Integrating Inflammatory and Epigenetic Signatures in IBD-Associated Colorectal Carcinogenesis: Models, Mechanisms, and Clinical Implications
Abstract
1. Introduction
1.1. The Impact of Chronic Inflammation in Carcinogenesis
1.2. Inflammatory Bowel Disease
2. Infectious Agents, Immune Responses, and the Inflammatory Basis of Gastrointestinal Carcinogenesis
2.1. Infection and Host Defense Mechanisms
2.1.1. The Physiology of Infection: A Brief Overview
2.1.2. Infection Stages
2.1.3. The Immune Response
2.1.4. Factors Influencing the Outcome of the Infection
2.1.5. Physiological Human-Bacteria Interactions
2.2. Pathogen Evasion and Immune Dysregulation
2.2.1. Antigens and Subversion of Immune Response
2.2.2. Bacterial Infection and Immune Dysregulation
2.3. Infectious Agents Linked to GI Inflammation and Cancer
2.3.1. Mycobacterium tuberculosis and GI Inflammation
2.3.2. Other Bacterial Drivers of GI Carcinogenesis
2.3.3. The Involvement of Microbes in the Mechanisms of Carcinogenesis
2.3.4. Effect of Bacterial Infection on Gastrointestinal Cancer
2.3.5. Colorectal Cancer
2.4. Chronic Inflammation as a Driver of Colorectal Carcinogenesis
2.5. Mechanistic and Historical Perspectives on Inflammation-Induced Cancer
Inflammation-Induced Cancers Associated with the GI Tract
- Persistent inflammation promotes epigenetic changes, including:
- DNA methylation of tumor suppressor genes (e.g., MLH1, CDKN2A/p16).
- Histone modifications such as hypoacetylation and trimethylation of histone H3 on lysine 27 (H3K27me3).
- Anti-inflammatory agents, such as mesalamine (5-ASA) and infliximab, which reduce inflammatory cytokine activity.
- Epigenetic drugs, including decitabine (a DNA methyltransferase inhibitor) and vorinostat (a histone deacetylase inhibitor), are being explored for their potential to reverse aberrant epigenetic states in cancer and inflammation.
- Tumor suppressors and mismatch repair genes: APC (5q22.2), MLH1 (3p22.2), MSH2 (2p21-16.3), MSH6 (2p16.3), PMS2 (7p22.1).
- Polymerases and modifiers: POLD 1 (19q13.33), POLE.
- Other associated loci: MUTYH (1p34.1), EPCAM (2p21), GREM1 (15q13.3).
3. Animal Models of IBD and Colitis-Associated Colorectal Cancer (CAC)
3.1. Murine Models of Gastrointestinal Cancer (GIC) Through Pathogen Infection
3.2. Non-Infectious Animal Models for Gastrointestinal Cancer (GIC)
3.2.1. Chemically Induced Models: AOM/DSS
3.2.2. Genetically Engineered Mouse Models (GEMMs)
3.2.3. Xenograft and Patient-Derived Xenograft (PDX) Models
4. Overview of Chromatin and Epigenetic Modulations
4.1. Integration of Epigenetic Alterations and Inflammatory Pathways
4.2. Epigenetic Alterations in Inflammation-Associated Pathologies of the GI Tract—An Overview
5. Epigenetic Mechanisms Linking Inflammation in IBD to Colorectal Carcinogenesis
An Overview of Inflammation-Driven Carcinogenic Transition in Humans and Murine Models
6. DNA Methylation and Histone Modifications During Tumorigenesis of the GI
6.1. DNA Methylation in IBD, CAC and CRC
6.2. Histone Modifications in Inflammation-Related Cancer Progression
6.2.1. Histone Acetylation in CRC
6.2.2. Histone Methylation in Inflammatory Signaling and CRC Progression
6.2.3. Histone Phosphorylation in CRC
7. Exploring the Role of Non-Coding RNAs in Epigenetic Regulation of IBD and CRC
7.1. MicroRNAs—Molecular Insights of microRNA Dysregulation or Aberrant Function and Its Involvement in IBD and CRC
7.2. MicroRNAs as Biomarkers in GI Diseases
7.3. LncRNAs in IBD and CRC—A General Overview
LncRNA as Biomarkers in GI Diseases
7.4. Other ncRNAs in GI Diseases
7.4.1. PiRNAs
7.4.2. CircRNAs
7.4.3. Functions and Implications of circRNAs
7.4.4. Circular RNA-Protein Interactions: Functional Significance and Binding Site Density
7.5. Inflammation-Driven ncRNA Modulation in CRC
8. Diagnostic and Therapeutic Implications: Biomarkers and Epigenetic Drugs
8.1. Translational Epigenetics and RNA-Based Therapeutics in IBD/CRC
8.2. Epigenetic Drugs and Clinical Trials in IBD-Associated CRC
8.3. RNA-Based Therapeutics in Preclinical and Clinical Use
8.4. Diagnostic and Prognostic Utility of Epigenetic and RNA Biomarkers
8.5. Comparative Human-Mouse Evidence
9. Concluding Remarks and Future Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AOM/DSS | azoxymethane/dextran sulfate sodium |
| APC | adenomatous polyposis coli |
| CAC | colitis-associated cancer |
| CD | Crohn’s disease |
| circRNA | circular RNA |
| CRC | colorectal cancer |
| CRP | C-reactive protein |
| EMT | epithelial–mesenchymal transition |
| GEMM | genetically engineered mouse models |
| GI | gastrointestinal |
| GIC | gastrointestinal cancer |
| GIT | gastrointestinal tract |
| GITB | gastrointestinal tuberculosis |
| IBD | inflammatory bowel disease |
| IECs | intestinal epithelial cells |
| ILCs | innate lymphoid cells |
| lincRNA | long intergenic RNA |
| lncRNA | long non-coding RNA |
| MTBC | Mycobacterium tuberculosis complex |
| ncRNA | non-coding RNA |
| NFκB | nuclear factor-kappa B |
| piRNA | PIWI-interacting RNA |
| PDX | patient-derived xenografts |
| RBP | RNA-binding protein |
| TB | tuberculosis |
| TF | transcription factor |
| TSS | transcription start site |
| UC | ulcerative colitis |
References
- Chen, G.Y.; Nunez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Callaway, J.B.; Ting, J.P. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 2015, 21, 677–687. [Google Scholar] [CrossRef]
- Zhang, S.; Meng, Y.; Zhou, L.; Qiu, L.; Wang, H.; Su, D.; Zhang, B.; Chan, K.-M.; Han, J. Targeting epigenetic regulators for inflammation: Mechanisms and intervention therapy. MedComm 2022, 3, e173. [Google Scholar] [CrossRef]
- Tan, S.Y.X.; Zhang, J.; Tee, W.W. Epigenetic Regulation of Inflammatory Signaling and Inflammation-Induced Cancer. Front. Cell Dev. Biol. 2022, 10, 931493. [Google Scholar] [CrossRef]
- Das, D.; Karthik, N.; Taneja, R. Crosstalk between inflammatory signaling and methylation in cancer. Front. Cell Dev. Biol. 2021, 9, 756458. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Mishra, S.R.; Mahapatra, K.K.; Behera, B.P.; Bhol, C.S.; Praharaj, P.P.; Panigrahi, D.P.; Patra, S.; Singh, A.; Patil, S.; Dhiman, R.; et al. Inflammasomes in cancer: Effect of epigenetic and autophagic modulations. Semin. Cancer Biol. 2022, 83, 399–412. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Hou, J.; Zhu, B.; Min, Z.; Zhang, M.; Song, D.; Cheng, Y.; Wang, X. Targeting roles of inflammatory microenvironment in lung cancer and metastasis. Cancer Metastasis Rev. 2015, 34, 319–331. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Vieujean, S.; Caron, B.; Haghnejad, V.; Jouzeau, J.Y.; Netter, P.; Heba, A.C.; Ndiaye, N.C.; Moulin, D.; Barreto, G.; Danese, S.; et al. Impact of the exposome on the epigenome in inflammatory bowel disease patients and animal models. Int. J. Mol. Sci. 2022, 23, 7611. [Google Scholar] [CrossRef]
- Zeng, Z.; Mukherjee, A.; Zhang, H. From genetics to epigenetics, roles of epigenetics in inflammatory bowel disease. Front. Genet. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Gerbeth, L.; Glauben, R. Histone Deacetylases in the inflamed intestinal epithelium-Promises of new therapeutic strategies. Front. Med. 2021, 8, 655956. [Google Scholar] [CrossRef]
- Jawad, N.; Direkze, N.; Leedham, S.J. Inflammatory bowel disease and colon cancer. Recent Results Cancer Res. 2011, 185, 99–115. [Google Scholar] [CrossRef]
- Thomson, P.D.; Smith, D.J., Jr. What is infection? Am. J. Surg. 1994, 167, S7–S11, discussion 10S–11S. [Google Scholar] [CrossRef]
- Qerqez, A.N.; Silva, R.P.; Maynard, J.A. Outsmarting pathogens with antibody engineering. Annu. Rev. Chem. Biomol. Eng. 2023, 14, 217–241. [Google Scholar] [CrossRef]
- van Seventer, J.M.; Hochberg, N.S. Principles of infectious diseases: Transmission, diagnosis, prevention, and control. In International Encyclopedia of Public Health, 2nd ed.; Quah, S.R., Ed.; Elsevier: Cambridge, MA, USA, 2017; pp. 22–39. [Google Scholar] [CrossRef]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. Innate Immunity. In Fenner and White’s Medical Virology, 5th ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 57–64. [Google Scholar] [CrossRef]
- Schertzer, J.D.; Lam, T.K.T. Peripheral and central regulation of insulin by the intestine and microbiome. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E234–E239. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target Ther. 2021, 6, 291. [Google Scholar] [CrossRef] [PubMed]
- Carretta, M.D.; Quiroga, J.; López, R.; Hidalgo, M.A.; Burgos, R.A. Participation of short-chain fatty acids and their receptors in gut inflammation and colon cancer. Front. Physiol. 2021, 12, 662739. [Google Scholar] [CrossRef]
- Madison, A.; Kiecolt-Glaser, J.K. Stress, depression, diet, and the gut microbiota: Human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr. Opin. Behav. Sci. 2019, 28, 105–110. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Abu-Ali, G.; Huttenhower, C. The healthy human microbiome. Genome Med. 2016, 8, 51. [Google Scholar] [CrossRef]
- Finlay, B.B.; McFadden, G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef] [PubMed]
- Martini, G.R.; Tikhonova, E.; Rosati, E.; DeCelie, M.B.; Sievers, L.K.; Tran, F.; Lessing, M.; Bergfeld, A.; Hinz, S.; Nikolaus, S.; et al. Selection of cross-reactive T cells by commensal and food-derived yeasts drives cytotoxic TH1 cell responses in Crohn’s disease. Nat. Med. 2023, 29, 2602–2614. [Google Scholar] [CrossRef] [PubMed]
- Giamarellos-Bourboulis, E.J.; Raftogiannis, M. The immune response to severe bacterial infections: Consequences for therapy. Expert Rev. Anti. Infect. Ther. 2012, 10, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Al-Zanbagi, A.B.; Shariff, M.K. Gastrointestinal tuberculosis: A systematic review of epidemiology, presentation, diagnosis and treatment. Saudi J. Gastroenterol. 2021, 27, 261–274. [Google Scholar] [CrossRef]
- Choudhury, A.; Dhillon, J.; Sekar, A.; Gupta, P.; Singh, H.; Sharma, V. Differentiating gastrointestinal tuberculosis and Crohn’s disease- a comprehensive review. BMC Gastroenterol. 2023, 23, 246. [Google Scholar] [CrossRef]
- Eraksoy, H. Gastrointestinal and abdominal tuberculosis. Gastroenterol. Clin. N. Am. 2021, 50, 341–360. [Google Scholar] [CrossRef]
- Malikowski, T.; Mahmood, M.; Smyrk, T.; Raffals, L.; Nehra, V. Tuberculosis of the gastrointestinal tract and associated viscera. J. Clin. Tuberc. Other Mycobact. Dis. 2018, 12, 1–8, Erratum in: J. Clin. Tuberc. Other Mycobact. Dis. 2020, 21, 100177. https://doi.org/10.1016/j.jctube.2020.100177. [Google Scholar] [CrossRef]
- Gopalaswamy, R.; Dusthackeer, V.N.A.; Kannayan, S.; Subbian, S. Extrapulmonary Tuberculosis—An update on the diagnosis, treatment and drug resistance. J. Resp. 2021, 1, 141–164. [Google Scholar] [CrossRef]
- Choi, E.; Coyle, W. Gastrointestinal tuberculosis. In Tuberculosis and Nontuberculous Mycobacterial Infections, 7th ed.; Schlossberg, D., Ed.; Wiley: Washington, DC, USA, 2017; pp. 411–432. [Google Scholar] [CrossRef]
- Dasgupta, A.; Singh, N.; Bhatia, A. Abdominal tuberculosis: A histopathological study with special reference to intestinal perforation and mesenteric vasculopathy. J. Lab. Physicians 2009, 1, 56–61. [Google Scholar] [CrossRef]
