Hormone Receptor Positive/HER2 Negative Breast Carcinoma: Association of PIK3CA Mutational Status with PD-L1 and Tumor Cell Microenvironment and Their Prognostic Significance
Abstract
1. Introduction
2. Results
2.1. Study Cohort
2.2. Association of Clinical and Pathological Characteristics with PIK3CA Mutational Status
2.3. CD4, CD8, CD68, CD163 Distribution and PD-L1 Expression in Non-Mutated and Mutated PIK3CA Carcinomas
2.4. Correlation of PD-L1 Expression Depending on PIK3CA Mutational Status
2.5. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Tumor Specimens
4.2. Immunohistochemical Staining
4.3. Immunohistochemical Evaluation
4.4. DNA Isolation from FFPE
4.5. PIK3CA Mutation Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DCIS | Ductal carcinoma in situ |
| DFS | Disease-free survival |
| DSS | Disease-specific survival |
| FFPE | Formalin fixed paraffin embedded |
| HR+/HER2− | Hormone receptor positive/human epidermal growth factor-2 negative |
| IHC | Immunohistochemistry |
| MAP | Mitogen activated protein |
| MDSCs | Myeloid-derived suppressor cells |
| NST | No special type |
| PD-L1 | Programmed cell death ligand 1 |
| ROC | Receiver operating characteristic |
| TAMs | Tumor associated macrophages |
| TILs | Tumor-infiltrating lymphocytes |
| TMAs | Tissue microarrays |
| TME | Tumor microenvironment |
| TNBC | Triple negative breast carcinomas |
References
- Zhang, W.; Lee, A.; Tiwari, A.K.; Yang, M.Q. Characterizing the Tumor Microenvironment and Its Prognostic Impact in Breast Cancer. Cells 2024, 13, 1518. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stanton, S.E.; Disis, M.L. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brummel, K.; Eerkens, A.L.; de Bruyn, M.; Nijman, H.W. Tumour-infiltrating lymphocytes: From prognosis to treatment selection. Br. J. Cancer 2023, 128, 451–458. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Valenza, C.; Taurelli Salimbeni, B.; Santoro, C.; Trapani, D.; Antonarelli, G.; Curigliano, G. Tumor Infiltrating Lymphocytes across Breast Cancer Subtypes: Current Issues for Biomarker Assessment. Cancers 2023, 15, 767. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Angelico, G.; Broggi, G.; Tinnirello, G.; Puzzo, L.; Vecchio, G.M.; Salvatorelli, L.; Memeo, L.; Santoro, A.; Farina, J.; Mulé, A.; et al. Tumor Infiltrating Lymphocytes (TILS) and PD-L1 Expression in Breast Cancer: A Review of Current Evidence and Prognostic Implications from Pathologist’s Perspective. Cancers 2023, 15, 4479. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Karnik, T.; Kimler, B.F.; Fan, F.; Tawfik, O. PD-L1 in breast cancer: Comparative analysis of 3 different antibodies. Hum. Pathol. 2018, 72, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Fang, J.; Peng, J.; Wang, X.; Xing, P.; Jia, K.; Hu, J.; Wang, D.; Ding, Y.; Wang, X.; et al. PD-1/PD-L1 immune checkpoint blockade in breast cancer: Research insights and sensitization strategies. Mol. Cancer 2024, 23, 266. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Szekely, B.; Bossuyt, V.; Li, X.; Wali, V.B.; Patwardhan, G.A.; Frederick, C.; Silber, A.; Park, T.; Harigopal, M.; Pelekanou, V.; et al. Immunological differences between primary and metastatic breast cancer. Ann. Oncol. 