Impact of Helicobacter pylori Virulence Genotypes cagA, vacA, oipA, and babA2 on Severity of Gastropathies in Brazilian Patients
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Characteristics of Participants
2.2. Endoscopic Findings and Their Association with H. pylori Infection
2.3. Association Between Virulence Genes and Demographic or Clinical Variables
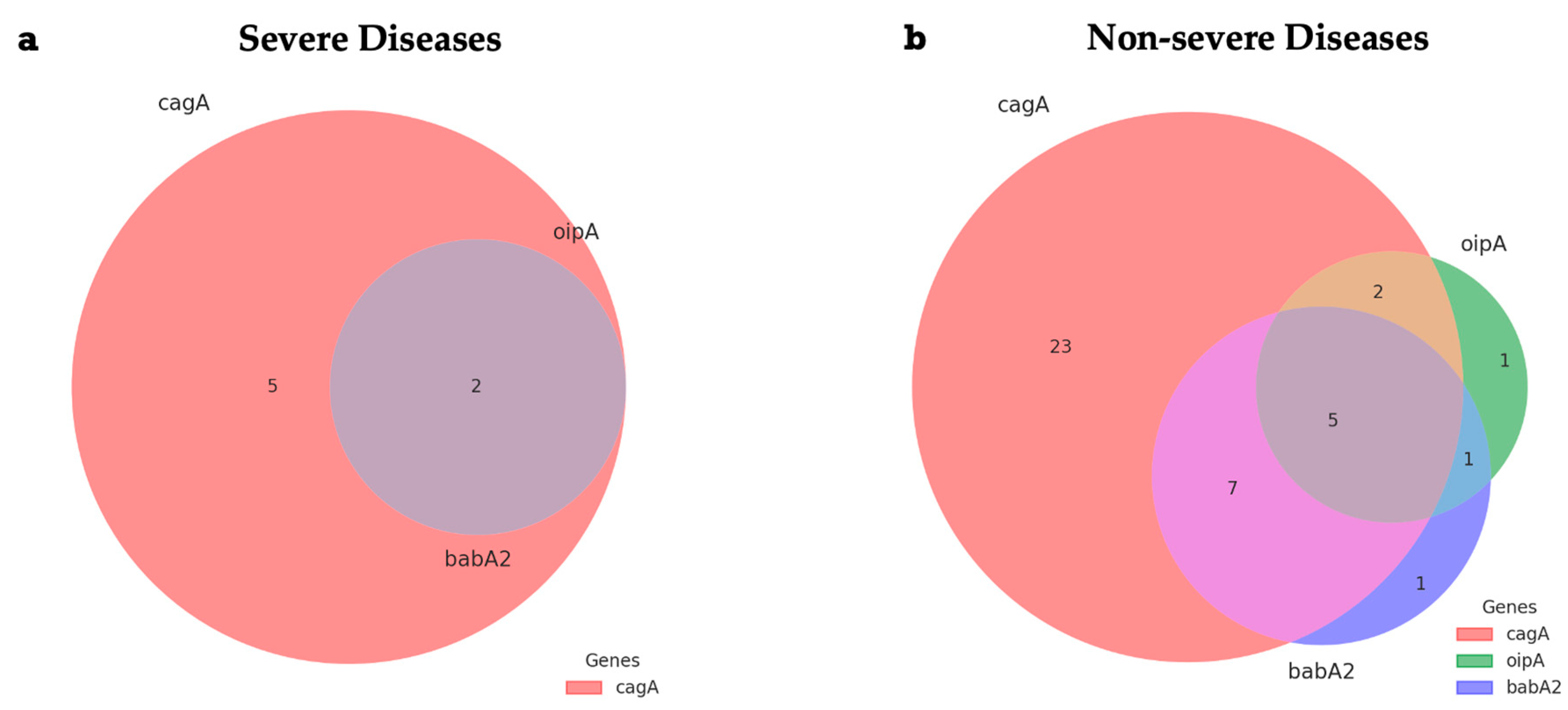
2.4. Distribution Patterns of Virulence Gene Combinations
3. Discussion
4. Materials and Methods
4.1. Population and Samples
4.2. Histology
4.3. DNA Extraction
4.4. DNA Amplification of H. pylori and Virulence Genes cagA, vacA, oipA, and babA2
4.5. Diagnosis of Gastroduodenal Diseases and Severity Criteria
4.6. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| H. pylori | Helicobacter pylori |
| cagA | Cytotoxin-associated A gene |
| vacA | Vacuolating cytotoxin A gene |
| oipA | Outer Inflammatory Protein A gene |
| babA2 | Blood-group antigen-binding adhesin gene |
| PCR | Polymerase chain reaction |
References
- Wattanawongdon, W.; Simawaranon Bartpho, T.; Tongtawee, T. Relationship between Helicobacter pylori virulence genes and gastroduodenal disease. J. Int. Med. Res. 2023, 51, 03000605231161465. [Google Scholar] [CrossRef] [PubMed]
- Barhoine, M.; Moustaoui, F.; Hammani, O.; Aghrouch, M.; Lemkhente, Z.; Belhabib, Z.; Bajaddoub, Z.; Touyar, A.; Aqoudad, N.; Rherissi, B.; et al. The Effect of Helicobacter pylori Gene Combinations of cagA, cagE, virB11, vacA and babA on the Outcome of Gastric Disease in a Southern Moroccan Population. Pathogens 2025, 14, 279. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.K.; Khan, M.B. Overview of Helicobacter pylori Infection, Prevalence, Risk Factors, and Its Prevention. Adv. Gut Microbiome Res. 2023, 2023, 9747027. [Google Scholar] [CrossRef]
- Zamani, M.; Ebrahimtabar, F.; Zamani, V.; Miller, W.H.; Alizadeh-Navaei, R.; Shokri-Shirvani, J.; Derakhshan, M.H. Systematic review with meta-analysis: The worldwide prevalence of Helicobacter pylori infection. Aliment. Pharmacol. Ther. 2018, 1, 868–876. [Google Scholar] [CrossRef]
- Sedarat, Z.; Taylor-Robinson, A.W. Helicobacter pylori Outer Membrane Proteins and Virulence Factors: Potential Targets for Novel Therapies and Vaccines. Pathogens 2024, 13, 392. [Google Scholar] [CrossRef]
- Hooi, J.K.; Ying Lai, W.; Khoon Ng, W.; Suen, M.M.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.; Wu, J.C.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-analysis. Gastroenterology 2017, 153, 420–429. [Google Scholar] [CrossRef]