- Maulahela, H.; Simadibrata, M.; Nelwan, E.J.; Rahadiani, N.; Renesteen, E.; Suwarti, S.W.T.; Anggraini, Y.W. Recent advances in the diagnosis of intestinal tuberculosis. BMC Gastroenterol. 2022, 22, 89. [Google Scholar] [CrossRef]
- Chatzicostas, C.; Koutroubakis, I.E.; Tzardi, M.; Roussomoustakaki, M.; Prassopoulos, P.; Kouroumalis, E.A. Colonic tuberculosis mimicking Crohn’s disease: Case report. BMC Gastroenterol. 2002, 2, 10. [Google Scholar] [CrossRef]
- Arhan, M.; Köksal, A.Ş.; Özin, Y.; Mesut, Z.; Kılıç, Y.; Tunç, B.; Ülker, A. Colonic tuberculosis or Crohn’s disease ? An important differential diagnosis. Acta Gastroenterol. Belg. 2013, 76, 59–61. [Google Scholar]
- González-Puga, C.; Palomeque-Jiménez, A.; García-Saura, P.L.; Pérez-Cabrera, B.; Jiménez-Ríos, J.A. Colonic tuberculosis mimicking Crohn’s disease: An exceptional cause of massive surgical rectal bleeding. Med. Mal. Infect. 2015, 45, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, G.; Buda, B.; Shah, J.K.; Ghimire, B.; Kansaker, P.B.S. Colonic tuberculosis masquerading as ascending colon carcinoma in a patient of FIGO Stage IIB cervical carcinoma following chemo-radiotherapy: A case report. Int. J. Surg. Case Rep. 2022, 93, 106943. [Google Scholar] [CrossRef]
- Luczynski, P.; Poulin, P.; Romanowski, K.; Johston, J.C. Tuberculosis and risk of cancer: A systematic review and meta-analysis. PLoS ONE 2022, 17, e0278661. [Google Scholar] [CrossRef] [PubMed]
- Saidu, A.S.; Okolocha, E.C.; Gamawa, A.A.; Babashani, M.; Bakari, N.A. Occurrence and distribution of bovine tuberculosis (Mycobacterium bovis) in slaughtered cattle in the abattoirs of Bauchi State, Nigeria. Vet. World 2015, 8, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Luboya, L.W.; Malangu, M.; Kaleka, M.; Ngulu, N.; Nkokele, B.; Maryabo, K.; Pourrut, X.; Vincent, T.; Gonzalez, J.P. An assessment of caprine tuberculosis prevalence in Lubumbashi slaughterhouse, Democratic Republic of Congo. Trop. Anim. Health Prod. 2017, 49, 875–878. [Google Scholar] [CrossRef]
- Engelmann, N.; Ondreka, N.; Michalik, J.; Neiger, R. Intra-abdominal Mycobacterium tuberculosis infection in a dog. J. Vet. Intern. Med. 2014, 28, 934–938. [Google Scholar] [CrossRef]
- Mentula, S.; Karkamo, V.; Skrzypczak, T.; Seppänen, J.; Hyyryläinen, H.L.; Haanperä, M.; Soini, H. Emerging source of infection—Mycobacterium tuberculosis in rescue dogs: A case report. Access Microbiol. 2020, 2, acmi000168. [Google Scholar] [CrossRef]
- Ribeiro, M.G.; Lima, M.C.F.; Franco, M.M.J.; Megid, J.; Soares, L.M.; Machado, L.H.A.; Miyata, M.; Pavan, F.R.; Heinemann, M.B.; Souza Filho, A.F.; et al. Pre-multidrug-resistant Mycobacterium tuberculosis infection causing fatal enteric disease in a dog from a family with history of human tuberculosis. Transbound. Emerg. Dis. 2017, 64, e4–e7. [Google Scholar] [CrossRef]
- Dhama, K.; Mahendran, M.; Tiwari, R.; Dayal Singh, S.; Kumar, D.; Singh, S.; Sawant, P.M. Tuberculosis in Birds: Insights into the Mycobacterium avium Infections. Vet. Med. Int. 2011, 2011, 712369. [Google Scholar] [CrossRef]
- Bertram, C.A.; Barth, S.A.; Glöckner, B.; Lübke-Becker, A.; Klopfleisch, R. Intestinal Mycobacterium avium Infection in Pet Dwarf Rabbits (Oryctolagus cuniculus). J. Comp. Pathol. 2020, 180, 73–78. [Google Scholar] [CrossRef]
- Vetere, A.; Bertocchi, M.; Pagano, T.B.; Di Ianni, F.; Nardini, G. First case of systemic fatal mycobacteriosis caused by Mycobacterium goodii in a pet Kenyan sand boa (Eryx colubrinus loveridgei). BMC Vet. Res. 2022, 18, 291. [Google Scholar] [CrossRef]
- Lindsay, S.A.; Gray, R. A Novel Presentation of tuberculosis with intestinal perforation in a free-ranging Australian sea lion (Neophoca cinerea). J. Wildl. Dis. 2021, 57, 220–224. [Google Scholar] [CrossRef]
- Zella, D.; Gallo, R.C. Viruses and bacteria associated with cancer: An overview. Viruses 2021, 13, 1039. [Google Scholar] [CrossRef]
- Clay, S.L.; Fonseca-Pereira, D.; Garrett, W.S. Colorectal cancer: The facts in the case of the microbiota. J. Clin. Investig. 2022, 132, e155101. [Google Scholar] [CrossRef] [PubMed]
- van Elsland, D.; Neefjes, J. Bacterial infections and cancer. EMBO Rep. 2018, 19, e46632. [Google Scholar] [CrossRef] [PubMed]
- Vogelmann, R.; Amieva, M.R. The role of bacterial pathogens in cancer. Curr. Opin. Microbiol. 2007, 10, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424, Erratum in: CA Cancer J. Clin. 2020, 70, 313. https://doi.org/10.3322/caac.21609. [Google Scholar] [CrossRef] [PubMed]
- Elsalem, L.; Jum’ah, A.A.; Alfaqih, M.A.; Aloudat, O. The Bacterial Microbiota of Gastrointestinal Cancers: Role in Cancer Pathogenesis and Therapeutic Perspectives. Clin. Exp. Gastroenterol. 2020, 13, 151–185. [Google Scholar] [CrossRef] [PubMed]
- Eaden, J.A.; Abrams, K.R.; Mayberry, J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef]
- Jess, T.; Rungoe, C.; Peyrin-Biroulet, L. Risk of colorectal cancer in patients with ulcerative colitis: A meta-analysis of population-based cohort studies. Clin. Gastroenterol. Hepatol. 2012, 10, 639–645. [Google Scholar] [CrossRef]
- Itzkowitz, S.H.; Yio, X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: The role of inflammation. Am. J. Physiol Gastrointest. Liver Physiol. 2004, 287, G7–G17. [Google Scholar] [CrossRef]
- Schroeder, J.H.; Howard, J.K.; Lord, G.M. Transcription factor-driven regulation of ILC1 and ILC3. Trends Immunol. 2022, 43, 564–579. [Google Scholar] [CrossRef]
- Karin, M.; Greten, F.R. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005, 5, 749–759. [Google Scholar] [CrossRef]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef]
- Bollrath, J.; Phesse, T.J.; von Burstin, V.A.; Putoczki, T.; Bennecke, M.; Bateman, T.; Nebelsiek, T.; Lundgren-May, T.; Canli, O.; Schwitalla, S.; et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell 2009, 15, 91–102. [Google Scholar] [CrossRef]
- Colotta, F.; Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 2009, 30, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhao, W.; Tang, S.; Chen, R.; Ji, M.; Yang, X. Role of ILC2s in solid tumors: Facilitate or inhibit? Front. Immunol. 2022, 13, 886045. [Google Scholar] [CrossRef]
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Fujiki, H. Gist of Dr. Katsusaburo Yamagiwa’s papers entitled “Experimental study on the pathogenesis of epithelial tumors” (I to VI reports). Cancer Sci. 2014, 105, 143–149. [Google Scholar] [CrossRef]
- Dvorak, H.F. Tumors: Wounds that do not heal-redux. Cancer Immunol. Res. 2015, 3, 111. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F. Tumors: Wounds that do not heal-a historical perspective with a focus on the fundamental roles of increased vascular permeability and clotting. Semin. Thromb. Hemost. 2019, 45, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Vanoli, A.; Parente, P.; Fassan, M.; Mastracci, L.; Grillo, F. Gut inflammation and tumorigenesis: Every site has a different tale to tell. Intern. Emerg. Med. 2023, 18, 2169–2179. [Google Scholar] [CrossRef]
- Grady, W.M.; Yu, M.; Markowitz, S.D. Epigenetic alterations in the gastrointestinal tract: Current and emerging use for biomarkers of cancer. Gastroenterology 2021, 160, 690–709. [Google Scholar] [CrossRef]
- Filho, A.M.; Laversanne, M.; Ferlay, J.; Colombet, M.; Piñeros, M.; Znaor, A.; Parkin, D.M.; Soerjomataram, I.; Bray, F. The GLOBOCAN 2022 cancer estimates: Data sources, methods, and a snapshot of the cancer burden worldwide. Int. J. Cancer 2025, 156, 1336–1346. [Google Scholar] [CrossRef]
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasaq, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Yang, F.; Li, X.F.; Cheng, L.N.; Li, X.L. Long non-coding RNA CRNDE promotes cell apoptosis by suppressing miR-495 in inflammatory bowel disease. Exp. Cell Res. 2019, 382, 111484. [Google Scholar] [CrossRef]
- Triantaphyllopoulos, K.A. Long non-coding RNAs and their “discrete” contribution to IBD and Johne’s disease-what stands out in the current picture? A comprehensive review. Int. J. Mol. Sci. 2023, 24, 13566. [Google Scholar] [CrossRef]
- Vetrano, S.; Borroni, E.M.; Sarukhan, A.; Savino, B.; Bonecchi, R.; Correale, C.; Arena, V.; Fantini, M.; Roncalli, M.; Malesci, A.; et al. The lymphatic system controls intestinal inflammation and inflammation-associated Colon Cancer through the chemokine decoy receptor D6. Gut 2010, 59, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Pelaseyed, T.; Hansson, G.C. Membrane mucins of the intestine at a glance. J. Cell Sci. 2020, 133, jcs240929. [Google Scholar] [CrossRef]
- Tang, W.; Liu, J.; Ma, Y.; Wei, Y.; Liu, J.; Wang, H. Impairment of intestinal barrier function induced by early weaning via autophagy and apoptosis associated with gut microbiome and metabolites. Front. Immunol. 2021, 12, 804870. [Google Scholar] [CrossRef] [PubMed]
- Bundgaard-Nielsen, C.; Baandrup, U.T.; Nielsen, L.P.; Sørensen, S. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer 2019, 19, 399. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Neufert, C.; Becker, C.; Neurath, M.F. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007, 2, 1998–2004. [Google Scholar] [CrossRef]
- Tanaka, T.; Kohno, H.; Suzuki, R.; Yamada, Y.; Sugie, S.; Mori, H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003, 94, 965–973. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Takeuchi, T.; Himuro, H.; Okada, T.; Mizoguchi, E. Genetically engineered mouse models for studying inflammatory bowel disease. J. Pathol. 2016, 238, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sui, H.; Fang, F.; Li, Q.; Li, B. The application of Apc Min/+ mouse model in colorectal tumor researches. J. Cancer Res. Clin. Oncol. 2019, 145, 1111–1122. [Google Scholar] [CrossRef]
- Moser, A.R.; Pitot, H.C.; Dove, W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990, 247, 322–324. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef]
- Rubio, K.; Dobersch, S.; Barreto, G. Functional interactions between scaffold proteins, noncoding RNAs, and genome loci induce liquid-liquid phase separation as organizing principle for 3-dimensional nuclear architecture: Implications in cancer. FASEB J. 2019, 33, 5814–5822. [Google Scholar] [CrossRef]
- Zhang, Y. Recent progress in the epigenetics and chromatin field. Cell Res. 2011, 21, 373–374. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef]
- Bentley, G.A.; Lewis-Bentley, A.; Finch, J.T.; Podjarny, A.D.; Roth, M. Crystal structure of the nucleosome core particle at 16 A resolution. J. Mol. Biol. 1984, 176, 5–75. [Google Scholar] [CrossRef]
- Richmond, T.J.; Finch, J.T.; Rushton, B.; Rhodes, D.; Klug, A. Structure of the nucleosome core particle at 7Å resolution. Nature 1984, 311, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, Y.; Xu, J.; Shao, Y.; Xing, D. Unlocking the potential of targeting histone-modifying enzymes for treating IBD and CRC. Clin. Epigenetics 2023, 15, 146. [Google Scholar] [CrossRef]