2018, 29, 2232–2239. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Cui, H.; Dai, L.; Chang, L.; Liu, D.; Yan, W.; Zhao, X.; Kang, H.; Ma, X. PIK3CA mutation-driven immune signature as a prognostic marker for evaluating the tumor immune microenvironment and therapeutic response in breast cancer. J. Cancer Res. Clin. Oncol. 2024, 150, 119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, Z.; Richmond, A. The Role of PI3K Inhibition in the Treatment of Breast Cancer, Alone or Combined With Immune Checkpoint Inhibitors. Front. Mol. Biosci. 2021, 8, 648663. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dunlap, J.; Le, C.; Shukla, A.; Patterson, J.; Presnell, A.; Heinrich, M.C.; Corless, C.L.; Troxell, M.L. Phosphatidylinositol-3-kinase and AKT1 mutations occur early in breast carcinoma. Breast Cancer Res. Treat. 2010, 120, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Bachman, K.E.; Argani, P.; Samuels, Y.; Silliman, N.; Ptak, J.; Szabo, S.; Konishi, H.; Karakas, B.; Blair, B.G.; Lin, C.; et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol. Ther. 2004, 3, 772–775. [Google Scholar] [CrossRef] [PubMed]
- Mollon, L.E.; Anderson, E.J.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Literature Review of the Prognostic and Predictive Value of PIK3CA Mutations in HR+/HER2− Metastatic Breast Cancer. Clin. Breast Cancer 2020, 20, e232–e243. [Google Scholar] [CrossRef] [PubMed]
- Dirican, E.; Akkiprik, M.; Özer, A. Mutation distributions and clinical correlations of PIK3CA gene mutations in breast cancer. Tumor Biol. 2016, 37, 7033–7045. [Google Scholar] [CrossRef] [PubMed]
- Beelen, K.; Opdam, M.; Severson, T.M.; Koornstra, R.H.; Vincent, A.D.; Wesseling, J.; Muris, J.J.; Berns, E.M.; Vermorken, J.B.; van Diest, P.J.; et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. 2014, 16, R13. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Loibl, S.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Denkert, C. PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane-Based Neoadjuvant Chemotherapy. Cancer Res. Treat. 2020, 52, 689–696. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Tran Dien, A.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Ugai, T.; Zhao, M.; Shimizu, T.; Akimoto, N.; Shi, S.; Takashima, Y.; Zhong, R.; Lau, M.C.; Haruki, K.; Arima, K.; et al. Association of PIK3CA mutation and PTEN loss with expression of CD274 (PD-L1) in colorectal carcinoma. Oncoimmunology 2021, 10, 1956173. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; van der Noort, V.; van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 90. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, W.; Ouyang, X.; Li, C.; Long, Y.; Chen, W.; Ji, Z.; Shen, X.; Xiang, L.; Yang, H. Targeting PI3Kα increases the efficacy of anti-PD-1 antibody in cervical cancer. Immunology 2023, 170, 419–438. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Phillips, W.A.; Loi, S. PIK3CA mutations in breast cancer: Reconciling findings from preclinical and clinical data. Breast Cancer Res. 2014, 16, 201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, M.H.; Cho, J.H.; Kwon, S.Y.; Jung, S.J.; Lee, J.H. Clinicopathological Characteristics of PIK3CA Mutation and Amplification in Korean Patients with Breast Cancers. Int. J. Med. Sci. 2020, 17, 1131–1135. