | GCF_001884065.1 | GCF_001884235.1 | GCF_018332515.1 | GCF_964017255.1 |
| Species | B. mycoides | Bacillus altaicus sp. nov | B. toyonensis | B. nitratireducens | B. proteolyticus | B. paramycoides | B. hominis | B. subtilis |
| BTyper3 taxon | B. mycoides | B. mycoides | B. toyonensis | B. mycoides | B. mycoides | B. paramycoides | B. mycoides | Unknown |
| Genome length, bp | 5,639,253 | 5,397,466 | 5,025,419 | 5,480,833 | 5,847,531 | 5,444,255 | 5,192,534 | 4,215,606 |
| Number of genes | 5817 | 5458 | 5198 | 5554 | 5950 | 5501 | 5239 | 4332 |
| GC content, % | 35.4 | 35.5 | 35.6 | 35.3 | 35.2 | 35.2 | 35.5 | 43.5 |
| ANI (DSM 2048T) | - | 95.3 | 90.8 | 93.9 | 93 | 91.5 | 93.7 | 78.1 |
| DDH (DSM 2048T) | - | 61.6 | 41.5 | 53.4 | 50.3 | 45.7 | 53.2 | 34.2 |
| 16S rRNA similarity (DSM 2048T) | - | 99.9 | 99.4 | 99.2 | 99.6 | 99.4 | 99.7 | 93.7 |
| ANI (al37.1T) | 95.3 | - | 90.2 | 93 | 92.5 | 91.7 | 94 | 76 |
| DDH (al37.1T) | 61.6 | - | 40.1 | 49.7 | 48.6 | 46.1 | 54.1 | 31.7 |
| 16S rRNA similarity (al37.1T) | 99.9 | - | 99.5 | 99.4 | 99.7 | 99.6 | 99.8 | 93.7 |
| gyrB similarity (al37.1T) | 97.77 | - | 90.15 | 91.19 | 91.14 | 93.99 | 95.9 | 70.98 |
| gyrB similarity (DSM 2048T) | - | 97.77 | 91.08 | 91.65 | 91.76 | 93.57 | 95.33 | 70.67 |
| MLST complex | 116 | Unknown | 111 | 769 | 765 | 780 | 3200 | 1 |
| Carbohydrates and Derivatives | Alcohols | Amino Acids | Organic Acids and Derivatives | Other Compounds |
|---|---|---|---|---|
| D-Maltose D-Trehalose D-Cellobiose D-Turanose N-Acetyl-β-D-Mannosamine β-Methyl-D-Glucoside N-Acetyl-D-Glucosamine N-Acetyl-D-Galactosamine α-D-Glucose D-Mannose D-Fructose D-Galactose L-Rhamnose L-Fucose D-Fucose D-Glucose-6-PO4 D-Fructose-6-PO4 3-Methyl Glucose D-Salicin | D-Arabitol myo-Inositol Glycerol | D-Serine L-Arginine L-Histidine L-Alanine L-Aspartic Acid L-Glutamic Acid L-Serine | L-Galactonic Acid Lactone D-Gluconic Acid D-Glucuronic Acid Glucuronamide Quinic Acid D-Saccharic Acid p-Hydroxy-Phenylacetic Acid Methyl Pyruvate D-Lactic Acid Methyl Ester L-Lactic Acid α-Ketoglutaric Acid L-Malic Acid Bromo-Succinic Acid α-Hydroxy-Butyric Acid β-Hydroxy-D, L-Butyric Acid α-Keto-Butyric Acid Propionic Acid Acetic Acid Formic Acid | Inosine Glycyl-L-Proline |
| Characteristic | B. altaicus sp. nov. al37.1T | B. mycoides DSM 2048T | B. toyonensis BCT-7112T |
|---|---|---|---|
| Cell length (μm) | 1.5–4.3 | 3.0–5.0 | 3.0–4.4 |
| Cell width (μm) | 0.6–1.8 | >1 | >1 |
| Rhyzoidal colony | − | + | − |
| Parasporal crystal | − | − | − |
| Temp range (°C) | +20–40 | +15–40 | +10–45 |
| Optimal temp (°C) | +30 | +30 | +35 |
| pH range | 6–9 | 5–9.5 | 5–9.5 |
| Optimal pH | 6.5 | 8 | 6.5 |
| NaCl range (%, wt/vol) | 0–1% | 0–4 | 0–5 |
| Optimal NaCl concentration (%, wt/vol) | 0.5 | 1 | 0 |
| Catalase test | + | + | + |
| Urease test | − | − | − |
| Cellular fatty acids (% of total) | |||
| 12:0 | 4.3 | 2.7 | N/D |
| 14:0 | 9.6 | 3.7 | 3.2 |
| 16:0 | 34.8 | 15.6 | 5.6 |
| 18:0 | 7.9 | 1.6 | N/D |
| 18:1 | 15.2 | N/D | N/D |
| Carbon source utilization | |||
| Glycerol | + | − | − |
| D-Mannose | + | − | − |
| D-Salicin | + | + | + |
| D-Cellobiose | + | Weakly positive | − |
| Sucrose | − | + | + |
| D-Trehalose | + | + | + |
| D-Turanose | + | − | + |
| D-Fructose | + | + | + |
| D-Maltose | + | + | + |
| Gentiobiose | − | − | − |
| L-Rhamnose | + | + | − |
| D-Sorbitol | − | + | − |
| D-Melibiose | − | + | − |
| D-Raffinose | − | + | − |
| D-Fucose | + | + | − |
| L-Fucose | + | + | − |
| D-Arabitol | + | + | − |
| D-Galactose | + | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shikov, A.E.; Romanenko, M.N.; Shmatov, F.M.; Belousov, M.V.; Solovchenko, A.; Chivkunova, O.; Savelev, G.K.; Kuznetsova, I.G.; Karlov, D.S.; Nizhnikov, A.A.; et al. Proposal of Bacillus altaicus sp. nov. Isolated from Soil in the Altai Region, Russia. Int. J. Mol. Sci. 2025, 26, 9517. https://doi.org/10.3390/ijms26199517
Shikov AE, Romanenko MN, Shmatov FM, Belousov MV, Solovchenko A, Chivkunova O, Savelev GK, Kuznetsova IG, Karlov DS, Nizhnikov AA, et al. Proposal of Bacillus altaicus sp. nov. Isolated from Soil in the Altai Region, Russia. International Journal of Molecular Sciences. 2025; 26(19):9517. https://doi.org/10.3390/ijms26199517
Chicago/Turabian StyleShikov, Anton E., Maria N. Romanenko, Fedor M. Shmatov, Mikhail V. Belousov, Alexei Solovchenko, Olga Chivkunova, Grigoriy K. Savelev, Irina G. Kuznetsova, Denis S. Karlov, Anton A. Nizhnikov, and et al. 2025. "Proposal of Bacillus altaicus sp. nov. Isolated from Soil in the Altai Region, Russia" International Journal of Molecular Sciences 26, no. 19: 9517. https://doi.org/10.3390/ijms26199517
APA StyleShikov, A. E., Romanenko, M. N., Shmatov, F. M., Belousov, M. V., Solovchenko, A., Chivkunova, O., Savelev, G. K., Kuznetsova, I. G., Karlov, D. S., Nizhnikov, A. A., & Antonets, K. S. (2025). Proposal of Bacillus altaicus sp. nov. Isolated from Soil in the Altai Region, Russia. International Journal of Molecular Sciences, 26(19), 9517. https://doi.org/10.3390/ijms26199517