- Singh, I.; Contreras, A.; Cordero, J.; Rubio, K.; Dobersch, S.; Gunther, S.; Jeratsch, S.; Mehta, A.; Krüger, M.; Graumann, J.; et al. MiCEE is a ncRNA-protein complex that mediates epigenetic silencing and nucleolar organization. Nat. Genet. 2018, 50, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2018, 10, 295–304. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Environmental epigenetics and a unified theory of the molecular aspects of evolution: A neo-lamarckian concept that facilitates neo-darwinian evolution. Genome Biol. Evol. 2015, 7, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Kealy, L.; Runting, J.; Thiele, D.; Scheer, S. An emerging maestro of immune regulation: How DOT1L orchestrates the harmonies of the immune system. Front. Immunol. 2024, 15, 1385319. [Google Scholar] [CrossRef]
- Dobersch, S.; Rubio, K.; Barreto, G. Pioneer factors and architectural proteins mediating embryonic expression signatures in cancer. Trends Mol. Med. 2019, 25, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhao, J.; Lv, Y.; Wang, W.; Feng, C.; Zou, W.; Su, L.; Jiao, J. Histone variants and histone modifications in neurogenesis. Trends Cell Biol. 2020, 30, 869–880. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Hake, S.B.; Xiao, A.; Allis, C.D. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br. J. Cancer 2007, 96, R31–R39. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Mattiroli, F.; Penengo, L. Histone Ubiquitination: An integrative signaling platform in genome stability. Trends Genet. 2021, 37, 566–581. [Google Scholar] [CrossRef]
- Andrews, A.J.; Luger, K. Nucleosome structure(s) and stability: Variations on a theme. Annu. Rev. Biophys. 2011, 40, 99–117. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifcations and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 93, 1074–1080. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Zhao, Y.T.; Ting, C.; Zhao, T.C. Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 2015, 20, 35–47. [Google Scholar] [CrossRef]
- Yang, Z.H.; Dang, Y.-Q.; Ji, G. Role of epigenetics in transformation of inflammation into colorectal cancer. World J. Gastroenterol. 2019, 25, 2863–2877. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Schubeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Saif, I.; Kasmi, Y.; Allali, K.; Ennaji, M.M. Prediction of DNA methylation in the promoter of gene suppressor tumor. Gene 2018, 651, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.V.; Schwartz, D.A. Epigenetic control of gene expression in the lung. Am. J. Respir. Crit. Care Med. 2011, 183, 1295–1301. [Google Scholar] [CrossRef]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Chakraborty, A.; Viswanathan, P. Methylation-demethylation dynamics: Implications of changes in acute kidney injury. Anal. Cell. Pathol. 2018, 2018, 8764384. [Google Scholar] [CrossRef]
- Cheishvili, D.; Boureau, L.; Szyf, M. DNA demethylation and invasive cancer: Implications for therapeutics. Br. J. Pharmacol. 2015, 172, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Wu, H.J.; Hsu, J.M.; Chang, S.S.; Labaff, A.M.; Li, C.W.; Wang, Y.; Hsu, J.L. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150. [Google Scholar]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Q.; Huang, Z.J.; Mao, N.; Lin, Z.T.; Cheng, L.; Sun, B.; Wang, G. CircRNAs: A new target for the diagnosis and treatment of digestive system neoplasms. Cell Death Dis. 2021, 12, 205. [Google Scholar] [CrossRef]
- Wang, F.; Nazarali, A.J.; Ji, S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016, 6, 1167–1176. [Google Scholar]
- Zhou, Z.; Wang, X.; Hu, Q.; Yang, Z. CircZfp609 contributes to cerebral infarction via sponging miR-145a-5p to regulate BACH1. Metab. Brain Dis. 2023, 38, 1971–1981. [Google Scholar] [CrossRef] [PubMed]
- Mowel, W.K.; Kotzin, J.J.; McCright, S.J.; Neal, V.D.; Henao-Mejia, J. Control of immune cell homeostasis and function by lncRNAs. Trends Immunol. 2018, 39, 55–69. [Google Scholar] [CrossRef]
- Lei, K.; Bai, H.; Wei, Z.; Xie, C.; Wang, J.; Li, J.; Chen, Q. The mechanism and function of circular RNAs in human diseases. Exp. Cell Res. 2018, 368, 147–158. [Google Scholar] [CrossRef]
- Aalto, A.P.; Pasquinelli, A.E. Small non-coding RNAs mount a silent revolution in gene expression. Curr. Opin. Cell Biol. 2012, 24, 333–340. [Google Scholar] [CrossRef]
- Li, J.; Zhong, Y.; Cai, S.; Zhou, P.; Yao, L. MicroRNA expression profiling in the colorectal normal-adenoma-carcinoma transition. Oncol. Lett. 2019, 18, 2013–2018. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, X.; Min, M.; Zou, L.; Shen, P.; Zhu, Y. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 44893–44909. [Google Scholar] [CrossRef]
- Kellermayer, R.; Zilbauer, M. The gut microbiome and the triple environmental hit concept of inflammatory bowel disease pathogenesis. J. Pediatr. Gastroenterol. Nutr. 2020, 71, 589–595. [Google Scholar] [CrossRef]
- Gargalionis, A.N.; Piperi, C.; Adamopoulos, C.; Papavassiliou, A.G. Histone modifications as a pathogenic mechanism of colorectal tumorigenesis. Int. J. Biochem. Cell Biol. 2012, 44, 1276–1289. [Google Scholar] [CrossRef]
- Tamgue, O.; Mezajou, C.F.; Ngongang, N.N.; Kameni, C.; Ngum, J.A.; Simo, U.S.F.; Tatang, F.J.; Akami, M.; Ngono, A.N. Non-coding RNAs in the etiology and control of major and neglected human tropical diseases. Front. Immunol. 2021, 12, 703936. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ghosh, S.; Banerjee, M.; Laha, S.; Bhattacharjee, D.; Sarkar, R.; Ray, S.; Banerjee, A.; Ghosh, R.; Halder, A.; et al. A combination of circulating microRNA-375-3p and chemokines CCL11, CXCL12, and G-CSF differentiate Crohn’s disease and intestinal tuberculosis. Sci. Rep. 2021, 11, 23303. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yang, Y.; Chen, L.; Zhang, Z.; Liu, L.; Zhang, C.; Mai, Q.; Chen, Y.; Chen, Z.; Lin, T.; et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbe 2022, 14, 2029997. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Q.; Zhang, X.; Zhang, K.; Chen, C.; Huang, Z.; Xu, Y. The gut and lung microbiota in pulmonary tuberculosis: Susceptibility, function, and new insights into treatment. Expert Rev. Anti-Infect. Ther. 2023, 21, 1355–1364. [Google Scholar] [CrossRef]
- Xu, J.; Xu, H.M.; Yang, M.F.; Liang, Y.J.; Peng, Q.Z.; Zhang, Y.; Tian, C.M.; Wang, L.S.; Yao, J.; Nie, Y.Q.; et al. New insights into the epigenetic regulation of inflammatory bowel disease. Front. Pharmacol. 2022, 13, 813659. [Google Scholar] [CrossRef]
- Lin, Y.; Qiu, T.; Wei, G.; Que, Y.; Wang, W.; Kong, Y.; Xie, T.; Chen, X. Role of histone post-translational modifications in inflammatory diseases. Front. Immunol. 2022, 13, 852272. [Google Scholar] [CrossRef]
- Goossens-Beumer, I.J.; Bernard, A.; van Hoesel, A.Q.; Zeestraten, E.C.; Putter, H.; Böhringer, S.; Liefers, G.J.; Morreau, H.; van de Velde, C.J.; Kuppen, P.J. Age-dependent clinical prognostic value of histone modifications in colorectal cancer. Transl. Res. 2015, 165, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Wen, B.; Liang, Y.; Yu, W.; Li, H. Histone modifications and their role in colorectal cancer. Pathol. Oncol. Res. 2020, 26, 2023–2033. [Google Scholar] [CrossRef]
- Sarvestani, S.K.; Signs, S.A.; Lefebvre, V.; Mack, S.; Ni, Y.; Morton, A.; Chan, E.R.; Li, X.; Fox, P.; Ting, A.; et al. Cancer-predicting transcriptomic and epigenetic signatures revealed for ulcerative colitis in patient-derived epithelial organoids. Oncotarget 2018, 9, 28717–28730. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, Z.; Zhao, C.; Li, Q.; Geng, Y.; Xiao, Y.; Chen, M.K.; Li, L.; Chen, Z.X.; Wu, M. Dynamic chromatin states coupling with key transcription factors in colitis-associated colorectal cancer. Adv. Sci. 2022, 9, e2200536. [Google Scholar] [CrossRef]
- Parang, B.; Barrett, C.W.; Williams, C.S. AOM/DSS Model of colitis-associated cancer. Methods Mol. Biol. 2016, 1422, 297–307. [Google Scholar] [CrossRef]
- Xiao, J.; Duan, Q.; Wang, Z.; Yan, W.; Sun, H.; Xue, P.; Fan, X.; Zeng, X.; Chen, J.; Shao, C.; et al. Phosphorylation of TOPK at Y74, Y272 by Src increases the stability of TOPK and promotes tumorigenesis of colon. Oncotarget 2016, 7, 24483–24494. [Google Scholar] [CrossRef]
- Cheung, P.; Tanner, K.G.; Cheung, W.L.; Sassone-Corsi, P.; Denu, J.M.; Allis, C.D. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 2000, 5, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Lu, X.; Liu, R.; Ai, N.; Cao, Z.; Li, Y.; Liu, J.; Yu, B.; Liu, K.; Wang, H.; et al. Histone cross-talk connects protein phosphatase 1α (PP1α) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. J. Biol. Chem. 2014, 289, 23154–23167. [Google Scholar] [CrossRef]
- Komar, D.; Juszcynski, P. Rebelled epigenome: Histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin. Epigenetics 2020, 12, 147. [Google Scholar] [CrossRef]
- Tarcic, O.; Pateras, I.S.; Cooks, T.; Shema, E.; Kanterman, J.; Ashkenazi, H.; Boocholez, H.; Hubert, A.; Rotkopf, R.; Baniyash, M.; et al. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep. 2016, 14, 1462–1476. [Google Scholar] [CrossRef]
- Jeusset, L.M.; McManus, K.J. Characterizing and exploiting the many roles of aberrant H2B monoubiquitination in cancer pathogenesis. Semin. Cancer Biol. 2021, 22, 1044–1579. [Google Scholar] [CrossRef]
- Worden, E.J.; Wolberger, C. Activation and regulation of H2B-ubiquitin-dependent histone methyltransferases. Curr. Opin. Struct. Biol. 2020, 59, 98–106. [Google Scholar] [CrossRef]
- Worden, E.J.; Zhang, X.; Wolberger, C. Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife 2020, 9, e53199. [Google Scholar] [CrossRef] [PubMed]
- Van Tongelen, A.; Loriot, A.; De Smet, C. Oncogenic roles of DNA hypomethylation through the activation of cancer-germline genes. Cancer Lett. 2017, 396, 130–137. [Google Scholar] [CrossRef]
- Vilain, A.; Vogt, N.; Dutrillaux, B.; Malfoy, B. DNA methylation and chromosome instability in breast cancer cell lines. FEBS Lett. 1999, 460, 231–234. [Google Scholar] [CrossRef][Green Version]
- Dokun, O.Y.; Florl, A.R.; Seifert, H.H.; Wolff, I.; Schulz, W.A. Relationship of SNCG, S100A4, S100A9 and LCN2 gene expression and DNA methylation in bladder cancer. Int. J. Cancer 2008, 123, 2798–2807. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.S.; Power, B.E.; Molloy, P.L. DNA hypomethylation and human diseases. Biochim. Biophys. Acta Rev. Cancer 2007, 1775, 138–162. [Google Scholar] [CrossRef]
- Porcellini, E.; Laprovitera, N.; Riefolo, M.; Ravaioli, M.; Garajova, I.; Ferracin, M. Epigenetic and epitranscriptomic changes in colorectal cancer: Diagnostic, prognostic, and treatment implications. Cancer Lett. 2018, 419, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Antelo, M.; Balaguer, F.; Shia, J.; Shen, Y.; Hur, K.; Moreira, L.; Cuatrecasas, M.; Bujanda, L.; Giraldez, M.D.; Takahashi, M.; et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. PLoS ONE 2012, 7, e45357. [Google Scholar] [CrossRef]
- Ahn, J.B.; Chung, W.B.; Maeda, O.; Shin, S.J.; Kim, H.S.; Chung, H.C.; Kim, N.K.; Issa, J.P. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer 2011, 117, 1847–1854. [Google Scholar] [CrossRef]
- Ogino, S.; Nosho, K.; Kirkner, G.J.; Kawasaki, T.; Chan, A.T.; Schernhammer, E.S.; Giovannucci, E.L.; Fuchs, C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer Inst. 2008, 100, 1734–1738. [Google Scholar] [CrossRef]