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zardavas, D.; Te Marvelde, L.; Milne, R.L.; Fumagalli, D.; Fountzilas, G.; Kotoula, V.; Razis, E.; Papaxoinis, G.; Joensuu, H.; Moynahan, M.E.; et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018, 36, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rozen, V.; Zhao, Y.; Wang, Z. Oncogenic activation of PIK3CA in cancers: Emerging targeted therapies in precision oncology. Genes Dis. 2024, 12, 101430. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, Y.H.; Zhang, J.X.; Chen, X.; Liu, F.; Cao, J.Z.; Chen, Y.; Chen, W.; Luo, J.H. Predictive Value of the TP53/PIK3CA/ATM Mutation Classifier for Patients With Bladder Cancer Responding to Immune Checkpoint Inhibitor Therapy. Front. Immunol. 2021, 12, 643282. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mishima, S.; Kawazoe, A.; Nakamura, Y.; Sasaki, A.; Kotani, D.; Kuboki, Y.; Bando, H.; Kojima, T.; Doi, T.; Ohtsu, A.; et al. Clinicopathological and molecular features of responders to nivolumab for patients with advanced gastric cancer. J. Immunother. Cancer 2019, 7, 24. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ruffell, B.; Au, A.; Rugo, H.S.; Esserman, L.J.; Hwang, E.S.; Coussens, L.M. Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. USA 2012, 109, 2796–2801. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goff, S.L.; Danforth, D.N. The Role of Immune Cells in Breast Tissue and Immunotherapy for the Treatment of Breast Cancer. Clin. Breast Cancer 2021, 21, e63–e73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zareinejad, M.; Mehdipour, F.; Roshan-Zamir, M.; Faghih, Z.; Ghaderi, A. Dual Functions of T Lymphocytes in Breast Carcinoma: From Immune Protection to Orchestrating Tumor Progression and Metastasis. Cancers 2023, 15, 4771. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jing, H.; Gao, Y.; Sun, Z.; Liu, S. Recent advances in novel tumor immunotherapy strategies based on regulating the tumor microenvironment and immune checkpoints. Front. Immunol. 2025, 16, 1529403. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pelekanou, V.; Villarroel-Espindola, F.; Schalper, K.A.; Pusztai, L.; Rimm, D.L. CD68, CD163, and matrix metalloproteinase 9 (MMP-9) co-localization in breast tumor microenvironment predicts survival differently in ER-positive and -negative cancers. Breast Cancer Res. 2018, 20, 154. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Caronni, N.; La Terza, F.; Vittoria, F.M.; Barbiera, G.; Mezzanzanica, L.; Cuzzola, V.; Barresi, S.; Pellegatta, M.; Canevazzi, P.; Dunsmore, G.; et al. IL-1β+ macrophages fuel pathogenic inflammation in pancreatic cancer. Nature 2023, 623, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch. Pathol. Lab. Med. 2010, 134, 907–922. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- WHO Classification of Tumours Editorial Board. Breast Tumours, 5th ed.; WHO Classification of Tumors Series; Lyon (France) International Agency for Research on Cancer: Lyon, France, 2019; Volume 2. [Google Scholar]