- Wondmagegn, Y.M.; Girmay, G.; Amare, G.A.; Assefa, M.; Tamir, M.; Abriham, Z.Y.; Setegn, A. Prevalence of intestinal parasites and Helicobacter pylori co-infection in people with gastrointestinal symptoms in Africa: A systematic review and meta-analysis. BMC Infect. Dis. 2025, 25, 20. [Google Scholar] [CrossRef]
- Teng, K.W.; Hsieh, K.S.; Hung, J.S.; Wang, C.J.; Liao, E.C.; Chen, P.C.; Lin, Y.H.; Wu, D.C.; Lin, C.H.; Wang, W.C.; et al. Helicobacter pylori employs a general protein glycosylation system for the modification of outer membrane adhesins. Gut Microbes 2022, 14, 2130650. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Yamaoka, Y. Helicobacter pylori BabA in adaptation for gastric colonization. World J. Gastroenterol. 2017, 23, 158–4169. [Google Scholar] [CrossRef]
- González-Stegmaier, R.; Aguila-Torres, P.; Villarroel-Espíndola, F. Historical and Molecular Perspectives on the Presence of Helicobacter pylori in Latin America: A Niche to Improve Gastric Cancer Risk Assessment. Int. J. Mol. Sci. 2024, 25, 1761. [Google Scholar] [CrossRef]
- Curado, M.P.; de Oliveira, M.M.; Fagundes, M.d.A. Prevalence of Helicobacter pylori infection in Latin America and the Caribbean populations: A systematic review and meta-analysis. Cancer Epidemiol. 2019, 60, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Bustos-Fraga, S.; Salinas-Pinta, M.; Vicuña-Almeida, Y.; de Oliveira, R.B.; Baldeón-Rojas, L. Prevalence of Helicobacter pylori genotypes: cagA, vacA (m1), vacA (s1), babA2, dupA, iceA1, oipA and their association with gastrointestinal diseases. A cross-sectional study in Quito-Ecuador. BMC Gastroenterol. 2023, 23, 197. [Google Scholar] [CrossRef]
- Roberts, J.R.; Tran, S.C.; Frick-Cheng, A.E.; Bryant, K.N.; Okoye, C.D.; McDonald, W.H.; Ohi, M.D. Subdomains of the Helicobacter pylori Cag T4SS outer membrane core complex exhibit structural independence. Life Sci. Alliance 2024, 7, 6. [Google Scholar]
- Farsimadan, M.; Heravi, F.S.; Emamvirdizadeh, A.; Moradi, S.; Iranpour, H.; Tabasi, E.; Cover, T.L.; Ohi, M.D. Evaluation of Helicobacter pylori Genotypes in Obese Patients with Gastric Ulcer, Duodenal Ulcer, and Gastric Cancer: An Observational Study. Dig. Dis. 2022, 40, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Thai, T.H.N.; Nguyen, H.P.; Nguyen, T.H.Y.; Nguyen, T.B.H.; Nguyen, T.H.; Nguyen, T.M.N.; Ha, T.M.T. Genetic diversity of the oipA gene among Helicobacter pylori isolates and clinical outcome in Vietnam. Infect. Genet. Evol. 2023, 112, 105438. [Google Scholar] [CrossRef]
- Hedayati, M.A.; Salavati, S. Transcriptional Profile of Helicobacter pylori Virulence Genes in Patients with Gastritis and Gastric Cancer. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 1309519. [Google Scholar] [CrossRef]