- Baba, Y.; Huttenhower, C.; Nosho, K.; Tanaka, N.; Shima, K.; Hazra, A.; Schernhammer, E.S.; Hunter, D.J.; Giovannucci, E.L.; Fuchs, C.S.; et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol. Cancer 2010, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Swets, M.; Zaalberg, A.; Boot, A.; van Wezel, T.; Frouws, M.A.; Bastiaannet, E.; Gelderblom, H.; van de Velde, C.J.; Kuppen, P.J. Tumor LINE-1 methylation level in association with survival of patients with stage II colon cancer. Int. J. Mol. Sci. 2016, 18, 36. [Google Scholar] [CrossRef]
- Mima, K.; Nowak, J.A.; Qian, Z.R.; Cao, Y.; Song, M.; Masugi, Y.; Shi, Y.; da Silva, A.; Gu, M.; Li, W.; et al. Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget 2016, 7, 55098–55109. [Google Scholar] [CrossRef]
- Kawakami, K.; Matsunoki, A.; Kaneko, M.; Saito, K.; Watanabe, G.; Minamoto, T. Long interspersed nuclear element-1 hypomethylation is a potential biomarker for the prediction of response to oral fluoropyrimidines in microsatellite stable and CpG island methylator phenotype-negative colorectal cancer. Cancer Sci. 2011, 102, 166–174. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Manzo, M.; Baubec, T. Dynamics and context-dependent roles of DNA methylation. J. Mol. Biol. 2017, 429, 1459–1475. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, S. Epigenetics and cancer. J. Appl. Physiol. 2010, 109, 598–605. [Google Scholar] [CrossRef]
- Nimmo, E.R.; Prentergast, J.G.; Aldhous, M.C.; Kennedy, N.A.; Henderson, P.; Drummond, H.E.; Ramsahoye, B.H.; Wilson, D.C.; Semple, C.A.; Satsangi, J. Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm. Bowel Dis. 2012, 18, 889–899. [Google Scholar] [CrossRef]
- Hasler, R.; Feng, Z.; Backdahl, L.; Spehlmann, M.E.; Franke, A.; Teschendorff, A.; Rakyan, V.K.; Down, T.A.; Wilson, G.A.; Feber, A.; et al. A functional methylome map of ulcerative colitis. Genome Res. 2012, 22, 2130–2137. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.; Zhang, H.; Greger, L.; Silva, A.L.; Massey, D.; Dawson, C.; Metz, A.; Ibrahim, A.; Parkes, M. Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 2128–2137. [Google Scholar] [CrossRef]
- Saito, S.; Kato, J.; Hiraoka, S.; Horii, J.; Suzuki, H.; Higashi, R.; Kaji, E.; Kondo, Y.; Yamamoto, K. DNA methylation of colon mucosa in ulcerative colitis patients: Correlation with inflammatory status. Inflamm. Bowel Dis. 2011, 17, 1955–1965. [Google Scholar] [CrossRef]
- Harris, R.A.; Nagy-Szakal, D.; Mir, S.A.; Frank, E.; Szigeti, R.; Kaplan, J.L.; Bronsky, J.; Opekun, A.; Ferry, G.D.; Winter, H.; et al. DNA methylation-associated colonic mucosal immune and defense responses in treatment-naïve pediatric ulcerative colitis. Epigenetics 2014, 9, 1131–1137. [Google Scholar] [CrossRef]
- Koizumi, K.; Alonso, S.; Miyaki, Y.; Okada, S.; Ogura, H.; Shiiya, N.; Konishi, F.; Taya, T.; Perucho, M.; Suzuki, K. Array-based identification of common DNA methylation alterations in ulcerative colitis. Int. J. Oncol. 2012, 40, 983–994. [Google Scholar] [CrossRef]
- Lin, Z.; Hegarty, J.P.; Yu, W.; Cappel, J.A.; Chen, X.; Faber, P.W.; Wang, Y.; Poritz, L.S.; Fan, J.B.; Koltun, W.A. Identification of disease-associated DNA methylation in B cells from Crohn’s disease and ulcerative colitis patients. Dig. Dis. Sci. 2012, 57, 3145–3153. [Google Scholar] [CrossRef]
- Foran, E.; Garrity-Park, M.M.; Mureau, C.; Newell, J.; Smyrk, T.C.; Limburg, P.J.; Egan, L.J. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol. Cancer Res. 2010, 8, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Y.; Arisawa, T.; Tahara, T.; Takahama, K.; Watanabe, M.; Hirata, I.; Nakano, H. Aberrant DNA methylation in ulcerative colitis without neoplasia. Hepato-gastroenterology 2008, 55, 62–65. [Google Scholar]
- Hsieh, C.J.; Klump, B.; Holzmann, K.; Borchard, F.; Gregor, M.; Porschen, R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998, 58, 942–945. [Google Scholar]
- Abu-Remaileh, M.; Bender, S.; Raddatz, G.; Ansari, I.; Cohen, D.; Gutekunst, J.; Musch, T.; Linhart, H.; Breiling, A.; Pikarsky, E.; et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015, 75, 2120–2130. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Harpaz, N.; Shibata, D.; Xu, Y.; Yin, J.; Mori, Y.; Zou, T.T.; Wang, S.; Desai, K.; Leytin, A.; et al. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer Res. 2002, 62, 1148–1151. [Google Scholar]
- Gerecke, C.; Scholtka, B.; Lowenstein, Y.; Fait, I.; Gottschalk, U.; Rogoll, D.; Melcher, R.; Kleuser, B. Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: Putative risk markers for colitis-associated cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P. CpG island methylator phenotype in cancer. Nat. Rev. Cancer 2004, 4, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Zhang, J.; Zhang, R.; Li, F.; Li, Y.; Jia, Y. Multiprobe Assay for clinical SEPT9 methylation based on the carbon dot-modified liquid-exfoliated graphene field effect transistor with a potential to present a methylation panorama. ACS Omega 2020, 5, 16228–16237. [Google Scholar] [CrossRef]
- Song, L.; Li, Y. SEPT9: A specific circulating biomarker for colorectal cancer. Adv. Clin. Chem. 2015, 72, 171–204. [Google Scholar] [CrossRef]
- Kondo, Y.; Issa, J.P. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 29–39. [Google Scholar] [CrossRef]
- Philipp, A.B.; Stieber, P.; Nagel, D.; Neumann, J.; Spelsberg, F.; Jung, A.; Lamerz, R.; Herbst, A.; Kolligs, F.T. Prognostic role of methylated free circulating DNA in colorectal cancer. Int. J. Cancer 2012, 131, 2308–2319. [Google Scholar] [CrossRef]
- Melotte, V.; Lentjes, M.H.; van den Bosch, S.M.; Hellebrekers, D.M.; de Hoon, J.P.; Wouters, K.A.; Daenen, K.L.; Partouns-Hendriks, I.E.; Stessels, F.; Louwagie, J.; et al. N-Myc downstream-regulated gene 4 (NDRG4): A candidate tumor suppressor gene and potential biomarker for colorectal cancer. J. Natl. Cancer Inst. 2009, 101, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.; Chia, J.A.; Greco, S.; Cozzi, S.J.; Buttenshaw, R.L.; Bond, C.E.; Simms, L.A.; Pike, T.; Young, J.P.; Jass, J.R.; et al. Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosomes Cancer 2008, 47, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Cai, W.; Shi, H.; Wang, Y.; Li, M.; Jiao, J.; Chen, M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: A meta-analysis. Br. J. Cancer 2013, 108, 2542–2548. [Google Scholar] [CrossRef]
- Jiang, W.; Wang, P.G.; Zhan, Y.; Zhang, D. Prognostic value of p16 promoter hypermethylation in colorectal cancer: A meta-analysis. Cancer Investig. 2014, 32, 43–52. [Google Scholar] [CrossRef]
- West, N.R.; McCuaig, S.; Franchini, F.; Powrie, F. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 2015, 15, 615–629. [Google Scholar] [CrossRef]
- Li, Y.; Deuring, J.; Peppelenbosch, M.P.; Kuipers, E.J.; de Haar, C.; van der Woude, C.J. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis 2012, 33, 1889–1896. [Google Scholar] [CrossRef]
- Clawson, G.A. Histone deacetylase inhibitors as cancer therapeutics. Ann. Transl. Med. 2016, 4, 287. [Google Scholar] [CrossRef]
- Timp, W.; Feinberg, A.P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 2013, 13, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Ashktorab, H.; Belgrave, K.; Hosseinkhah, F.; Brim, H.; Nouraie, M.; Takkikto, M.; Hewitt, S.; Lee, E.L.; Dashwood, R.H.; Smoot, D. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Dig. Dis. Sci. 2009, 54, 2109–2117. [Google Scholar] [CrossRef] [PubMed]
- Tamagawa, H.; Oshima, T.; Shiozawa, M.; Morinaga, S.; Nakamura, Y.; Yoshihara, M.; Sakuma, Y.; Kameda, Y.; Akaike, M.; Masuda, M.; et al. The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol. Rep. 2012, 27, 637–642. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Hong, Y.; Zhao, Y.; Ismail, T.M.; Wong, Y.; Eu, K.W. Histone H3 (lys-9) deacetylation is associated with transcriptional silencing of E-cadherin in colorectal cancer cell lines. Cancer Investig. 2008, 26, 575–582. [Google Scholar] [CrossRef]
- Peláez, I.M.; Kalogeropoulou, M.; Ferraro, A.; Voulgari, A.; Pankotai, T.; Boros, I.; Pintzas, A. Oncogenic RAS alters the global and gene-specific histone modification pattern during epithelial-mesenchymal transition in colorectal carcinoma cells. Int. J. Biochem. Cell Biol. 2010, 42, 911–920. [Google Scholar] [CrossRef]
- Zuo, X.; Morris, J.S.; Shureiqi, I. Chromatin modification requirements for 15-lipoxygenase-1 transcriptional reactivation in colon cancer cells. J. Biol. Chem. 2008, 283, 31341–31347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.X.; Fang, J.Y.; Lu, R.; Qiu, D.K. Expression of p21(WAF1) is related to acetylation of histone H3 in total chromatin in human colorectal cancer. World J. Gastroenterol. 2007, 13, 2209–2213. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Chen, H. Transcriptional silencing of N-Myc downstream-regulated gene 1 (NDRG1) in metastatic colon cancer cell line SW620. Clin. Exp. Metastasis 2011, 28, 127–135. [Google Scholar] [CrossRef]
- Ishii, M.; Wen, H.; Corsa, C.A.; Liu, T.; Coelho, A.L.; Allen, R.M.; Carson, W.F., 4th; Cavassani, K.A.; Li, X.; Lukacs, N.W.; et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009, 114, 3244–3254. [Google Scholar] [CrossRef]
- De Santa, F.; Totaro, M.; Prosperini, E.; Notarbartolo, S.; Testa, G.; Natoli, G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007, 130, 1083–1094. [Google Scholar] [CrossRef]
- Bayarsaihan, D. Epigenetic mechanisms in inflammation. J. Dent. Res. 2011, 90, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Nagarsheth, N.; Peng, D.; Kryczek, I.; Wu, K.; Li, W.; Zhao, E.; Zhao, L.; Wei, S.; Frankel, T.; Vatan, L.; et al. PRC2 epigenetically silences Th1-type chemokines to suppress effector T-cell trafficking in colon cancer. Cancer Res. 2016, 76, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, I.; Riveiro, M.; Paradis, V.; Faivre, S.; de Parga, P.M.; Raymond, E. Insights on the CXCL12-CXCR4 axis in hepatocellular carcinoma carcinogenesis. Am. J. Transl. Res. 2014, 6, 340–352. [Google Scholar]
- Liu, H.; Liu, Y.; Liu, W.; Zhang, W.; Xu, J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat. Commun. 2015, 6, 8494, Erratum in: Nat. Commun. 2021, 12, 6487. https://doi.org/10.1038/s41467-021-26555-7. [Google Scholar] [CrossRef] [PubMed]
- Enroth, S.; Alvaro, R.I.; Andersson, R.; Wallerman, O.; Wanders, A.; Påhlman, L.; Komorowski, J.; Wadelius, C. Cancer associated epigenetic transitions identified by genome-wide histone methylation binding profiles in human colorectal cancer samples and paired normal mucosa. BMC Cancer 2011, 11, 450. [Google Scholar] [CrossRef]
- Nguyen, C.T.; Weisenberger, D.J.; Velicescu, M.; Gonzales, F.A.; Lin, J.C.; Liang, G.; Jones, P.A. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002, 62, 6456–6461. [Google Scholar] [PubMed]
- An, X.; Lan, X.; Feng, Z.; Li, X.; Su, Q. Histone modification: Biomarkers and potential therapies in colorectal cancer. Ann. Hum. Genet. 2023, 87, 274–284. [Google Scholar] [CrossRef]