- Guo, H.; Ding, Q.; Gong, Y.; Gilcrease, M.Z.; Zhao, M.; Zhao, J.; Sui, D.; Wu, Y.; Chen, H.; Liu, H.; et al. Comparison of three scoring methods using the FDA-approved 22C3 immunohistochemistry assay to evaluate PD-L1 expression in breast cancer and their association with clinicopathologic factors. Breast Cancer Res. 2020, 22, 69. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Matsumoto, H.; Thike, A.A.; Li, H.; Yeong, J.; Koo, S.L.; Dent, R.A.; Tan, P.H.; Iqbal, J. Increased CD4 and CD8-positive T cell infiltrate signifies good prognosis in a subset of triple-negative breast cancer. Breast Cancer Res. Treat. 2016, 156, 237–247. [Google Scholar] [CrossRef] [PubMed]

| Clinicopathological Parameters | Number of Patients (%) |
|---|---|
| Age (years) ≤50 >50 | 27 (22.0) 96 (78.0) |
| Histological grade | |
| 1 | 50 (40.7) |
| 2 | 61 (49.4) |
| 3 | 4 (3.3) |
| N/A | 8 (6.4) |
| Tumor size (cm) <2 ≥2 N/A | 39 (31.7) 79 (64.2) 5 (4.1) |
| Multifocality Absent Present N/A | 105 (85.4) 14 (11.4) 4 (3.2) |
| Exulcerated tumor Absent Present N/A | 112 (91.1) 7 (5.7) 4 (3.2) |
| Breast Right Left Both | 63 (51.6) 54 (44.3) 5 (4.1) |
| Inflammatory carcinoma Absent Present N/A | 116 (94.3) 2 (1.6) 5 (4.1) |
| Histological type NST Spec.type (lobular) Other spec. type (papillary, mucinous, cribriform) Mixed NST + mucinous Mixed NST + lobular | 90 (73.2) 20 (16.3) 8 (6.5) 4 (3.3) 1 (0.7) |
| Histological subtype Luminal A Luminal B Multifocality | 65 (52.8) 57 (46.3) 1 (0.9) |
| Clinical stage 1 2 3 4 N/A | 32 (26.0) 39 (31.7) 28 (22.8 21 (17.1) 3 (2.4) |
| Lymphovascular invasion Absent Present N/A | 36 (29.3) 82 (66.7) 5 (4.0) |
| Perineural invasion Absent Present N/A | 59 (48.8) 55 (45.5) 7 (5.7) |
| Necrosis Absent Present N/A | 88 (71.5) 33 (26.9) 2 (1.6) |
| Calcifications Absent Present N/A | 82 (66.7) 39 (31.7) 2 (1.6) |
| Ki-67 <20% ≥20% | 72 (58.5) 51 (41.5) |
| Lymph node status Negative Positive N/A | 51 (41.5) 54 (43.9) 18 (14.6) |
| Extra-nodal tumor spreading Absent Present N/A | 21 (38.9) 32 (59.3) 1 (1.8) |
| Metastasis present at the diagnosis Absent Present N/A | 99 (80.5) 22 (17.9) 2 (1.6) |
| Metastasis and recurrence Absent Present, local Present, distant metastasis N/A | 63 (51.2) 8 (6.5) 46 (37.4) 6 (4.9) |
| Months until the appearance of recurrence or metastasis Median (range) | 41.5 (3–180) |
| Follow up (months) Median (range) | 73 (12–240) |
| Number of died patients N (%) | 38 (30.9) |
| PIK3CA mutations Wild type Mutations Invalid | 59 (48.0) 53 (43.1) 11 (8.9) |
| PIK3CA mutations Wild type Exon 9 Exon 20 N/A | 59 (48.0) 20 (16.3) 33 (26.8) 11 (8.9) |
| PD-L1 <1% ≥1% N/A | 85 (69.1) 12 (9.8) 26 (21.1) |
| CD4 median (range) CD8 median (range) CD68 median (range) CD163 median (range) | 8.0 (0.0–232.0) 10 (0.0–142.0) 7 (1.0–75.0) 6 (0.0–68.0) |
| Characteristics (HR+/HER-2-) | PIK3CA | p-Value | PIK3CA | p-Value | |||
|---|---|---|---|---|---|---|---|
| Wt N (%) | Mt N (%) | Wt N (%) | Ex9 N (%) | Ex20 N (%) | |||
| Age (years) ≤50 >50 | 14 (23.7) 45 (76.3) | 12 (22.6) 41 (77.4) | 1.00 ¶ | 14 (23.7) 45 (76.3) | 6 (30.0) 14 (70.0) | 6 (18.2) 27 (81.8) | 0.608 ¶ |