- Zhao, Q.; Song, C.; Wang, K.; Li, D.; Yang, Y.; Liu, D.; Wang, L.; Zhou, N.; Xie, Y. Prevalence of Helicobacter pylori babA, oipA, sabA and homB genes in isolates from Chinese patients with different gastroduodenal diseases. Med. Microbiol. Immunol. 2020, 209, 565–577. [Google Scholar] [CrossRef]
- Abadi, M.S.S.; Ashrafi-Dehkordi, K.; Ahmadi, R.; Rahimian, G.; Mirzaei, Y.; Fereidani, R.; Shohan, M.; Azadegan-Dehkordi, F. Frequency of virulence-associated genotypes of Helicobacter pylori and their correlation with clinical outcome and histological parameters in infected patients. Heliyon 2021, 7, e07610. [Google Scholar] [CrossRef]
- Qasim, M.T.; Mohammed, Z.I. The Association of Helicobacter pylori Infection and Virulence Factors in Gastric Cancer in Thi-Qar, Iraq. Asian Pac. J. Cancer Biol. 2024, 9, 541–545. [Google Scholar] [CrossRef]
- Othman, K.I.; Balaky, S.T.J. Investigating the potential association of Helicobacter pylori cagA, vacA s1/s2, iceA1, iceA2, babA2, sabA and oipA genotypes with gastric disease severity. Cell. Mol. Biol. 2025, 71, 88–95. [Google Scholar] [CrossRef]
- Farzi, N.; Yadegar, A.; Aghdaei, H.A.; Yamaoka, Y.; Zali, M.R. Genetic diversity and functional analysis of oipA gene in association with other virulence factors among Helicobacter pylori isolates from Iranian patients with different gastric diseases. Infect. Genet. Evol. 2018, 60, 26–34. [Google Scholar] [CrossRef]
- Liu, D.; Peng, J.; Xie, J.; Xie, Y. Comprehensive analysis of the function of helicobacter-associated ferroptosis gene YWHAE in gastric cancer through multi-omics integration, molecular docking, and machine learning. Apoptosis 2024, 29, 439–456. [Google Scholar] [CrossRef]
- Mohamed, S.H.; Hamed, M.; Alamoudi, H.A.; Jastaniah, Z.; Alakwaa, F.M.; Reda, A. Multi-omics analysis of Helicobacter pylori–associated gastric cancer identifies hub genes as a novel therapeutic biomarker. Brief. Bioinform. 2025, 26, bbaf241. [Google Scholar] [CrossRef]
- Chen, R.; Li, Y.; Chen, X.; Chen, J.; Song, J.; Yang, X.; Ye, L.; Wu, Z.; Xie, P.; Zhong, Q.; et al. dupA+ H. pylori reduces diversity of gastric microbiome and increases risk of erosive gastritis. Front. Cell. Infect. Microbiol. 2023, 13, 1103909. [Google Scholar] [CrossRef]
- Razuka-Ebela, D.; Polaka, I.; Parshutin, S.; Santare, D.; Ebela, I.; Murillo, R.; Herrero, R.; Tzivian, L.; Leja, M. Sociodemographic, Lifestyle and Medical Factors Associated with Helicobacter Pylori Infection. J. Gastrointest. Liver Dis. 2020, 29, 319–327. [Google Scholar] [CrossRef]