- Qiu, F.; Wang, Y.; Chu, X.; Wang, J. ASF1A regulates H4 Y72 phosphorylation and promotes autophagy in colon cancer cells via a kinase activity. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2754–2763. [Google Scholar] [CrossRef]
- Ghate, N.B.; Kim, S.; Spiller, E.; Kim, S.; Shin, Y.; Rhie, S.K.; Smbatyan, G.; Lenz, H.J.; Mumenthaler, S.M.; An, W. VprBP directs epigenetic gene silencing through histone H2A phosphorylation in colon cancer. Mol. Oncol. 2021, 15, 2801–2817. [Google Scholar] [CrossRef]
- Lee, Y.C.; Yin, T.C.; Chen, Y.T.; Chai, C.Y.; Wang, J.Y.; Liu, M.C.; Lin, Y.C.; Kan, J.Y. High expression of phospho-H2AX predicts a poor prognosis in colorectal cancer. Anticancer Res. 2015, 35, 2447–2453. [Google Scholar]
- Yu, D.; Li, Z.; Gan, M.; Zhang, H.; Yin, X.; Tang, S.; Wan, L.; Tian, Y.; Zhang, S.; Zhu, Y.; et al. Decreased expression of dual specificity phosphatase 22 in colorectal cancer and its potential prognostic relevance for stage IV CRC patients. Tumour Biol. 2015, 36, 8531–8535. [Google Scholar] [CrossRef]
- Chen, T.; Li, J.; Xu, M.; Zhao, Q.; Hou, Y.; Yao, L.; Zhong, Y.; Chou, P.C.; Zhang, W.; Zhou, P.; et al. PKCε phosphorylates MIIP and promotes colorectal cancer metastasis through inhibition of RelA deacetylation. Nat. Commun. 2017, 8, 939. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Tian, R.; Wang, S.; Guo, Q.; Luo, M.; Zhou, W.; Liu, G.; Jiang, H.; Jiang, Q. LncRNA2Target v2.0: A comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019, 47, D140–D144. [Google Scholar] [CrossRef]
- Ghoussaini, M.; Mountjoy, E.; Carmona, M.; Peat, G.; Schmidt, E.M.; Hercules, A.; Fumis, L.; Miranda, A.; Carvalho-Silva, D.; Buniello, A.; et al. Open Targets Genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021, 49, D1311–D1320. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Volders, P.J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Yang, M.; Lu, H.; Liu, J.; Wu, S.; Kim, P.; Zhou, X. lncRNAfunc: A knowledgebase of lncRNA function in human cancer. Nucleic Acids Res. 2022, 50, D1295–D1306. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ji, B.; Liu, K.; Hu, G.; Wang, F.; Chen, Q.; Yu, R.; Huang, P.; Ren, J.; Guo, C.; et al. EVLncRNAs 2.0: An updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2021, 49, D86–D91. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhou, G.; Chen, P.; Wang, Y.; Han, J.; Chen, M.; He, Y.; Zhang, S. Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis. 2020, 11, 456. [Google Scholar] [CrossRef]
- Cummins, J.M.; He, Y.; Leary, R.J.; Pagliarini, R.; Diaz, L.A., Jr.; Sjoblom, T.; Barad, O.; Bentwich, Z.; Szafranska, A.E.; Labourier, E.; et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 2006, 103, 3687–3692. [Google Scholar] [CrossRef]
- Lanza, G.; Ferracin, M.; Gafa, R.; Veronese, A.; Spizzo, R.; Pichiorri, F.; Liu, C.G.; Calin, G.A.; Croce, C.M.; Negrini, M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer 2007, 6, 54. [Google Scholar] [CrossRef]
- Garajova, I.; Ferracin, M.; Porcellini, E.; Palloni, A.; Abbati, F.; Biasco, G.; Brandi, G. Non-coding RNAs as predictive biomarkers to current treatment in metastatic colorectal cancer. Int. J. Mol. Sci. 2017, 18, 1547. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Condorelli, A.G.; Battaglia, R.; Tamburello, L.; Barbagallo, D.; Di Pietro, C.; Purrello, M. Non-coding landscapes of colorectal cancer. World J. Gastroenterol. 2015, 21, 11709–11739. [Google Scholar] [CrossRef]
- Lujambio, A.; Ropero, S.; Ballestar, E.; Fraga, M.F.; Cerrato, C.; Setien, F.; Casado, S.; Suarez-Gauthier, A.; Sanchez-Cespedes, M.; Git, A.; et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007, 67, 1424–1429. [Google Scholar] [CrossRef]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef] [PubMed]
- Davalos, V.; Moutinho, C.; Villanueva, A.; Boque, R.; Silva, P.; Carneiro, F.; Esteller, M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012, 31, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Agirre, X.; Bitarte, N.; Ramirez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 2009, 125, 2737–2743. [Google Scholar] [CrossRef]
- Coskun, M.; Bjerrum, J.T.; Seidelin, J.B.; Troelsen, J.T.; Olsen, J.; Nielsen, O.H. miR-20b, miR-98, miR-125b-1 *, and let-7e * as new potential diagnostic biomarkers in ulcerative colitis. World J. Gastroenterol. 2013, 19, 4289–4299. [Google Scholar] [CrossRef]
- Baud, V.; Karin, M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2005, 8, 33–40. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yang, J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.; Samowitz, W.S.; Wolff, R.K.; Stevens, J.R.; Herrick, J.S. The NF-kappaB signalling pathway in colorectal cancer: Associations between dysregulated gene and miRNA expression. J. Cancer Res. Clin. Oncol. 2018, 144, 269–283. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar] [CrossRef]
- Wang, H.; Nie, L.; Wu, L.; Liu, Q.; Guo, X. NR2F2 inhibits Smad7 expression and promotes TGF-beta-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem. Biophys. Res. Commun. 2017, 485, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; He, X.; Zhou, R.; Jia, G.; Qiao, Q. STAT3 induces colorectal carcinoma progression through a novel miR-572-MOAP-1 pathway. Onco. Targets Ther. 2018, 11, 3475–3484. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jaw, J.J.; Stutzman, N.C.; Zou, Z.; Sun, P.D. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 2012, 91, 299–309. [Google Scholar] [CrossRef]
- Zhang, L.L.; Zhang, L.F.; Shi, Y.B. MiR-24 inhibited the killing effect of natural killer cells to colorectal cancer cells by downregulating Paxillin. Biomed. Pharmacother. 2018, 101, 257–263. [Google Scholar] [CrossRef]
- Alamdari-Palangi, V.; Vahedi, F.; Shabaninejad, Z.; Dokeneheifard, S.; Movehedpour, A.; Taheri-Anganeh, M.; Savardashtaki, A. MicroRNA in inflammatory bowel disease at a glance. Eur. J. Gastroenterol. Hepatol. 2021, 32, 140–148. [Google Scholar] [CrossRef]
- Correia, C.N.; Nalpas, N.C.; McLoughlin, K.E.; Browne, J.A.; Gordon, S.V.; MacHugh, D.; Shaughnessy, R.G. Circulating microRNAs as Potential Biomarkers of Infectious Disease. Front. Immunol. 2017, 8, 118. [Google Scholar] [CrossRef]
- Rashid, H.; Hossain, B.; Siddiqua, T.; Kabir, M.; Noor, Z.; Ahmed, M.; Haque, R. Fecal microRNAs as potential biomarkers for screening and diagnosis of intestinal diseases. Front. Mol. Biosci. 2020, 7, 181. [Google Scholar] [CrossRef] [PubMed]
- Thorlacius-Ussing, G.; Schnack Nielsen, B.; Andersen, V.; Holmstrom, K.; Pedersen, A.E. Expression and localization of mir-21 and mir-126 in mucosal tissue from patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2017, 23, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- Shi, C.; Liang, Y.; Yang, J.; Xia, Y.; Chen, H.; Han, H.; Yang, Y.; Wu, W.; Gao, R.; Qin, H. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS ONE 2013, 8, e66814. [Google Scholar] [CrossRef]
- Wang, L.G.; Gu, J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012, 36, e61–e67. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, A.; Li, J.; Fu, J.; Wang, G.; Yang, Y.; Sun, J. Fecal miR-29a and miR-224 as the noninvasive biomarkers for colorectal cancer. Cancer Biomark. 2016, 16, 259–264. [Google Scholar] [CrossRef]
- Iborra, M.; Bernuzzi, F.; Correale, C.; Vetrano, S.; Fiorino, G.; Beltran, B.; Marabita, F.; Locati, M.; Spinelli, A.; Nos, P.; et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 2013, 173, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Li, L.; Yu, Q.; Chao, K.; Zhou, G.; Qiu, Y.; Feng, R.; Huang, S.; He, Y.; et al. Circulating microRNA146b-5p is superior to C-reactive protein as a novel biomarker for monitoring inflammatory bowel disease. Aliment. Pharmacol. Ther. 2019, 49, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.M.; Wu, J.; Zhang, H.; Shi, G.; Chen, Z.T. Circulating miR-125a but not miR-125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn’s disease. World J. Gastroenterol. 2017, 23, 7888–7898. [Google Scholar] [CrossRef]
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5. [Google Scholar] [CrossRef]
- Zahm, A.M.; Thayu, M.; Hand, N.J.; Horner, A.; Leonard, M.B.; Friedman, J.R. Circulating microRNA is a biomarker of pediatric Crohn disease. J. Pediatr. Gastroenterol. Nutr. 2011, 53, 26–33. [Google Scholar] [CrossRef]
- Zahm, A.M.; Hand, N.J.; Tsoucas, D.M.; Le Guen, C.L.; Baldassano, R.N.; Friedman, J.R. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J. Crohns Colitis 2014, 8, 1108–1117. [Google Scholar] [CrossRef]
- Heier, C.R.; Fiorillo, A.A.; Chaisson, E.; Gordish-Dressman, H.; Hathout, Y.; Damsker, J.M.; Conklin, L.S. Identification of pathway-specific serum biomarkers of response to glucocorticoid and infliximab treatment in children with inflammatory bowel disease. Clin. Transl. Gastroenterol. 2016, 7, e192. [Google Scholar] [CrossRef] [PubMed]
- Batra, S.K.; Heier, C.R.; Diaz-Calderon, L.; Tully, C.B.; Fiorillo, A.A.; van den Anker, J.; Conklin, L.S. Serum miRNAs are pharmacodynamic biomarkers associated with therapeutic response in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2020, 26, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Murugesan, S.; Ibrahim, N.; Elawad, M.; Al Khodor, S. Predictive biomarkers for anti-TNF alpha therapy in IBD patients. J. Transl. Med. 2024, 22, 284. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Xie, Y.; Fu, Y.; Zhou, Q.; Ma, Z.; Ma, J.; Chen, J. The signaling axis of microRNA-31/interleukin-25 regulates Th1/Th17-mediated inflammation response in colitis. Mucosal. Immunol. 2017, 10, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chao, K.; Ng, S.C.; Bai, A.H.; Yu, Q.; Yu, J.; Zhang, S. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016, 17, 58. [Google Scholar] [CrossRef]
- Jin, X.; Chen, D.; Zheng, R.H.; Zhang, H.; Chen, Y.P.; Xiang, Z. MiRNA-133a-UCP2 pathway regulates inflammatory bowel disease progress by influencing inflammation, oxidative stress and energy metabolism. World J. Gastroenterol. 2017, 23, 76–86. [Google Scholar] [CrossRef]
- Schonauen, K.; Le, N.; von Arnim, U.; Schulz, C.; Malfertheiner, P.; Link, A. Circulating and fecal microRNAs as biomarkers for inflammatory bowel diseases. Inflamm. Bowel Dis. 2018, 24, 1547–1557. [Google Scholar] [CrossRef]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.; Ho, H.K. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol. Rep. 2014, 32, 97–104. [Google Scholar] [CrossRef]
- Duran-Sanchon, S.; Moreno, L.; Auge, J.M.; Serra-Burriel, M.; Cuatrecasas, M.; Moreira, L.; Martín, A.; Serradesanferm, A.; Pozo, À.; Costa, R.; et al. Identification and validation of microRNA profiles in fecal samples for detection of colorectal cancer. Gastroenterology 2020, 158, 947–957.e4. [Google Scholar] [CrossRef]