| Clinical stage I II III IV | 12 (20.3) 19 (32.2) 17 (28.8) 11 (18.6) | 15 (30.0) 17 (34.0) 8 (16.0) 10 (20.0) | 0.390 * | 12 (20.3) 19 (32.2) 17 (28.8) 11 (18.6) | 4 (21.1) 5 (26.3) 4 (21.1) 6 (31.6) | 11 (35.5) 12 (38.7) 4 (12.9) 4 (12.9) | 0.319 * |
| Tumor size <2 cm ≥2 cm | 18 (30.5) 41 (69.5) | 17 (35.4) 31 (64.6) | 0.679 ¶ | 18 (30.5) 41 (69.5) | 5 (29.4) 12 (70.6) | 12 (38.7) 19 (61.3) | 0.697 ¶ |
| Multifocality Present Absent | 48 (82.8) 10 (17.2) | 47 (94.0) 3 (6.0) | 0.084 ¶ | 10 (17.2) 48 (82.8) | 1 (5.3) 18 (94.7) | 2 (6.5) 29 (93.5) | 0.199 ¶ |
| Exulcerated Present Absent | 54 (93.1) 4 (6.9) | 47 (94.0) 3 (6.0) | 1.00 ¶ | 4 (6.9) 54 (93.1) | 1 (5.3) 18 (94.7) | 2 (6.5) 29 (93.5) | 0.969 ¶ |
| Inflammatory carcinoma Present Absent | 55 (96.5) 2 (3.5) | 50 (100) 0 (0) | 0.497 ¶ | 2 (3.5) 55 (96.5) | 0 (0) 19 (100) | 0 (0) 31 (100) | 0.409 ¶ |
| Bilateral tumor Present Absent | 54 (91.5) 5 (8.5) | 52 (100) 0 (0) | 0.059 ¶ | 5 (8.5) 54 (91.5) | 0 (0) 20 (100) | 0 (0) 32 (100) | 0.099 ¶ |
| Histological type Ductal NST Lobular Other spec. type Mixed (duct.+lob.) | 44 (74.6) 9 (15.3) 5 (8.5) 1 (1.7) | 42 (79.2) 9 (17.0) 2 (3.8) 0 (0) | 0.569 * | 44 (74.6) 9 (15.3) 5 (8.5) 1 (1.7) | 14 (70.0) 6 (30.0) (0) 0 (0) | 28 (84.8) 3 (9.1) 2 (6.1) 0 (0) | 0.370 * |
| Histological subtype Luminal A Luminal B | 27 (45.8) 32 (54.2) | 31 (58.5) 22 (41.5) | 0.191 ¶ | 27 (45.8) 32 (54.2) | 10 (50.0) 10 (50.0) | 21 (63.6) 12 (36.4) | 0.254 ¶ |
| Histological grade 1 2 3 | 21 (36.8) 33 (57.9) 3 (5.3) | 24 (51.1) 23 (48.9) 0 (0) | 0.131 * | 21 (36.8) 33 (57.9) 3 (5.3) | 5 (29.4) 12 (70.6) 0 (0) | 19 (63.3) 11 (36.7) 0 (0) | 0.055 * |
| Lymph node status Negative Positive | 22 (43.1) 29 (56.9) | 23 (53.5) 20 (46.5) | 0.408 ¶ | 22 (43.1) 29 (56.9) | 7 (43.7) 9 (56.2) | 16 (59.3) 11 (40.7) | 0.373 ¶ |
| Extranodal tumor spreading Absent Present | 11 (37.9) 18 (62.1) | 7 (36.8) 12 (63.2) | 1.00 ¶ | 11 (37.9) 18 (62.1) | 3 (33.3) 6 (66.7) | 4 (40.0) 6 (60.0) | 0.953 ¶ |
| Metastasis present at diagnosis Absent Present | 46 (79.3) 12 (20.7) | 42 (80.8) 10 (19.2) | 1.00 ¶ | 46 (79.3) 12 (20.7) | 14 (70.0) 6 (30.0) | 28 (87.5) 4 (12.5) | 0.302 ¶ |
| Metastasis and recurrence Absent Present | 26 (46.4) 30 (53.6) | 31 (59.6) 21 (40.4) | 0.183 ¶ | 26 (46.4) 30 (53.6) | 9 (45.0) 11 (55.0) | 22 (68.7) 10 (31.2) | 0.097 ¶ |
| Ki-67 <20% ≥20% | 31 (52.5) 28 (47.5) | 35 (66.0) 18 (34.0) | 0.179 ¶ | 31 (52.5) 28 (47.5) | 10 (50.0) 10 (50.0) | 25 (75.8) 8 (24.2) | 0.041 ¶ |
| Variable | PIK3CA | p-Value | ||
|---|---|---|---|---|
| Wt | Mutation | |||
| CD4 median (range) CD4 Low (≤6) High (>6) | 8.0 (0.0–94.0) 21 (41.2) 30 (58.8) | 9.0 (0.0–232.0) 14 (34.1) 27 (65.9) | 0.436 0.524 | |
| CD8 median (range) CD8 Low (≤8) High (>8) | 10.0 (0.0–100.0) 17 (34.7) 32 (65.3) | 14.5 (1.0–142.0) 15 (35.7) 27 (64.3) | 0.655 1.00 | |
| CD68 median (range) CD68 Low (≤23) High (>23) | 8.0 (1.0–30.0) 43 (84.3) 8 (15.7) | 8.0 (1.0–75.0) 37 (92.5) 3 (7.5) | 0.794 0.336 | |
| CD163 median (range) CD163 Low (≤12) High (>12) | 8.0 (1.0–68.0) 32 (72.7) 12 (27.3) | 6.0 (0.0–41.0) 32 (82.1) 7 (17.9) | 0.143 0.433 | |
| PD-L1 <1% ≥1% | 42 (93.3) 3 (6.7) | 34 (81.0) 8 (19.0) | 0.110 | |
| PD-L1 <1% ≥1% | 42 (93.3) 3 (6.7) | Ex9 14 (93.3) 1 (6.7) | Ex20 20 (74.1) 7 (25.9) | 0.044 |