- Abdullah, M.; Greenfield, L.K.; Bronte-Tinkew, D.; Capurro, M.I.; Rizzuti, D.; Jones, N.L. VacA promotes CagA accumulation in gastric epithelial cells during Helicobacter pylori infection. Sci. Rep. 2019, 9, 38. [Google Scholar] [CrossRef]
- Yang, K.; Shi, L.; Chen, J. Analysis of the relationship between oipA, babA2, babB genotypes and Helicobacter pylori-related gastric precancerous lesions. J. Pak. Med. Assoc. 2023, 73, 249–252. [Google Scholar] [CrossRef]
- Fan, J.; Zhu, J.; Xu, H. Strategies of Helicobacter pylori in evading host innate and adaptive immunity: Insights and prospects for therapeutic targeting. Front. Cell. Infect. Microbiol. 2024, 14, 1342913. [Google Scholar] [CrossRef]
- Osman, H.A.; Hasan, H.; Suppian, R.; Hassan, S.; Andee, D.Z.; Majid, N.A.; Zilfalil, B.A. Prevalence of Helicobacter pylori cagA, babA2, and dupA genotypes and correlation with clinical outcome in Malaysian patients with dyspepsia. Turk. J. Med. Sci. 2015, 45, 940–946. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Kodama, T.; Gutierrez, O.; Kim, J.G.; Kashima, K.; Graham, D.Y. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: Studies in four different countries. J. Clin. Microbiol. 1999, 37, 2274–2279. [Google Scholar] [CrossRef]
- Sallas, M.L.; Melchiades, J.L.; Zabaglia, L.M.; Moreno, J.R.d.P.; Orcini, W.A.; Chen, E.S.; Smith, M.D.A.C.; Payão, S.L.M.; Rasmussen, L.T. Prevalence of Helicobacter pylori vacA, cagA, dupA and oipA Genotypes in Patients with Gastric Disease. Adv. Microbiol. 2017, 7, 1–9. [Google Scholar] [CrossRef][Green Version]
- Cadamuro, A.C.T.; Rossi, A.F.T.; Maniezzo, N.M.; Silva, A.E. Helicobacter pylori infection: Host immune response, implications on gene expression and microRNAs. World J. Gastroenterol. 2014, 20, 1424. [Google Scholar] [CrossRef] [PubMed]
- Cover, T.L.; Lacy, D.B.; Ohi, M.D. The Helicobacter pylori Cag Type IV Secretion System. Trends Microbiol. 2020, 28, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Elbehiry, A.; Marzouk, E.; Abalkhail, A.; Sindi, W.; Alzahrani, Y.; Alhifani, S.; Alshehri, T.; Anajirih, N.A.; ALMutairi, T.; Alsaedi, A.; et al. Pivotal role of Helicobacter pylori virulence genes in pathogenicity and vaccine development. Front. Med. 2024, 11, 1523991. [Google Scholar] [CrossRef]
- Coelho, L.G.V.; Marinho, J.R.; Genta, R.; Ribeiro, L.T.; Passos Mdo, C.F.; Zaterka, S.; Assumpção, P.P.; Barbosa, A.J.A.; Barbuti, R.; Braga, L.L.; et al. IV Brazilian Consensus Conference on Helicobacter Pylori Infection. Arq. Gastroenterol. 2018, 55, 97–121. [Google Scholar] [CrossRef]