- Duran-Sanchon, S.; Moreno, L.; Gomez-Matas, J.; Auge, J.M.; Serra-Burriel, M.; Cuatrecasas, M.; Moreira, L.; Serradesanferm, A.; Pozo, À.; Grau, J.; et al. Fecal microRNA-based algorithm increases effectiveness of fecal immunochemical test-based screening for colorectal cancer. Clin. Gastroenterol. Hepatol. 2021, 19, 323–330.e1. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, Y.; Wang, F.; Moyer, M.P.; Wei, Q.; Zhang, P.; Yang, Z.; Liu, W.; Zhang, H.; Chen, N.; et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut 2016, 65, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, C.; Sharbati, J.; Kutz-Lohroff, B.; Huber, O.; Einspanier, R.; Sharbati, S. TFF3-dependent resistance of human colorectal adenocarcinoma cells HT-29/B6 to apoptosis is mediated by miR-491-5p regulation of lncRNA PRINS. Cell Death Discov. 2017, 3, 16106. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Zhang, J.; Li, M.; Feng, Y.; Wang, X.; Yao, S.; Jin, G.; Du, J.; Han, W.; Yin, Y.; et al. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin. Cancer Res. 2018, 24, 4808–4819. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Liao, L.; Yin, F.; Kuang, H.; Zhou, X.; Wang, Y. LncRNA AB073614 induces epithelial- mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway. Cancer Biomark. 2018, 21, 849–858. [Google Scholar] [CrossRef]
- Li, C.Y.; Liang, G.Y.; Yao, W.Z.; Sui, J.; Shen, X.; Zhang, Y.Q.; Peng, H.; Hong, W.W.; Ye, Y.C.; Zhang, Z.Y.; et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int. J. Oncol. 2016, 48, 1965–1976. [Google Scholar] [CrossRef]
- Tufail, M. HOTAIR in colorectal cancer: Structure, function, and therapeutic potential. Med. Oncol. 2023, 40, 259. [Google Scholar] [CrossRef]
- Svoboda, M.; Slyskova, J.; Schneiderova, M.; Makovicky, P.; Bielik, L.; Levy, M.; Lipska, L.; Hemmelova, B.; Kala, Z.; Protivankova, M.; et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 2014, 35, 1510–1515. [Google Scholar] [CrossRef]
- Zhang, Y.; Bu, D.; Huo, P.; Wang, Z.; Rong, H.; Li, Y.; Liu, J.; Ye, M.; Wu, Y.; Jiang, Z.; et al. ncFANs v2.0: An integrative platform for functional annotation of non-coding RNAs. Nucleic Acids Res. 2021, 49, W459–W468. [Google Scholar] [CrossRef]
- Zeng, L.; Zhao, K.; Liu, J.; Liu, M.; Cai, Z.; Sun, T.; Li, Z.; Liu, R. Long noncoding RNA GAS5 acts as a competitive endogenous RNA to regulate GSK-3β and PTEN expression by sponging miR-23b-3p in Alzheimer’s disease. Neural Regen. Res. 2024, 21, 392–405. [Google Scholar] [CrossRef]
- Zhi, H.; Li, X.; Wang, P.; Gao, Y.; Gao, B.; Zhou, D.; Zhang, Y.; Guo, M.; Yue, M.; Shen, W.; et al. Lnc2Meth: A manually curated database of regulatory relationships between long non-coding RNAs and DNA methylation associated with human disease. Nucleic Acids Res. 2018, 46, D133–D138. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z.; Zhang, Y.; Zhou, L.; Xu, Q.; Li, L.; Zeng, L.; Xue, J.; Niu, H.; Zhong, J.; et al. Mammalian PIWI-piRNA-target complexes reveal features for broad and efficient target silencing. Nat. Struct. Mol. Biol. 2024, 31, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Kumegawa, K.; Maruyama, R.; Yamamoto, E.; Ashida, M.; Kitajima, H.; Tsuyada, A.; Niinuma, T.; Kai, M.; Yamano, H.O.; Sugai, T.; et al. A genomic screen for long noncoding RNA genes epigenetically silenced by aberrant DNA methylation in colorectal cancer. Sci. Rep. 2016, 6, 26699. [Google Scholar] [CrossRef]
- Diaz-Lagares, A.; Crujeiras, A.B.; Lopez-Serra, P.; Soler, M.; Setien, F.; Goyal, A.; Sandoval, J.; Hashimoto, Y.; Martinez-Cardús, A.; Gomez, A.; et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E7535–E7544. [Google Scholar] [CrossRef]
- Hibi, K.; Nakamura, H.; Hirai, A.; Fujikake, Y.; Kasai, Y.; Akiyama, S.; Ito, K.; Takagi, H. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996, 56, 480–482. [Google Scholar]
- Tian, F.; Tang, Z.; Song, G.; Pan, Y.; He, B.; Bao, Q.; Wang, S. Loss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patients. Mol. Med. Rep. 2012, 5, 1536–1540. [Google Scholar] [CrossRef]
- Hidaka, H.; Higashimoto, K.; Aoki, S.; Mishima, H.; Hayashida, C.; Maeda, T.; Koga, Y.; Yatsuki, H.; Joh, K.; Noshiro, H.; et al. Comprehensive methylation analysis of imprinting-associated differentially methylated regions in colorectal cancer. Clin. Epigenetics 2018, 10, 150. [Google Scholar] [CrossRef]
- Yarani, R.; Mirza, A.H.; Kaur, S.; Pociot, F. The emerging role of lncRNAs in inflammatory bowel disease. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Ming, Y.; Zhang, Y. LncRNA Mirt2 is downregulated in ulcerative colitis and regulates IL-22 expression and apoptosis in colonic epithelial cells. Gastroenterol. Res. Pract. 2019, 2019, 8154692. [Google Scholar] [CrossRef]
- Li, F.; Liu, H.; Fu, J.; Fan, L.; Lu, S.; Zhang, H.; Liu, Z. Knockdown of long non-coding RNA NEAT1 relieves inflammation of ulcerative colitis by regulating the miR-603/FGF9 pathway. Exp. Ther. Med. 2022, 23, 131. [Google Scholar] [CrossRef]
- Nie, J.; Zhao, Q. Lnc-ITSN1-2, derived from RNA sequencing, correlates with increased disease risk, activity and promotes CD4(+) T cell activation, proliferation and Th1/Th17 cell differentiation by serving as a ceRNA for IL-23R via sponging mir-125a in inflammatory bowel disease. Front. Immunol. 2020, 11, 852. [Google Scholar] [CrossRef]
- Hur, K.; Kim, S.H.; Kim, J.M. Potential implications of long noncoding RNAs in autoimmune diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef]
- Elamir, A.; Shaker, O.; Kamal, M.; Khalefa, A.; Abdelwahed, M.; Abd El Reheem, F.; Ahmed, T.; Hassan, E.; Ayoub, S. Expression profile of serum LncRNA THRIL and MiR-125b in inflammatory bowel disease. PLoS ONE 2022, 17, e0275267. [Google Scholar] [CrossRef]
- Tian, Y.; Cui, L.; Lin, C.; Wang, Y.; Liu, Z.; Miao, X. LncRNA CDKN2B-AS1 relieved inflammation of ulcerative colitis via sponging miR-16 and miR-195. Int. Immunopharmacol. 2020, 88, 106970. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Dong, Y.; Lin, G.; Cao, Y. Long noncoding RNA antisense noncoding RNA in the INK4 locus correlates with risk, severity, inflammation and infliximab efficacy in Crohn’s disease. Am. J. Med. Sci. 2019, 357, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.W.; Wang, P.Y.; Liu, Y.C.; Sun, L.; Zhu, J.; Zuo, S.; Ma, J.; Li, T.Y.; Zhang, J.L.; Chen, G.W.; et al. Effect of long noncoding RNA H19 overexpression on intestinal barrier function and its potential role in the pathogenesis of ulcerative colitis. Inflamm. Bowel Dis. 2016, 22, 2582–2592. [Google Scholar] [CrossRef]
- Liu, R.; Tang, A.; Wang, X.; Chen, X.; Zhao, L.; Xiao, Z.; Shen, S. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int. J. Mol. Med. 2018, 42, 2903–2913. [Google Scholar] [CrossRef]
- Lucafo, M.; Di Silvestre, A.; Romano, M.; Avian, A.; Antonelli, R.; Martelossi, S.; Naviglio, S.; Tommasini, A.; Stocco, G.; Ventura, A.; et al. Role of the long non-coding RNA growth arrest-specific 5 in glucocorticoid response in children with inflammatory bowel disease. Basic Clin. Pharmacol. Toxicol. 2018, 122, 87–93. [Google Scholar] [CrossRef]
- Nemati Bajestan, M.; Piroozkhah, M.; Chaleshi, V.; Ghiasi, N.E.; Jamshidi, N.; Mirfakhraie, R.; Balaii, H.; Shahrokh, S.; Asadzadeh Aghdaei, H.; Salehi, Z.; et al. Expression analysis of long noncoding RNA-MALAT1 and interleukin-6 in inflammatory bowel disease patients. Iran. J. Allergy Asthma Immunol. 2023, 22, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Du, Y.; Ma, Y.; Gu, L.; Zhou, J.; Deng, D. MALAT1-miR663a negative feedback loop in colon cancer cell functions through direct miRNA-lncRNA binding. Cell Death Dis. 2018, 9, 857. [Google Scholar] [CrossRef]
- Wang, S.; Hou, Y.; Chen, W.; Wang, J.; Xie, W.; Zhang, X.; Zeng, L. KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Mol. Med. Rep. 2018, 17, 2195–2202. [Google Scholar] [CrossRef]
- Cao, L.; Tan, Q.; Zhu, R.; Ye, L.; Shi, G.; Yuan, Z. LncRNA MIR4435-2HG suppression regulates macrophage M1/M2 polarization and reduces intestinal inflammation in mice with ulcerative colitis. Cytokine 2023, 170, 156338. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xiong, Z.; Li, W.; Lin, Y.; Huang, W.; Zhang, S. FHIP1A-DT is a potential novel diagnostic, prognostic, and therapeutic biomarker of colorectal cancer: A pan-cancer analysis. Biochem. Biophys. Res. Commun. 2023, 679, 191–204. [Google Scholar] [CrossRef]
- Guo, K.; Yao, J.; Yu, Q.; Li, Z.; Huang, H.; Cheng, J.; Zhu, Y. The expression pattern of long non coding PVT1 in tumor tissues and in extracellular vesicles of colorectal cancer correlates with cancer progression. Tumour Biol. 2017, 39, 1010428317699122. [Google Scholar] [CrossRef]
- Mani, S.R.; Juliano, C.E. Untangling the Web: The diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 2013, 80, 632–664. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Lin, H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell Biol. 2009, 10, 116–125. [Google Scholar] [CrossRef]
- Stefani, G.; Slack, F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar] [CrossRef]
- Saxe, J.P.; Lin, H. Small noncoding RNAs in the germline. Cold Spring Harb. Perspect. Biol. 2011, 3, a002717. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Watanabe, T.; Lin, H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol. Cell 2014, 56, 18–27. [Google Scholar] [CrossRef]
- Moyano, M.; Stefani, G. piRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 2015, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.; Franco, B.; Robine, N.; Lai, E.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.; Bestor, T.; Hannon, G. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef]
- Watanabe, T.; Tomizawa, S.; Mitsuya, K.; Totoki, Y.; Yamamoto, Y.; Kuramochi-Miyagawa, S.; Iida, N.; Hoki, Y.; Murphy, P.; Toyoda, A.; et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011, 332, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S.; Siomi, M.C.; Siomi, H. piRNA clusters and open chromatin structure. Mobile DNA 2014, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.T.; He, N.; Chuva de Sousa Lopes, S.M.; van der Westerlaken, L.A.; Zischler, H.; Butter, F.; Roelen, B.A.; et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef]
- Aravin, A.; Hannon, G.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef]
- Muhammad, A.; Waheed, R.; Khan, N.A.; Jiang, H.; Song, X. piRDisease v1.0: A manually curated database for piRNA associated diseases. Database 2019, 2019, baz052. [Google Scholar] [CrossRef]
- Ghosh, B.; Sarkar, A.; Mondal, S.; Bhattacharya, N.; Khatua, S.; Ghosh, Z. piRNAQuest V.2: An updated resource for searching through the piRNAome of multiple species. RNA Biol. 2022, 19, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, S.; Zhang, H.; Guan, W.; Zeng, B.; Wei, Y.; Chi-Fung Chan, G.; Li, W. piRPheno: A manually curated database to prioritize and analyze human disease related piRNAs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, P.; Si, X.; Skogerbø, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014, bau110. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, P.; Lu, Y.; Li, Y.; Zheng, Y.; Kan, Y.; Chen, R.; He, S. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2019, 47, D175–D180. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.; Baird, A.M.; Brady, L.; Lim, M.; Gray, S.G.; McDermott, R.; Finn, S.P. Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 101. [Google Scholar] [CrossRef]
- Hao, S.; Cong, L.; Qu, R.; Liu, R.; Zhang, G.; Li, Y. Emerging roles of circular RNAs in colorectal cancer. Onco. Targets Ther. 2019, 12, 4765–4777. [Google Scholar] [CrossRef]