| Variable PIK3CA Wt | Metastasis/Recurrence | p-Value | |
|---|---|---|---|
| No | Yes | ||
| CD4 | |||
| Low (≤6) | 5 (23.8) | 15 (55.6) | 0.039 |
| High (>6) | 16 (76.2) | 12 (44.4) | |
| CD8 | |||
| Low (≤8) | 6 (27.3) | 9 (37.5) | 0.539 |
| High (>8) | 16 (72.7) | 15 (62.5) | |
| CD68 | |||
| Low (≤23) | 17 (81.0) | 24 (88.9) | 0.683 |
| High (>23) | 4 (19.0) | 3 (11.1) | |
| CD163 | |||
| Low (≤12) | 12 (63.2) | 19 (86.4) | 0.144 |
| High (>12) | 7 (36.8) | 3 (13.6) | |
| PD-L1 | |||
| <1% | 17 (89.5) | 22 (95.7) | 0.581 |
| ≥1% | 2 (10.5) | 1 (4.3) | |
| PIK3CA mt | |||
| CD4 | |||
| Low (≤6) | 7 (26.9) | 6 (42.9) | 0.480 |
| High (>6) | 19 (73.1) | 8 (57.1) | |
| CD8 | |||
| Low (≤8) | 8 (28.6) | 6 (46.2) | 0.307 |
| High (>8) | 20 (71.4) | 7 (53.8) | |
| CD68 | |||
| Low (≤23) | 25 (96.2) | 11 (84.6) | 0.253 |
| High (>23) | 1 (3.8) | 2 (15.4) | |
| CD163 | |||
| Low (≤12) | 18 (72.0) | 14 (100) | 0.036 |
| High (>12) | 7 (28.0) | 0 (0) | |
| PDL-1 | |||
| <1% | 21 (75.0) | 12 (92.3) | 0.398 |
| ≥1% | 7 (25.0) | 1 (7.7) | |
| Whole study group | |||
| CD4 | |||
| Low (≤6) | 17 (31.5) | 22 (50.0) | 0.096 |
| High (>6) | 37 (68.5) | 22 (50.0) | |
| CD8 | |||
| Low (≤8) | 17 (29.8) | 17 (42.5) | 0.279 |
| High (>8) | 40 (70.2) | 23 (57.5) | |
| CD68 | |||
| Low (≤23) | 46 (90.2) | 38 (88.4) | 1.00 |
| High (>23) | 5 (9.8) | 5 (11.6) | |
| CD163 | |||
| Low (≤12) | 32 (64.0) | 36 (92.3) | 0.002 |
| High (>12) | 18 (36.0) | 3 (7.7) | |
| PD-L1 | |||
| <1% | 43 (81.1) | 37 (94.9) | 0.065 |
| ≥1% | 10 (18.9) | 2 (5.1) | |
| Variable | CD4 | CD8 | CD68 | CD163 | |
|---|---|---|---|---|---|
| PD-L1 wt | rs | 0.025 | 0.063 | 0.053 | −0.317 |
| P | 0.872 | 0.693 | 0.730 | 0.052 | |
| PD-L1 mt | rs | 0.285 | 0.462 | 0.398 | 0.617 |
| P | 0.078 | 0.0027 | 0.0134 | <0.0001 | |
| PD-L1 whole group | rs | 0.203 | 0.312 | 0.238 | 0.205 |
| P | 0.051 | 0.0026 | 0.023 | 0.061 | |
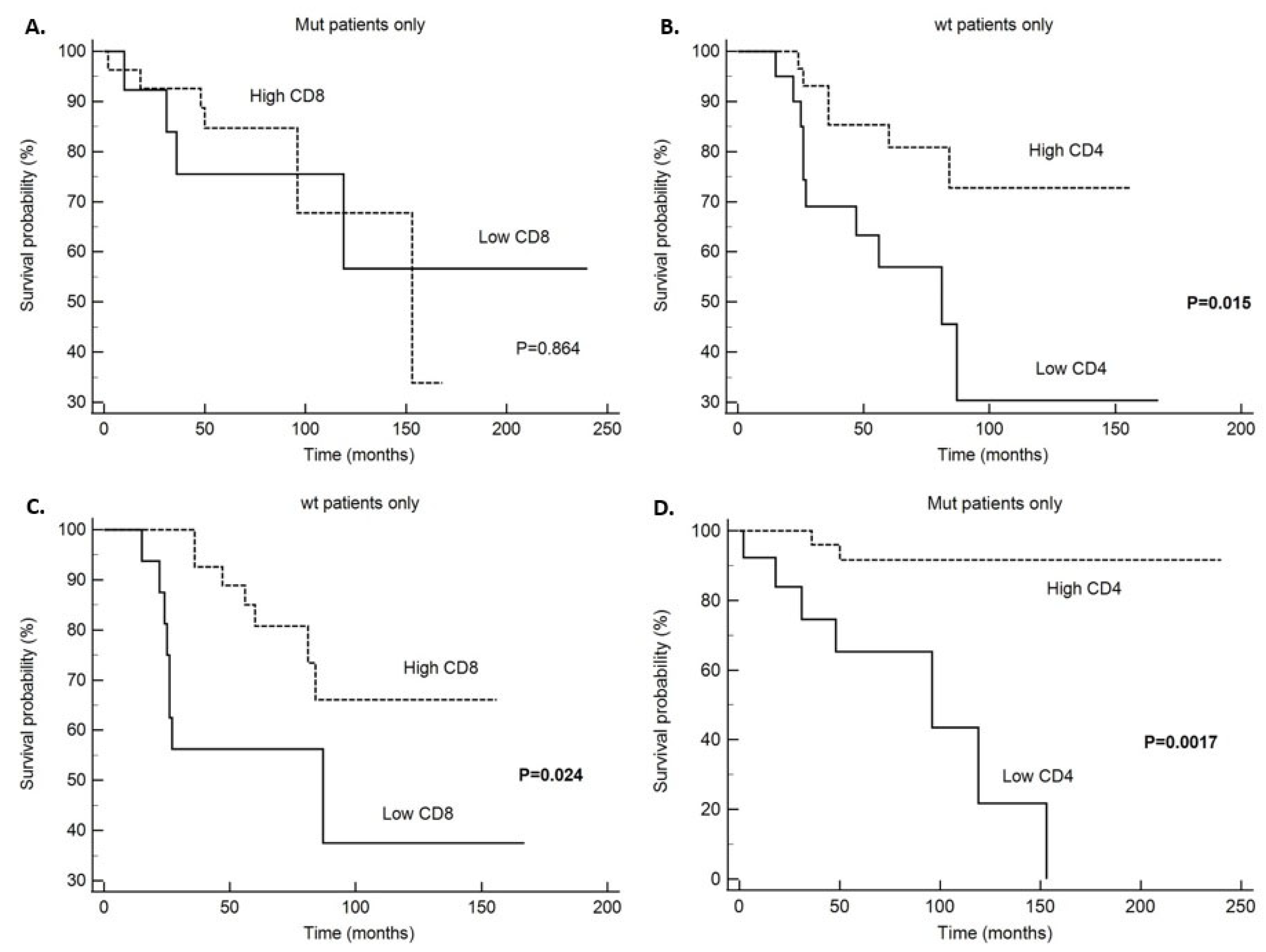
| Variable | N | Died from Underlying Disease (N) | Five-Year Survival (%) | Average Value ± SD | 95% CI | χ2 | Log-Rank Test (p Value) |
|---|---|---|---|---|---|---|---|
| Patients without PIK3CA mutation (wt) | |||||||
| CD4 | |||||||
| Low (≤6) | 20 | 10 | 63 | 86.7 ± 16.2 | 54.9–118.6 | 5.92 | 0.015 |
| High (>6) | 30 | 6 | 85 | 127.5 ± 10.2 | 107.5–147.6 | ||
| CD8 | |||||||
| Low (<8) | 16 | 8 | 56 | 89.3 ± 18.9 | 52.1–126.4 | 5.09 | 0.024 |
| High (≥8) | 32 | 7 | 85 | 124.3 ± 10.1 | 104.5–144.2 | ||
| CD68 | |||||||
| Low (≤23) | 42 | 15 | 75 | 111.0 ± 11.2 | 89.1–132.9 | 1.256 | 0.263 |
| High (>23) | 8 | 1 | 85 | 132.3 ± 16.4 | 100.1–164.4 | ||
| CD163 | |||||||
| Low (≤12) | 31 | 10 | 76 | 99.3 ± 13.3 | 73.3–125.4 | 0.722 | 0.395 |
| High (>12) | 12 | 3 | 75 | 120.5 ± 14.8 | 91.4–149.5 | ||
| PD-L1 | |||||||
| <1% | 41 | 13 | 77 | 119.2 ± 10.7 | 98.1–140.2 | 0.553 | 0.457 |
| ≥1% | 3 | 0 | 100 | 55.0 ± 0.0 | 55.0–55.0 | ||
| Patients with PIK3CA mutation (mt) | |||||||
| CD4 | |||||||
| Low (≤6) | 13 | 7 | 63 | 89.1 ± 17.9 | 54.4–123.8 | 9.88 | 0.0017 |
| High (>6) | 26 | 2 | 85 | 223.6 ± 11.2 | 201.7–245.4 | ||
| CD8 | |||||||
| Low (<8) | 13 | 4 | 75 | 164.8 ± 30.1 | 105.9–223.7 | 0.029 | 0.864 |
| High (≥8) | 27 | 6 | 85 | 129.6 ± 13.4 | 103.4–155.8 | ||