- Braga, L.L.B.C.; Batista, M.H.R.; De Azevedo, O.G.R.; Da Silva Costa, K.C.; Gomes, A.D.; Rocha, G.A.; Queiroz, D.M.M. oipA “on” status of Helicobacter pylori is associated with gastric cancer in North-Eastern Brazil. BMC Cancer 2019, 19, 48. [Google Scholar] [CrossRef]
- Silva, L.L.L.; Oliveira, A.K.S.; Gama, A.R.; Pontes, J.C.; Ramos, A.F.P.L.; Silva, A.M.T.C.; Cardoso, D.M.M.; Blanco, A.J.V.; Rasmussem, L.T.; Carneiro, L.C.; et al. Prevalence of Helicobacter pylori cagA, dupA and vacA genotypes and their association with the severity of gastropathies in patients with dyspepsia. Genet. Mol. Res 2021, 20, GMR18883. [Google Scholar] [CrossRef]
- Oliveira, A.K.S.; Silva, L.L.d.L.; Miguel, M.P.; Blanco, A.J.V.; Carneiro, L.C.; Barbosa, M.S. Helicobacter pylori cagA virulence gene and severe esogastroduodenal diseases: Is there an association? Arq. Gastroenterol. 2021, 58, 468–475. [Google Scholar] [CrossRef]
- Stolte, M.; Meining, A. The updated Sydney System: Classification and grading of gastritis as the basis of diagnosis and treatment. Can. J. Gastroenterol. Hepatol. 2001, 15, 591–598. [Google Scholar] [CrossRef]
- Scholte, G.H.A.; Van Doom, L.J.; Quint, W.G.V.; Lindeman, J. Polymerase chain reaction for the detection of Helicobacter pylori in formaldehyde-sublimate fixed, paraffin-embedded gastric biopsies. Diagn. Mol. Pathol. 1997, 6, 238–243. [Google Scholar] [CrossRef]
- Dadashzadeh, K.; Peppelenbosch, M.P.; Adamu, A.I. Helicobacter pylori Pathogenicity Factors Related to Gastric Cancer. Can. J. Gastroenterol. Hepatol. 2017, 2017, 7942489. [Google Scholar] [CrossRef]
- Mansour, K.B.; Fendri, C.; Zribi, M.; Masmoudi, A.; Labbene, M.; Fillali, A.; Mami, N.B.; Najjar, T.; Meherzi, A.; Sfar, T.; et al. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 10. [Google Scholar] [CrossRef]
- Van Doorn, L.J.; Figueiredo, C.; Rossau, R.; Jannes, G.; Van Asbroeck, M.; Sousa, J.C.; Carneiro, F.; Quint, W.G.V. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J. Clin. Microbiol. 1998, 36, 1271–1276. [Google Scholar] [CrossRef]
- Paredes-Osses, E.; Sáez, K.; Sanhueza, E.; Hebel, S.; González, C.; Briceño, C.; Cancino, A.C. Association between cagA, vacAi and dupA genes of Helicobacter pylori and gastroduodenal pathologies in Chilean patients. Folia Microbiol. 2017, 62, 437–444. [Google Scholar] [CrossRef]

| Total n = 106 | H. pylori n (%) | p * | ||
|---|---|---|---|---|
| No n = 37 | Yes n = 69 | |||
| Age Group | ||||
| <40 years | 28 (26.4%) | 5 (13.5%) | 23 (33.3%) | 0.087 |
| 40 to 50 years | 19 (17.9%) | 8 (21.6%) | 11 (15.9%) | |
| 51 years or more | 59 (55.7%) | 24 (64.9%) | 35 (50.7%) | |
| Sex | ||||
| Female | 82 (77.4%) | 29 (78.4%) | 53 (76.8%) | 0.856 |