- Li, J.; Ni, S.; Zhou, C.; Ye, M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag. Res. 2018, 10, 2777–2784. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, B.; Lin, Y.; Yang, Y.; Zhang, Z.; Wang, Q.; Zhang, H.; Jiang, K.; Ye, Y.; Wang, S.; et al. Hsa_circ_0000231 promotes colorectal cancer cell growth through upregulation of CCND2 by IGF2BP3/miR-375 dual pathway. Cancer Cell Int. 2022, 22, 27. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Cui, K.; Liu, B.; Zhou, M.; Cao, Y.; Zhang, J.; Bian, Z.; Fei, B.; Huang, Z. Hsa_circ_0062682 promotes serine metabolism and tumor growth in colorectal cancer by regulating the miR-940/PHGDH axis. Front. Cell Dev. Biol. 2021, 9, 770006. [Google Scholar] [CrossRef]
- Long, F.; Lin, Z.; Li, L.; Ma, M.; Lu, Z.; Jing, L.; Li, X.; Lin, C. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol. Cancer 2021, 20, 26. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, C.; Su, Y.; Wang, W.; Yang, S.; Zhou, Q.; Wang, G.; Li, Z.; Song, J.; Zhang, Z.; et al. Regulatory mechanisms and clinical perspectives of circRNA in digestive system neoplasms. J. Cancer 2019, 10, 2885–2891. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Li, Y.; Zhang, H.; Hu, J.; Liao, J.; Su, Y.; Li, Q.; Chen, B.; Li, C.; Wang, Z.; et al. ExoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022, 50, D118–D128. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Liao, J.; Liang, J.; Chen, X.P.; Zhang, B.; Chu, L. Circular RNA HIPK3: A key circular RNA in a variety of human cancers. Front. Oncol. 2020, 10, 773. [Google Scholar] [CrossRef]
- Viralippurath Ashraf, J.; Sasidharan Nair, V.; Saleh, R.; Elkord, E. Role of circular RNAs in colorectal tumor microenvironment. Biomed. Pharmacother. 2021, 137, 111351. [Google Scholar] [CrossRef]
- Liang, X.; Long, L.; Guan, F.; Xu, Z.; Huang, H. Research status and potential applications of circRNAs affecting colorectal cancer by regulating ferroptosis. Life Sci. 2024, 352, 122870. [Google Scholar] [CrossRef]
- Camandona, A.; Gagliardi, A.; Licheri, N.; Tarallo, S.; Francescato, G.; Budinska, E.; Carnogurska, M.; Zwinsová, B.; Martinoglio, B.; Franchitti, L.; et al. Multiple regulatory events contribute to a widespread circular RNA downregulation in precancer and early stage of colorectal cancer development. Biomark. Res. 2025, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Lu, Y. Roles of circRNAs in the progression of colorectal cancer: Novel strategies for detection and therapy. Cancer Gene Ther. 2024, 31, 831–841. [Google Scholar] [CrossRef]
- Li, L.; Wei, C.; Xie, Y.; Su, Y.; Liu, C.; Qiu, G.; Liu, W.; Liang, Y.; Zhao, X.; Huang, D.; et al. Expanded insights into the mechanisms of RNA-binding protein regulation of circRNA generation and function in cancer biology and therapy. Genes Dis. 2024, 12, 101383. [Google Scholar] [CrossRef]
- Hussen, B.M.; Abdullah, S.R.; Mohammed, A.A.; Rasul, M.F.; Hussein, A.M.; Eslami, S.; Glassy, M.C.; Taheri, M. Advanced strategies of targeting circular RNAs as therapeutic approaches in colorectal cancer drug resistance. Pathol. Res. Pract. 2024, 260, 155402. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Lai, C.Y.; Yeh, K.Y.; Liu, B.F.; Chang, T.M.; Chang, C.H.; Liao, Y.F.; Liu, Y.W.; Her, G.M. MicroRNA-21 plays multiple oncometabolic roles in colitis-associated carcinoma and colorectal cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α signaling pathways in zebrafish. Cancers 2021, 13, 5565. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Jabłońska, E.; Białopiotrowicz, E.; Szydłowski, M.; Prochorec-Sobieszek, M.; Juszczyński, P.; Szumera-Ciećkiewicz, A. DEPTOR is a microRNA-155 target regulating migration and cytokine production in diffuse large B-cell lymphoma cells. Exp. Hematol. 2020, 88, 56–67.e2. [Google Scholar] [CrossRef]
- Uthman, Y.A.; Ibrahim, K.G.; Abubakar, B.; Bello, M.B.; Malami, I.; Imam, M.U.; Qusty, N.; Cruz-Martins, N.; Batiha, G.E.; Abubakar, M.B. MALAT1: A promising therapeutic target for the treatment of metastatic colorectal cancer. Biochem. Pharmacol. 2021, 190, 114657. [Google Scholar] [CrossRef]
- Yan, Y.; Su, M.; Qin, B. CircHIPK3 promotes colorectal cancer cells proliferation and metastasis via modulating of miR-1207-5p/FMNL2 signal. Biochem. Biophys. Res. Commun. 2020, 524, 839–846. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Lin, Y.C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Hatley, M.E.; Patrick, D.M.; Garcia, M.R.; Richardson, J.A.; Bassel-Duby, R.; van Rooij, E.; Olson, E.N. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 2010, 18, 282–293. [Google Scholar] [CrossRef]
- Wang, P.; Zhuang, L.; Zhang, J.; Fan, J.; Luo, J.; Chen, H.; Wang, K.; Liu, L.; Chen, Z.; Meng, Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol. Oncol. 2013, 7, 334–345. [Google Scholar] [CrossRef]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Q.; Qi, Y.; Hao, Y.; Gao, Y.; Zhi, H.; Zhang, Y.; Sun, Y.; Zhang, Y.; Xin, M.; et al. LncACTdb 3.0: An updated database of experimentally supported ceRNA interactions and personalized networks contributing to precision medicine. Nucleic Acids Res. 2022, 50, D183–D189. [Google Scholar] [CrossRef]
- Pillich, R.T.; Chen, J.; Churas, C.; Fong, D.; Gyori, B.M.; Ideker, T.; Karis, K.; Liu, S.N.; Ono, K.; Pico, A.; et al. NDEx IQuery: A multi-method network gene set analysis leveraging the network data exchange. Bioinformatics 2023, 39, btad118. [Google Scholar] [CrossRef] [PubMed]
- Persson, E.; Castresana-Aguirre, M.; Buzzao, D.; Guala, D.; Sonnhammer, E.L.L. FunCoup 5: Functional association networks in all domains of life, supporting directed links and tissue-specificity. J. Mol. Biol. 2021, 433, 166835. [Google Scholar] [CrossRef] [PubMed]
- Ilango, S.; Paital, B.; Jayachandran, P.; Padma, P.R.; Nirmaladevi, R. Epigenetic alterations in cancer. Front. Biosci. 2020, 25, 1058–1109. [Google Scholar] [CrossRef]
- Li, Y.; Seto, E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef] [PubMed]
- Gajjela, B.K.; Zhou, M.M. Bromodomain inhibitors and therapeutic applications. Curr. Opin. Chem. Biol. 2023, 75, 102323. [Google Scholar] [CrossRef]
- Martínez-Gutierrez, A.; Carbajal-Lopez, B.; Bui, T.M.; Mendoza-Rodriguez, M.; Campos-Parra, A.D.; Calderillo-Ruiz, G.; Cantú-De Leon, D.; Madrigal-Santillán, E.O.; Sumagin, R.; Pérez-Plasencia, C.; et al. A microRNA panel that regulates proinflammatory cytokines as diagnostic and prognosis biomarkers in colon cancer. Biochem. Biophys. Rep. 2022, 30, 101252. [Google Scholar] [CrossRef]
- Xie, X.; Liu, P.; Wu, H.; Li, H.; Tang, Y.; Chen, X.; Xu, C.; Liu, X.; Dai, G. MiR-21 antagonist alleviates colitis and angiogenesis via the PTEN/PI3K/AKT pathway in colitis mice induced by TNBS. Ann. Transl. Med. 2022, 10, 413. [Google Scholar] [CrossRef]
- Carter, J.V.; Galbraith, N.J.; Yang, D.; Burton, J.F.; Walker, S.P.; Galandiuk, S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2017, 116, 762–774. [Google Scholar] [CrossRef]
- Kalkusova, K.; Taborska, P.; Stakheev, D.; Smrz, D. The role of miR-155 in antitumor immunity. Cancers 2022, 14, 5414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, C.; Guo, Z.; Da, B.; Zhu, W.; Li, Q. miR-223 improves intestinal inflammation through inhibiting the IL-6/STAT3 signaling pathway in dextran sodium sulfate-induced experimental colitis. Immun. Inflamm. Dis. 2021, 9, 319–327, Erratum in: Immun. Inflamm. Dis. 2025, 13, e70209. https://doi.org/10.1002/iid3.70209. [Google Scholar] [CrossRef]
- Zhao, W.; Song, M.; Zhang, J.; Kuerban, M.; Wang, H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14131–14140. [Google Scholar]
- Liu, J.; Guo, B. RNA-based therapeutics for colorectal cancer: Updates and future directions. Pharmacol. Res. 2020, 152, 104550. [Google Scholar] [CrossRef]
- Fitzgerald, K.; Frank-Kamenetsky, M.; Shulga-Morskaya, S.; Liebow, A.; Bettencourt, B.R.; Sutherland, J.E.; Hutabarat, R.M.; Clausen, V.A.; Karsten, V.; Cehelsky, J.; et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: A randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 2014, 383, 60–68. [Google Scholar] [CrossRef]
- Hitchins, M.P.; Vogelaar, I.P.; Brennan, K.; Haraldsdottir, S.; Zhou, N.; Martin, B.; Alvarez, R.; Yuan, X.; Kim, S.; Guindi, M.; et al. Methylated SEPTIN9 plasma test for colorectal cancer detection may be applicable to Lynch syndrome. BMJ Open Gastroenterol. 2019, 6, e000299. [Google Scholar] [CrossRef]
- Sui, C.; Ma, J.; Chen, Q.; Yang, Y. The variation trends of SFRP2 methylation of tissue, feces, and blood detection in colorectal cancer development. Eur. J. Cancer Prev. 2016, 25, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Gonneaud, A.; Gagné, J.M.; Turgeon, N.; Asselin, C. The histone deacetylase Hdac1 regulates inflammatory signalling in intestinal epithelial cells. J. Inflamm. 2014, 11, 43. [Google Scholar] [CrossRef] [PubMed]
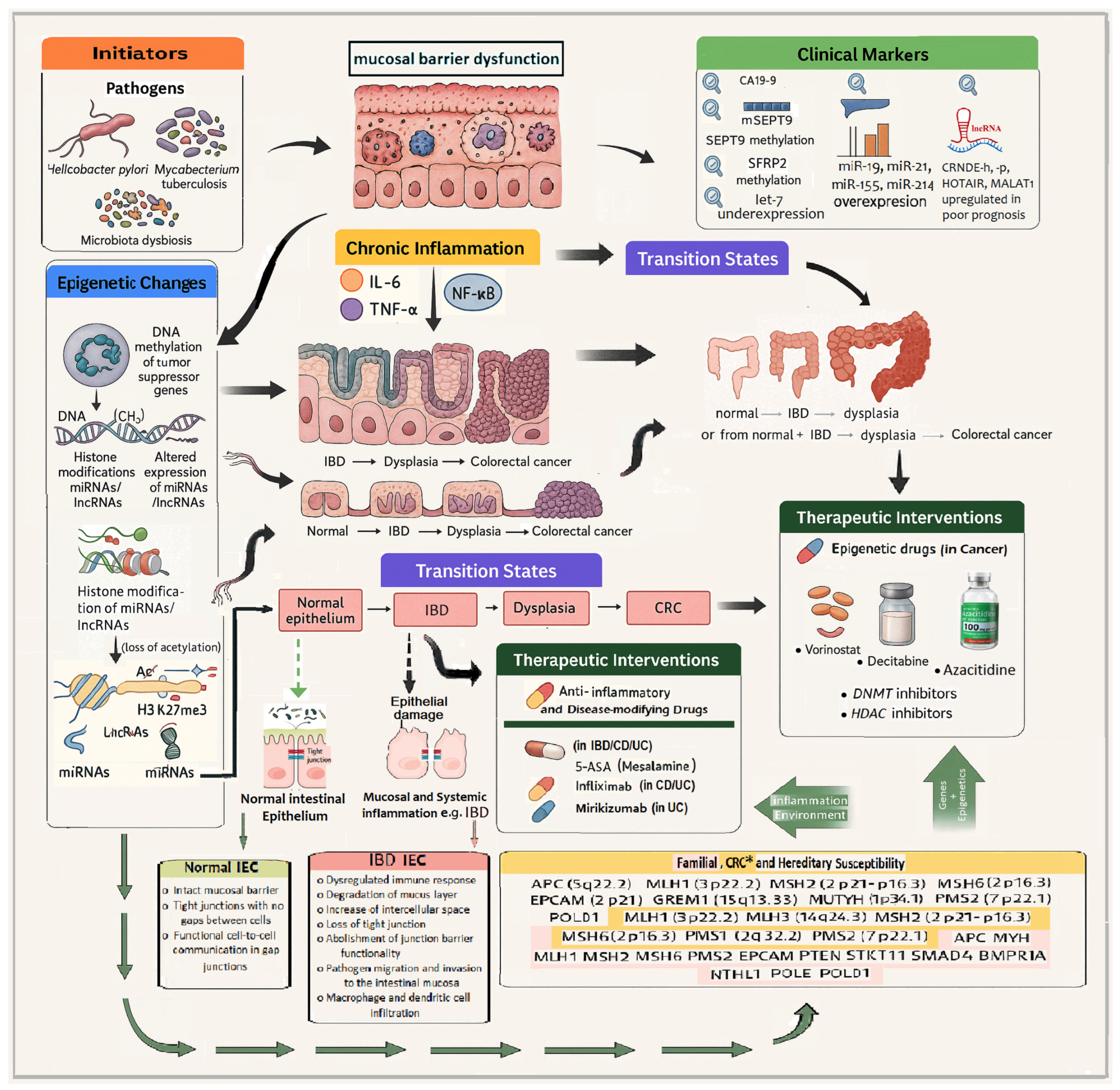
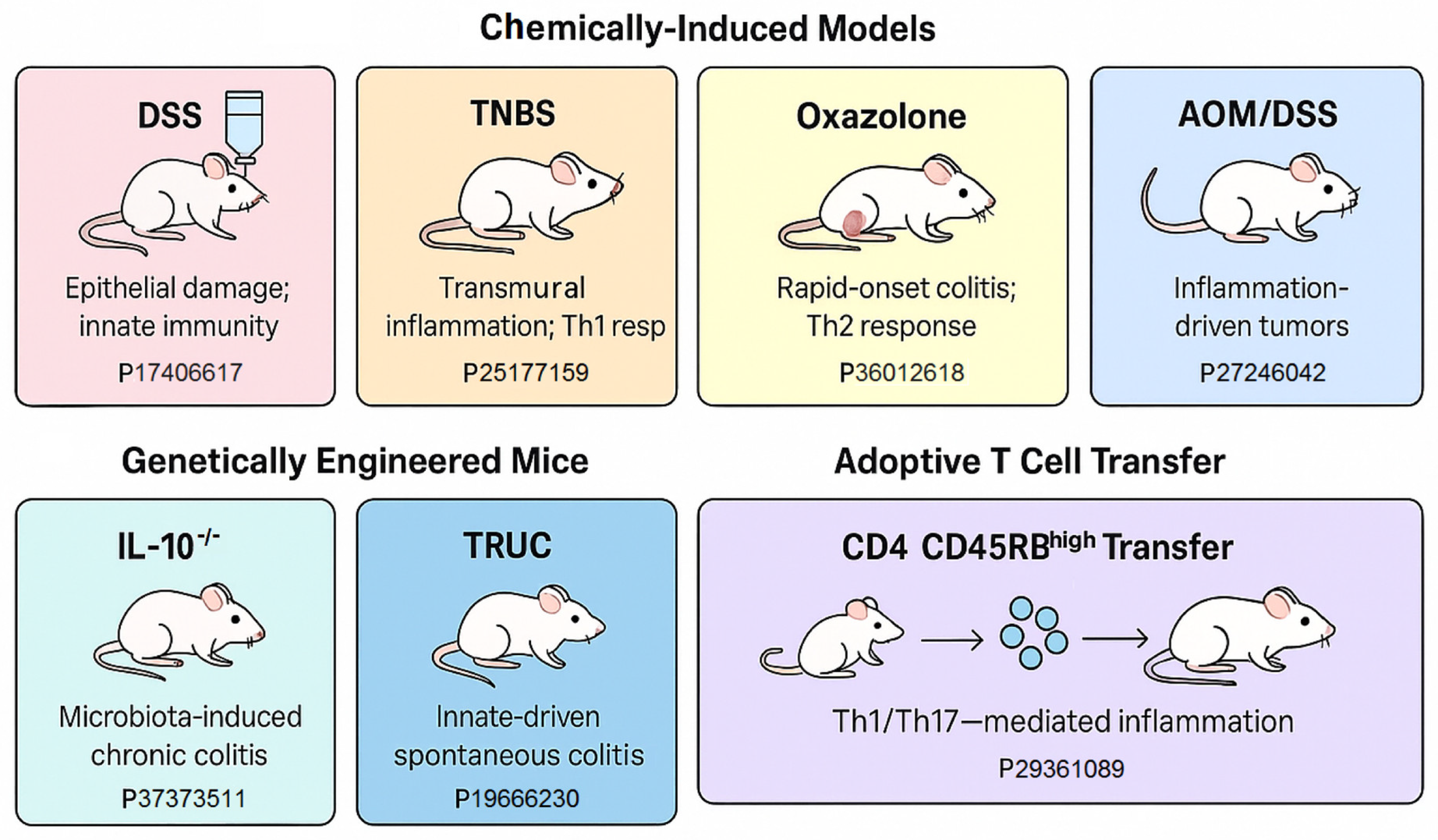
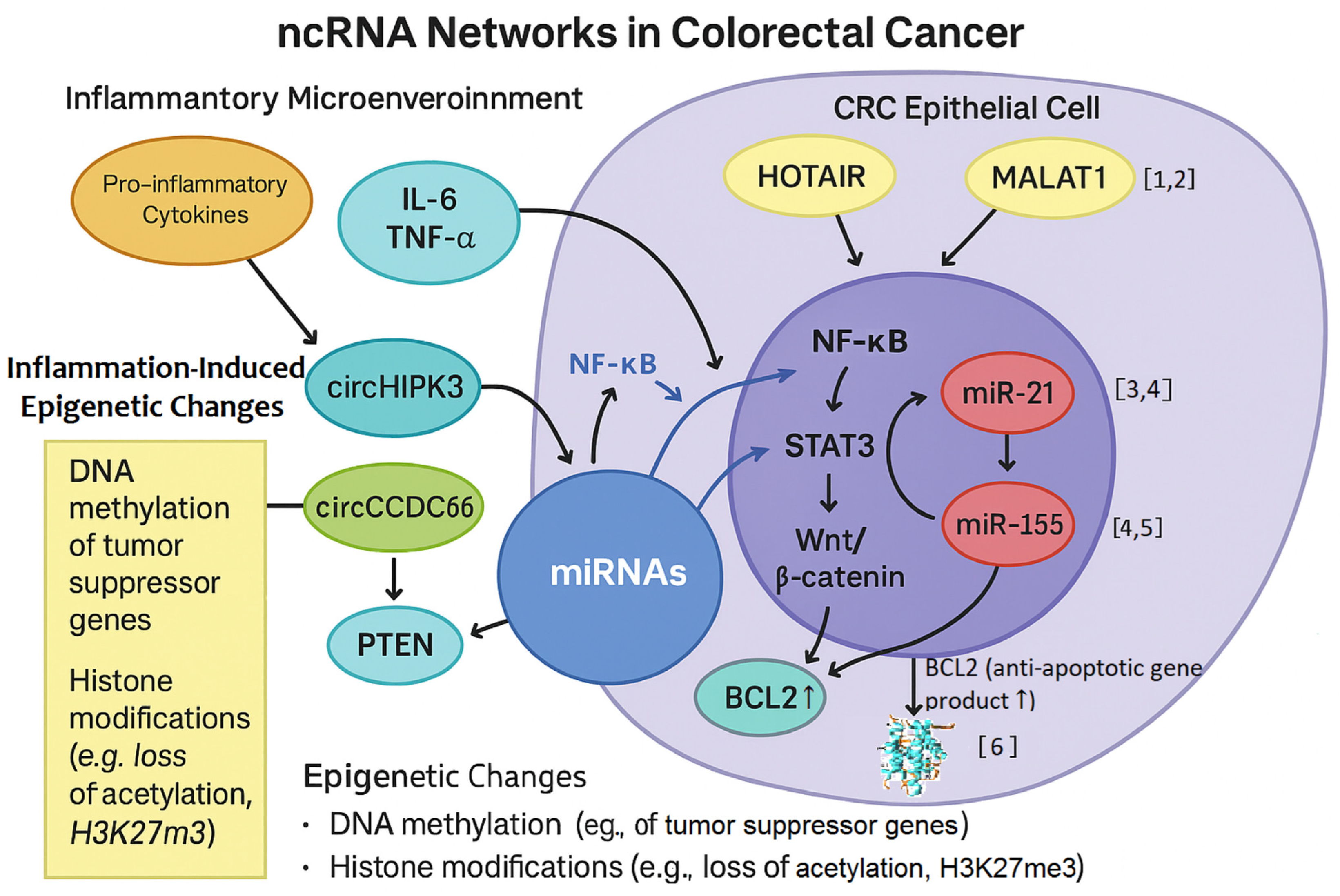
| Model | Method | Immune System Involvement | Advantages | Limitations | Reference (PMID) |
|---|---|---|---|---|---|
| DSS | Oral DSS in drinking water | Innate | Rapid, simple, epithelial injury | No adaptive immune involvement | 17406617 36012618 34440615 |
| AOM/DSS | AOM injection + DSS cycles | Innate + DNA damage | Models CAC pathogenesis | Less suitable for sporadic CRC | 27246042 |
| TNBS | Rectal instillation of TNBS | Adaptive (Th1) | Crohn’s-like inflammation | Variability, toxic risk | 25177159 34440615 |
| Oxazolone | Chemical | Adaptive (Th2) | Mimics UC; rapid onset | Short-lived; strain-dependent | 36012618 |
| IL-10−/− | Genetic deletion of IL-10 | Adaptive | Spontaneous colitis | Microbiota dependence | 37373511 |
| TRUC (T-bet−/− × RAG−/−) | Double knockout | Innate | Innate immunity-driven model progressing to colonic dysplasia and rectal adenocarcinoma | Complex breeding, microbiota sensitive | 19666230 |
| APCMin/+ | Genetic (multiple intestinal neoplasia) | Sporadic Intestinal Tumors | FAP model; Wnt pathway activation | Small intestine focus | 30887153 |
| CD4+ CD45RBhigh T Cell Transfer to RAG−/− Mice | Adaptive cell transfer | Adaptive | T-cell-driven colitis | Requires expertise, chronic model | 29361089 34440615 25989337 |
| Disease Type | Sample Type | Methylated Markers | Methylation Status | Reference (PMID) |
|---|---|---|---|---|
| IBD | Rectal biopsies | THRAP2, FANCC and GBGT1 | ↑ | 22419656 |
| CD | Blood | WDR8 and ITGB2 | ↑ | 27886173 |
| CD | Rectal biopsies | DOK2 and TNFSF4 | ↓ | 22419656 |
| CD | Blood | VMP1 | ↓ | 27886173 |
| UC | Rectal biopsies | CARD9 and CDH1 | ↑ | 22419656 |
| UC | Blood | WDR8 | ↑ | 27886173 |
| UC | Rectal biopsies | ICAM3, DOK2 and TNFSF4 | ↓ | 22419656 |
| UC | Blood | VMP1 | ↓ | 27886173 |
| UC | Colon biopsies | EBI3 | ↓ | 22419656 |
| CRC | Rectal biopsies | TGFB2, SLIT2, HS3ST2, TMEFF2, | ↑ | 27886173 |
| CRC | Colon biopsies | FOXE1, SYNE1 | ↑ | 22419656 |
| CRC | Colonic mucosa | APC, CDH13, MGMT, RUNX3 and MLH1 | ↑ | 27886173 |
| CRC | Colon biopsies | ITGA4 | ↑ | 34069352 |
| Disease Type | Sample Type | Histone Modifying Enzymes/Histone Markers | Modification Type/Status | Reference (PMID) |
|---|---|---|---|---|