| CD68 | |||||||
| Low (≤23) | 36 | 9 | 79 | 158.3 ± 22.9 | 113.2–203.3 | 1.219 | 0.269 |
| High (>23) | 3 | 0 | 100 | 168.0 ± 0.0 | 168.0–168.0 | ||
| CD163 | |||||||
| Low (≤12) | 32 | 8 | 84 | 164.2 ± 21.6 | 121.9–206.5 | 0.045 | 0.831 |
| High (>12) | 7 | 1 | 87 | 86.9 ± 5.7 | 75.7 – 98.0 | ||
| PD-L1 | |||||||
| <1% | 32 | 9 | 80 | 148.8 ± 23.0 | 103.7–193.9 | 0.859 | 0.354 |
| ≥1% | 8 | 1 | 89 | 153.3 ± 13.8 | 126.2–180.3 | ||
| Whole study group | |||||||
| CD4 | |||||||
| Low (≤6) | 39 | 20 | 62 | 85.8 ± 10.5 | 65.3–106.4 | 13.29 | 0.0003 |
| High (>6) | 61 | 11 | 85 | 196.6 ± 12.1 | 172.9- 220.2 | ||
| CD8 | |||||||
| Low (<8) | 34 | 15 | 62 | 129.7 ± 20.7 | 89.2–170.2 | 4.19 | 0.041 |
| High (≥8) | 65 | 16 | 85 | 128.5 ± 8.3 | 112.2–144.7 | ||
| CD68 | |||||||
| Low (≤23) | 86 | 28 | 72 | 148.6 ± 14.6 | 120.1–177.1 | 2.14 | 0.143 |
| High (>23) | 11 | 1 | 90 | 153.8 ± 13.5 | 127.4–180.2 | ||
| CD163 | |||||||
| Low (≤12) | 69 | 22 | 77 | 144.5 ± 16.0 | 113.1–175.9 | 0.829 | 0.363 |
| High (>12) | 23 | 5 | 87 | 124.7 ± 9.9 | 105.2–144.2 | ||
| PD-L1 | |||||||
| <1% | 82 | 27 | 75 | 144.4 ± 15.2 | 114.7–174.2 | 2.741 | 0.098 |
| ≥1% | 12 | 1 | 90 | 156.2 ± 11.2 | 134.3–178.1 | ||
| PIK3CA specific mutation | |||||||
| Wt | 58 | 19 | 72 | 132.6 ± 11.7 | 109.7–155.6 | 1.788 | 0.409 |
| Exon 9 | 20 | 4 | 85 | 140.8 ± 12.8 | 115.8–165.9 | ||
| Exon 20 | 31 | 8 | 80 | 159.4 ± 29.9 | 110.6–208.2 | ||
| PIK3CA mutation | |||||||
| Wt | 58 | 19 | 72 | 132.6 ± 11.7 | 109.7–155.6 | 1.697 | 0.195 |
| Mutation | 51 | 12 | 81 | 173.5 ± 16.7 | 140.7–206.3 | ||
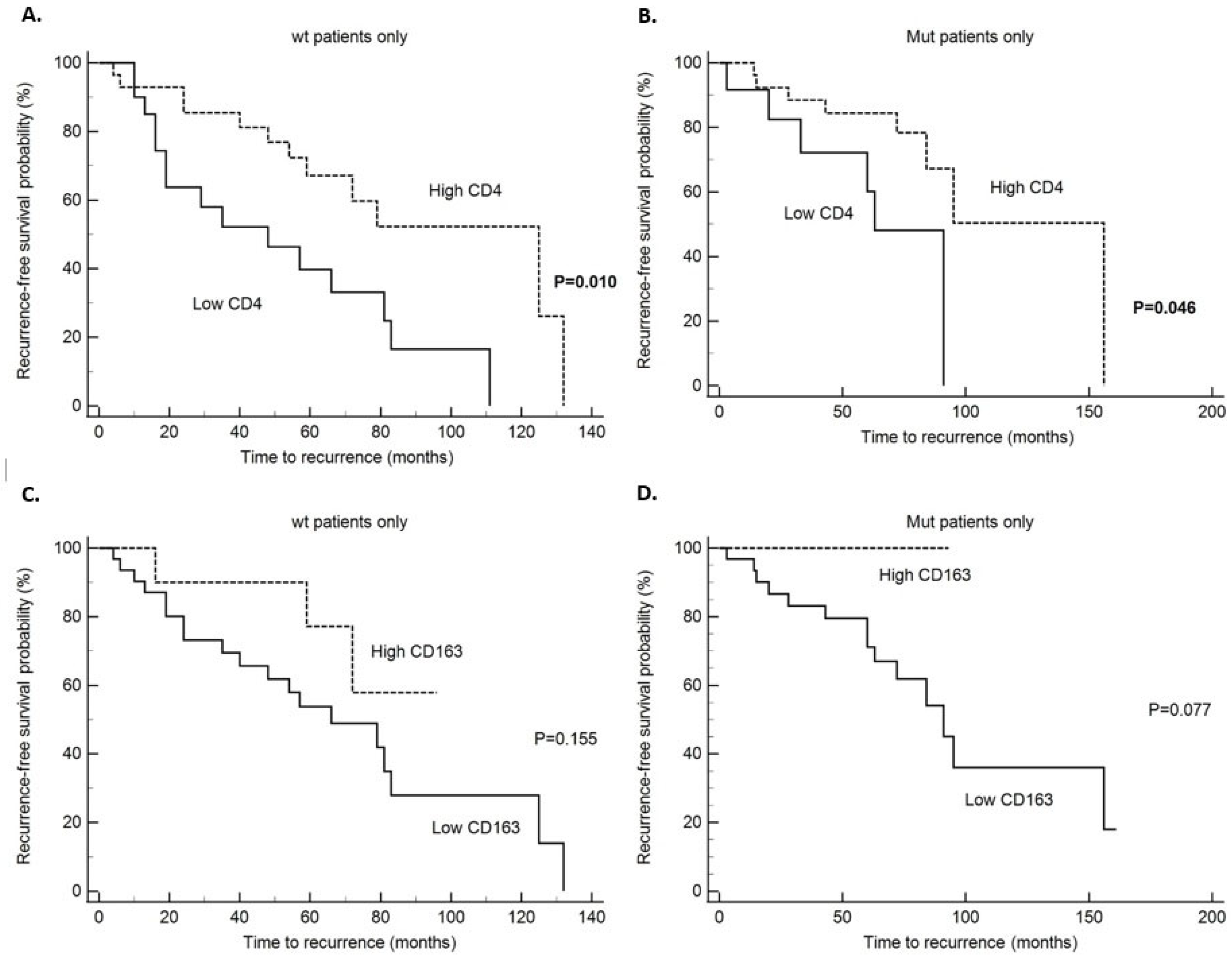
| Variable | N | Disease Recurrence (N) | Five Year DFS (%) | Mean Value ± SD | 95% CI | χ2 | Log-Rank Test (p-Value) |
|---|---|---|---|---|---|---|---|
| Patients without PIK3CA mutation (wt) | |||||||
| CD4 | 6.64 | 0.01 | |||||
| Low (≤6) | 20 | 15 | 45 | 51.95 ± 8.84 | 34.63–69.3 | ||
| High (>6) | 28 | 12 | 78.5 | 89.82 ± 7.25 | 70.35–109.3 | ||
| CD8 | 1.84 | 0.175 | |||||
| Low (<8) | 15 | 9 | 50 | 51.68 ± 8.90 | 34.24–69.12 | ||
| High (≥8) | 31 | 15 | 65 | 84.37 ± 8.78 | 67.16–101.59 | ||
| CD68 | 0.007 | 0.932 | |||||
| Low (≤23) | 41 | 24 | 55 | 72.4 ± 7.81 | 57.09–87.7 | ||
| High (>23) | 7 | 3 | 41 | 66.21 ± 12.34 | 42.02–90.4 | ||
| CD163 | 2.02 | 0.155 | |||||
| Low (≤12) | 31 | 19 | 55 | 69.3 ± 9.1 | 51.5–87.1 | ||
| High (>12) | 10 | 3 | 78 | 78.61 ± 8.56 | 61.8–95.4 | ||
| PD-L1 | 0.07 | 0.786 | |||||
| <1% | 39 | 22 | 59 | 72.45 ± 7.38 | 57.99–86.91 | ||
| ≥1% | 3 | 1 | 65 | 40.0 ± 12.25 | 15.99–64.01 | ||
| Patients with PIK3CA mutation (mt) | |||||||
| CD4 | 3.98 | 0.0458 | |||||
| Low (≤6) | 13 | 6 | 72 | 64.1 ± 10.4 | 43.7–84.4 | ||
| High (>6) | 26 | 8 | 85 | 112.3 ± 14.4 | 84.1–140.4 | ||
| CD8 | 0.313 | 0.576 | |||||
| Low (<8) | 13 | 6 | 75 | 101.6 ± 18.92 | 64.52–138.7 | ||
| High (≥8) | 27 | 7 | 88 | 83.41 ± 4.62 | 74.32–92.45 | ||
| CD68 | 1.714 | 0.190 | |||||
| Low (≤23) | 36 | 11 | 82 | 109.2 ± 12.29 | 85.13–133.29 | ||
| High (>23) | 3 | 2 | 66 | 64.67 ± 8.99 | 47.03–82.30 | ||
| CD163 | 3.136 | 0.077 | |||||
| Low (≤12) | 32 | 14 | 80 | 96.1 ± 11.77 | 73.02–119.17 | ||
| High (>12) | 7 | 0 | 100 | 93.0 ± 0.0 | 93.0–93.0 | ||