| IBD | Intestinal tissue | KAT2B | Acetylation ↓ | 26802082 |
| UC/CAC | Colon Tissue | H3K27ac | Acetylation ↑ | 29983891 |
| CD | Colonic biopsy | H2Bub1 | Ubiquitination ↓ | 34088983 |
| IBD | Colon Tissue | HDAC | Acetylation ↑ | 38903915 |
| IBD | Colon Tissue | HDAC8 | Acetylation ↑ | 36558966 |
| CRC | Primary cancer tissue | H3K9me2 | Methylation ↑ | 22076537 |
| Disease Type | Sample Type | miRNAs | Gene Expression Change | Reference (PMID) |
|---|---|---|---|---|
| UC | Serum | miR-29a, miR-196b, miR-127-3p | ↑ | 23607522 |
| CD | Serum | miR-140-3p, miR-127-3p | ↑ | 23607522 |
| UC | Serum | miR-150 | ↓ | 23607522 |
| CD | Blood | miRNA-125a | ↓ | 29209130 |
| UC | Blood | miRNA-19a | ↑ | 25886994 |
| UC | Blood | miRNA-146a | ↓ | 25886994 |
| CD | Blood | miR-31 | ↓ | 25886994 |
| CD | Blood | miR-101 | ↑ | 25886994 |
| IBD | Serum | miR-146b-5p | ↑ | 30734320 |
| IBD | Feces | miR-223, miR-1246 | ↑ | 32850969 |
| IBD | Serum | miR-16, miR-21, and miR-223 | ↑ | 29668922 |
| IBD | Feces | miR-223, miR-155 | ↑ | 29668922 |
| IBD | Serum/Feces | miR-21, miR-142-3p, miR-146a and let-7i | ↑ | 24613022 |
| IBD | IECs | miR-192, miR-194, miR-200b, miR-375 | ↓ | 24613022 |
| IBD | Serum | miR-192, miR-195, miR-20a, miR-30a, miR-484 and let-7b | ↑ | 21546856 |
| IBD | Serum | miR-146a, miR-146b, miR-320a, miR-126 and let-7c | ↓ | 32793975 |
| IBD | Colonic tissue | miR-133a | ↓ | 28104982 |
| CRC | Feces | miR-135b, miR-223 and miR-451 | ↑ | 24841830 |
| CRC | Feces | miR-29a, miR-223 and miR-224 | ↓ | 26756616 |
| CRC | Feces | miR-421, miR130b-3p miR27a-3P | ↑ | 31622624 |
| Disease Type | Sample Type | lncRNAs | Gene Expression Change | Reference (PMID) |
|---|---|---|---|---|
| UC | Plasma | Mirt2 | ↓ | 31687015 |
| CRC | Blood | HOTAIR | ↑ | 21862635, 24583926 |
| CRC | COAD tissue | FHIP1A-DT | ↓ | 37703762 |
| UC | Plasma | IFNG-AS1 | ↑ | 34970354 |
| UC | Plasma | ITSN1-2 | ↑ | 32547537 |
| IBD | Serum | THRIL | ↑ | 36206229 |
| UC | Blood | CDKN2B-AS1 | ↓ | 33182065 |
| CD | Blood | CDKN2B-AS1 | ↑ | 30665494 |
| UC | Colonic tissue | H19 | ↑ | 27661667 |
| IBD | Colonic tissue | CRNDE | ↑ | 31251902 |
| IBD | Serum | NEAT1 | ↑ | 30132508 |
| UC | Blood | GAS5 | ↑ | 28722800 |
| IBD/CRC | Intestinal tissue | MALAT1 | ↑ | 38085149 |
| IBD | Plasma/Tissue | KIF9-AS1 and LINC01272 | ↑ | 29207070 |
| IBD | Plasma/Tissue | DIO3OS | ↓ | 29207070 |
| UC | Human intestinal epithelial Caco2 cells and murine macrophage RAW264.7 cells | MIR4435-2HG | ↑ | 37597495 |
| CRC | Colon/Rectal biopsies | RVT1 | ↑ | 28381186 |
| Condition | DNA Methylation | Histone Acetylation | Histone Methylation | Non-Coding RNAs (miRNAs/lncRNAs) | References (PMID) |
|---|---|---|---|---|---|
| Normal Colon | Homeostatic balance of methylation | Balanced H3/H4 acetylation regulating gene expression | Physiological levels of H3K4me3 and H3K27me3 | Normal expression of regulatory miRNAs and lncRNAs | 26220502 |
| Inflammatory Bowel Disease (IBD) | ↑ Promoter hypermethylation of anti-inflammatory genes | ↑ H3K27ac | ↑ H3K4me3, ↑ H3K9me3 | ↑ Inflammatory miRNAs (e.g., miR-155), altered lncRNAs | 27886173, 37936149, 32464322 |
| Colitis (IBD) | Global hypomethylation; hypermethylation of SOCS3 | ↓ H3/H4 acetylation; HDAC overexpression | ↑ H3K9me3 (heterochromatinization); ↓ H3K27me3 | ↑ miR-155, miR-21; ↓ let-7; ↑ lncRNA HOTAIR | 22739025, 30137272 |
| Dysplasia | ↑ CIMP phenotype, ↑ Promoter hypermethylation of MLH1, CDKN2A, APC | Loss of histone acetylation at tumor suppressor loci | ↑ H3K27me3, H3K9me3 by EZH2; ↓ H3K4me3 at differentiation genes | ↑ MALAT1, CRNDE, ↓ MEG3; ↑ oncogenic lncRNAs, miRNAs, ↓ tumor-suppressive circRNAs | 16804544, 33930428 |
| Colorectal Cancer | Global hypomethylation; ↑ Promoter CpG island hypermethylation | Aberrant HDAC recruitment; hypoacetylation at TSGs; ↓ H4K16ac | ↑ H3K9me2/3; ↑ EZH2-mediated H3K27me3; ↓ H3K4me3 | ↑ Oncogenic miRNAs (e.g., miR-21, miR-135b); ↑ lncRNAs NEAT1, PVT1, CCAT1; ↓ lncRNAs like GAS5 | 15765097, 32464322 |
| Name | SNP Expression Info | Function | Database (#) | PubMed (PMID) |
|---|---|---|---|---|
| piR-hsa-679 | rs34383331, base change: A > T | May be involved in the development of CRC | piRBase | 25740697 |
| piR-hsa-7400 | rs2070766f, base change: C > G | -“- | piRBase | 25740697 |
| piR-hsa-21417 | rs2070766f, base change: C > G | -“- | piRBase | 25740697 |
| piR-hsa-29786 | rs2070766f, base change: C > G | -“- | piRBase | 25740697 |
| piR-hsa-21517 | rs11776042, base change: T > C | May be involved in the development of CRC | piRBase | 25740697 |
| piR-hsa-29056 | rs9368782, base change: A > G | -“- | piRBase | 25740697 |
| piR-hsa-2363 | rs12483859, base change: A > G | -“- | piRBase | 25740697 |
| piR-hsa-8401 | rs10433310, base change: C > T | -“- | piRBase | 25740697 |
| piR-hsa-3789 | rs12910401, base change: G > A | -“- | piRBase | 25740697 |
| piR-hsa-1245 | up-regulated | It is a novel oncogene and a potential prognostic biomarker in colorectal cancer | piRBase | 29382334 |
| piR-hsa-1282 | up-regulated | It interacts with HSF1 to promote Ser326 phosphorylation and HSF1 activation, enhancing CRC cell proliferation and suppressing cell apoptosis | piRBase | 28618124 |
| piR-hsa-17444 | up-regulated | Formation of PIWIL2/STAT3/phosphorylated-SRC (p-SRC) complex, which activates STAT3 signaling and promotes proliferation, metastasis and chemoresistance of CRC cells | piRBase | 30555542 |
| piR-hsa-1077 (*) | up-regulated | Ontology ID: EFO_1001951 | piRNAdb, piRPheno v2.0 | 16751776 |
| circRNA Name | Synonyms | Predicted Interacting RBP (Nο of Binding Sites) (†) | Methods | Expression Pattern | PubMed ID |
|---|---|---|---|---|---|
| hsa_circ_0020397 | hsa_circRNA_100722 | EIF4A3(32); HuR(3); IGF2BP1(2); AGO2(2); SFRS1(1); PTB(1); LIN28A(1); IGF2BP2(1); FUS(1) | qRT-PCR; dual luciferase reporter assay; in vitro knockdown; in vitro overexpression; Western blot; etc. | up-regulated | 28707774 |
| circ-BANP | hsa_circRNA_101902; hsa_circ_0003098 | EIF4A3(12); HuR(1); AGO2(1) | Microarray; RT-PCR; qRT-PCR; in vitro knockdown; ISH; Western blot; etc. | up-regulated | 28103507 |
| hsa_circ_0000069 | hsa_circRNA_100213; hsa_circ_001061 | EIF4A3(10); AGO2(9); IGF2BP3(7); PTB(6); IGF2BP2(6); IGF2BP1(4); HuR(2); FMRP(2); SFRS1(1); FXR2(1) | qRT-PCR; in vitro knockdown; etc. | up-regulated | 28003761 |
| hsa_circ_001569 | N/A | N/A | in vitro knockdown; in vitro overexpression; qRT-PCR; Western blot; luciferase reporter assay, etc. | up-regulated | 27058418 |
| hsa_circ_0001451 | hsa_circ_001988 | EIF4A3(6); HuR(2); LIN28A(1); IGF2BP3(1); IGF2BP2(1); DGCR8(1); AGO2(1) | RNA-seq; qRT-PCR | down-regulated | 26884878 |
| circHlPK3 | circ_0000284, hsa_circ_0000284 | IGF2BP1 (5), HuR (ELAVL1) (8), FUS (4) | RNA-seq; Microarray | up-regulated | 33536039 |
| circCCDC66 | circ_0001313 | HuR (ELAVL1) (6), PTBP1 (7), FUS (5) | RNA-seq; Microarray; droplet digital PCR | down-regulated | 33536039 |
| circZFR | hsa_circRNA_103809; hsa_circ_0072088 | FMRP(21); EIF4A3(7); AGO2(7); IGF2BP3(6); HuR(6); IGF2BP1(4); AGO1(4); ZC3H7B(1); U2AF65(1); PTB(1); LIN28B(1); IGF2BP2(1) | Microarray; qRT-PCR | down-regulated | 28349836 |
| circPTK2 | hsa_circRNA_104700; hsa_circ_0005273 | AGO2(5); EIF4A3(2); IGF2BP3(1); IGF2BP2(1); FUS(1) | Microarray; qRT-PCR | down-regulated | 28349836 |
| CDR1as | Cdr1as; ciRS-7; hsa_circRNA_105055; hsa_circ_0001946 | AGO2(43); FUS(26); IGF2BP1(11); IGF2BP2(10); IGF2BP3(9); AGO1(6); TNRC6(2); TDP43(2) | qRT-PCR; in vitro overexpression; in vivo overexpression; IHC; Western blot; etc. | up-regulated | 28174233 |
| ncRNA Type | Name | Function in CRC | References (PMID) |
|---|---|---|---|
| lncRNA | HOTAIR | Recruits PRC2 to silence tumor suppressor genes (e.g., CDKN1A); promotes invasion and metastasis. | 21862635; 28701486 |
| lncRNA | MALAT1 | Enhances β-catenin nuclear translocation; regulates alternative splicing and epithelial–mesenchymal transition (EMT). | 12970751; 34144008 |
| circRNA | circHIPK3 | Acts as a miR-1207 sponge, which is downregulated in CRC; enhances formin-like 2 (FMNL2) in CRC; contributes to chemoresistance and proliferation. | 32046858 |
| circRNA | circCCDC66 | Functions as a ceRNA; sequesters tumor-suppressive miRNAs (e.g., miR-33b); enhances c-MYC and YAP1 pathways. | 28249903 |
| miRNA | miR-21 | Overexpressed in CRC; targets PTEN, PDCD4, suppressing apoptosis; regulated by NF-κB and IL-6 inflammatory stimuli. | 17968323; 20797623; 34771727 |
| miRNA | miR-155 | Induced by NF-κB and STAT3; promotes tumor cell survival and immune evasion by targeting SOCS1 and TP53INP1. | 17242365; 17911593; 32702393 |
| mRNA target | PTEN | Tumor suppressor inhibited by miR-21; regulates PI3K/AKT signaling. | 32104279 |
| mRNA target | BCL2 | Anti-apoptotic protein regulated by multiple miRNAs (e.g., miR-15, miR-16); supports resistance to cell death. | 16166262; 28984869 |
| Drug/RNA | Type | Target | Disease Phase | Function/Application Notes | Reference (PMID) |
|---|---|---|---|---|---|
| OTX015 | BET inhibitor | MYC, NF-κB pathways | Phase I/II clinical trials | Solid tumors, anti-inflammatory profile | 37207401 |
| 5-Azacytidine | DNMT inhibitor | DNA methylation reversal | Approved (hematologic); off-label, investigational in CRC | Reverses methylation silencing | 24583822; 26317465; 33359448 |
| Anti-miR-21 Oligonucleotides | Antisense oligo | miR-21 suppression | Preclinical studies | Targets PDCD4, inhibits tumor invasion | 17968323 |
| miR-92a sponge | miRNA decoy | miR-92a suppression | Preclinical studies | Biomarker and therapeutic target | 28957811; 33620640 |
| Romidepsin | HDAC inhibitor | HDAC1/2 (Histone deacetylation) | Approved (T-cell lymphoma); clinical trials for CRC | Suppresses inflammation and tumor growth | 27599530 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Triantaphyllopoulos, K.A.; Ragia, N.D.; Panagiotopoulou, M.-C.E.; Sourlingas, T.G. Integrating Inflammatory and Epigenetic Signatures in IBD-Associated Colorectal Carcinogenesis: Models, Mechanisms, and Clinical Implications. Int. J. Mol. Sci. 2025, 26, 9498. https://doi.org/10.3390/ijms26199498
Triantaphyllopoulos KA, Ragia ND, Panagiotopoulou M-CE, Sourlingas TG. Integrating Inflammatory and Epigenetic Signatures in IBD-Associated Colorectal Carcinogenesis: Models, Mechanisms, and Clinical Implications. International Journal of Molecular Sciences. 2025; 26(19):9498. https://doi.org/10.3390/ijms26199498
Chicago/Turabian StyleTriantaphyllopoulos, Kostas A., Nikolia D. Ragia, Maria-Chara E. Panagiotopoulou, and Thomae G. Sourlingas. 2025. "Integrating Inflammatory and Epigenetic Signatures in IBD-Associated Colorectal Carcinogenesis: Models, Mechanisms, and Clinical Implications" International Journal of Molecular Sciences 26, no. 19: 9498. https://doi.org/10.3390/ijms26199498
APA StyleTriantaphyllopoulos, K. A., Ragia, N. D., Panagiotopoulou, M.-C. E., & Sourlingas, T. G. (2025). Integrating Inflammatory and Epigenetic Signatures in IBD-Associated Colorectal Carcinogenesis: Models, Mechanisms, and Clinical Implications. International Journal of Molecular Sciences, 26(19), 9498. https://doi.org/10.3390/ijms26199498