| PD-L1 | 1.943 | 0.163 | |||||
| <1% | 32 | 12 | 80 | 97.75 ± 12.8 | 72.58–122.9 | ||
| ≥1% | 8 | 1 | 100 | 92.0 ± 3.65 | 84.84–99.16 | ||
| Whole study group | |||||||
| CD4 | 8.09 | 0.005 | |||||
| Low (≤6) | 39 | 22 | 43 | 61.9 ± 6.6 | 49.1–74.8 | ||
| High (>6) | 58 | 22 | 78 | 98.4 ± 8.5 | 81.8–115.0 | ||
| CD8 | 1.493 | 0.222 | |||||
| Low (≤8) | 33 | 17 | 60 | 85.1 ± 12.4 | 60.8–109.2 | ||
| High (>8) | 63 | 23 | 73 | 90.8 ± 5.2 | 73.6–103.0 | ||
| CD68 | 0.367 | 0.545 | |||||
| Low (≤23) | 84 | 38 | 68 | 86.9 ± 7.2 | 72.9–101.0 | ||
| High (>23) | 10 | 5 | 50 | 66.3 ± 9.2 | 48.3–84.3 | ||
| CD163 | 6.874 | 0.009 | |||||
| Low (≤12) | 68 | 36 | 60.5 | 80.3 ± 7.5 | 65.7–94.9 | ||
| High (>12) | 21 | 3 | 89 | 87.7 ± 4.6 | 78.8–96.7 | ||
| PD-L1 | 3.229 | 0.072 | |||||
| <1% | 79 | 37 | 73 | 83.9 ± 7.5 | 69.3–98.5 | ||
| ≥1% | 12 | 2 | 91 | 85.7 ± 7.2 | 71.6–99.8 | ||
| PIK3CA specific mutation | 6.375 | 0.041 | |||||
| Wt | 56 | 30 | 65 | 77.9 ± 7.7 | 62.9–93.0 | ||
| Exon 9 | 20 | 11 | 72 | 85.8 ± 13.5 | 59.5–112.2 | ||
| Exon 20 | 31 | 10 | 82 | 125.0 ± 14.0 | 97.7 ± 152.4 | ||
| PIK3CA mutation | 4.971 | 0.026 | |||||
| Wt | 56 | 30 | 62 | 77.9 ± 7.7 | 62.9–93.0 | ||
| Mutation | 51 | 21 | 79.5 | 107.4 ± 10.9 | 85.9–128.8 | ||
| Disease Specific Survival | |||
|---|---|---|---|
| Variables (Cut Off) | HR | 95% CI | p-Value |
| CD4 (≤6) | 0.51 | 0.19–1.30 | 0.159 |
| CD8 (<8) | 0.61 | 0.23–0.1.59 | 0.313 |
| PD-L1 | 0.20 | 0.02–1.62 | 0.133 |
| Disease-free survival | |||
| CD4 (≤6) | 0.54 | 0.24–1.21 | 0.134 |
| CD163 (>12) | 0.35 | 0.09–1.24 | 0.351 |
| PD-L1 | 0.83 | 0.18–3.76 | 0.835 |
| PIK3CA mutation | 0.45 | 0.21–0.99 | 0.0488 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopac, D.; Babarović, E.; Hagen, J.; Valković Zujić, P.; Grebić, D.; Hadžisejdić, I. Hormone Receptor Positive/HER2 Negative Breast Carcinoma: Association of PIK3CA Mutational Status with PD-L1 and Tumor Cell Microenvironment and Their Prognostic Significance. Int. J. Mol. Sci. 2025, 26, 9489. https://doi.org/10.3390/ijms26199489
Lopac D, Babarović E, Hagen J, Valković Zujić P, Grebić D, Hadžisejdić I. Hormone Receptor Positive/HER2 Negative Breast Carcinoma: Association of PIK3CA Mutational Status with PD-L1 and Tumor Cell Microenvironment and Their Prognostic Significance. International Journal of Molecular Sciences. 2025; 26(19):9489. https://doi.org/10.3390/ijms26199489
Chicago/Turabian StyleLopac, Danijel, Emina Babarović, Justin Hagen, Petra Valković Zujić, Damir Grebić, and Ita Hadžisejdić. 2025. "Hormone Receptor Positive/HER2 Negative Breast Carcinoma: Association of PIK3CA Mutational Status with PD-L1 and Tumor Cell Microenvironment and Their Prognostic Significance" International Journal of Molecular Sciences 26, no. 19: 9489. https://doi.org/10.3390/ijms26199489
APA StyleLopac, D., Babarović, E., Hagen, J., Valković Zujić, P., Grebić, D., & Hadžisejdić, I. (2025). Hormone Receptor Positive/HER2 Negative Breast Carcinoma: Association of PIK3CA Mutational Status with PD-L1 and Tumor Cell Microenvironment and Their Prognostic Significance. International Journal of Molecular Sciences, 26(19), 9489. https://doi.org/10.3390/ijms26199489