| Male | 24 (22.6%) | 8 (21.6%) | 16 (23.2%) | |
| Education Level ** | ||||
| Illiterate | 7 (6.6%) | 3 (8.1%) | 4 (5.8%) | 0.463 |
| Primary education | 55 (51.9%) | 20 (54.1%) | 35 (50.7%) | |
| Secondary education | 35 (33.0%) | 13 (35.1%) | 22 (31.9%) | |
| Higher education | 9 (8.5%) | 1 (2.7%) | 8 (11.6%) | |
| Monthly Family Income | ||||
| <1 minimum wage | 13 (12.3%) | 3 (8.1%) | 10 (14.5%) | 0.471 |
| 1 to 2 minimum wages | 36 (34.0%) | 12 (32.4%) | 24 (34.8%) | |
| 3 to 5 minimum wages | 33 (31.1%) | 10 (27.0%) | 23 (33.3%) | |
| 5 or more minimum wages | 4 (3.8%) | 2 (5.4%) | 2 (2.9%) | |
| Unknown | 20 (18.9%) | 10 (27.0%) | 10 (14.5%) | |
| Total n = 106 | H. pylori n (%) | p * | ||
|---|---|---|---|---|
| No n = 37 | Yes n = 69 | |||
| Severe Diseases | ||||
| Gastric adenocarcinoma | 2 (1.9%) | 0 (0.0%) | 2 (2.9%) | 0.296 |
| Gastric atrophy | 11 (10.4%) | 7 (18.9%) | 4 (5.8%) | 0.035 |
| Intestinal metaplasia | 6 (5.7%) | 4 (10.8%) | 2 (2.9%) | 0.093 |
| Non-severe Diseases | ||||
| Esophagus | ||||
| Erosive esophagitis | 15 (14.2%) | 5 (13.5%) | 10 (14.5%) | 0.890 |
| Stomach | ||||
| Anthematous gastritis | 43 (40.6%) | 14 (37.8%) | 29 (42.0%) | 0.675 |
| Chronic gastritis | 1 (0.9%) | 1 (2.7%) | 0 (0.0%) | 0.170 |
| Erosive gastritis | 32 (30.2%) | 11 (29.7%) | 21 (30.4%) | 0.940 |
| Gastric ulcer | 4 (3.8%) | 3 (8.1%) | 1 (1.4%) | 0.086 |
| General gastritis | 74 (69.8%) | 25 (67.6%) | 49 (71.0%) | 0.713 |
| Xanthelasma (gastric) | 1 (0.9%) | 0 (0.0%) | 1 (1.4%) | 0.462 |
| Duodenum | ||||
| Anthematous duodenitis | 4 (3.8%) | 0 (0.0%) | 4 (5.8%) | 0.135 |
| Duodenal ulcer | 1 (0.9%) | 0 (0.0%) | 1 (1.4%) | 0.462 |
| Duodenitis | 1 (0.9%) | 1 (2.7%) | 0 (0.0%) | 0.170 |
| Erosive duodenitis | 10 (9.4%) | 2 (5.4%) | 8 (11.6%) | 0.299 |
| General duodenitis | 15 (14.2%) | 3 (8.1%) | 12 (17.4%) | 0.191 |
| Total n = 69 | Diagnostic | p * | |||
|---|---|---|---|---|---|
| Normal 18 (17.0%) | Non-Severe 72 (67.9%) | Severe 16 (15.1%) | |||
| Genes | |||||
| cagA | 55 (79.7%) | 11 (84.6%) | 37 (75.5%) | 7 (100.0%) | 0.285 |
| vacA | 37 (53.6%) | 7 (53.8%) | 27 (55.1%) | 3 (42.9%) | 0.831 |
| oipA | 11 (15.9%) | 0 (0.0%) | 9 (18.4%) | 2 (28.6%) | 0.173 |
| babA2 | 18 (26.1%) | 2 (15.4%) | 14 (28.6%) | 2 (28.6%) | 0.621 |
| Gene Combinations | |||||
| cagA/vacA/oipA/babA2 | 5 (7.2%) | 0 (0.0%) | 5 (10.2%) | 0 (0.0%) | 0.333 |
| cagA/oipA | 9 (13.0%) | 0 (0.0%) | 7 (14.3%) | 2 (28.6%) | 0.173 |
| cagA/babA2 | 15 (21.7%) | 1 (7.7%) | 12 (24.5%) | 2 (28.6%) | 0.383 |
| cagA/oipA/babA2 | 7 (10.1%) | 0 (0.0%) | 5 (10.2%) | 2 (28.6%) | 0.130 |
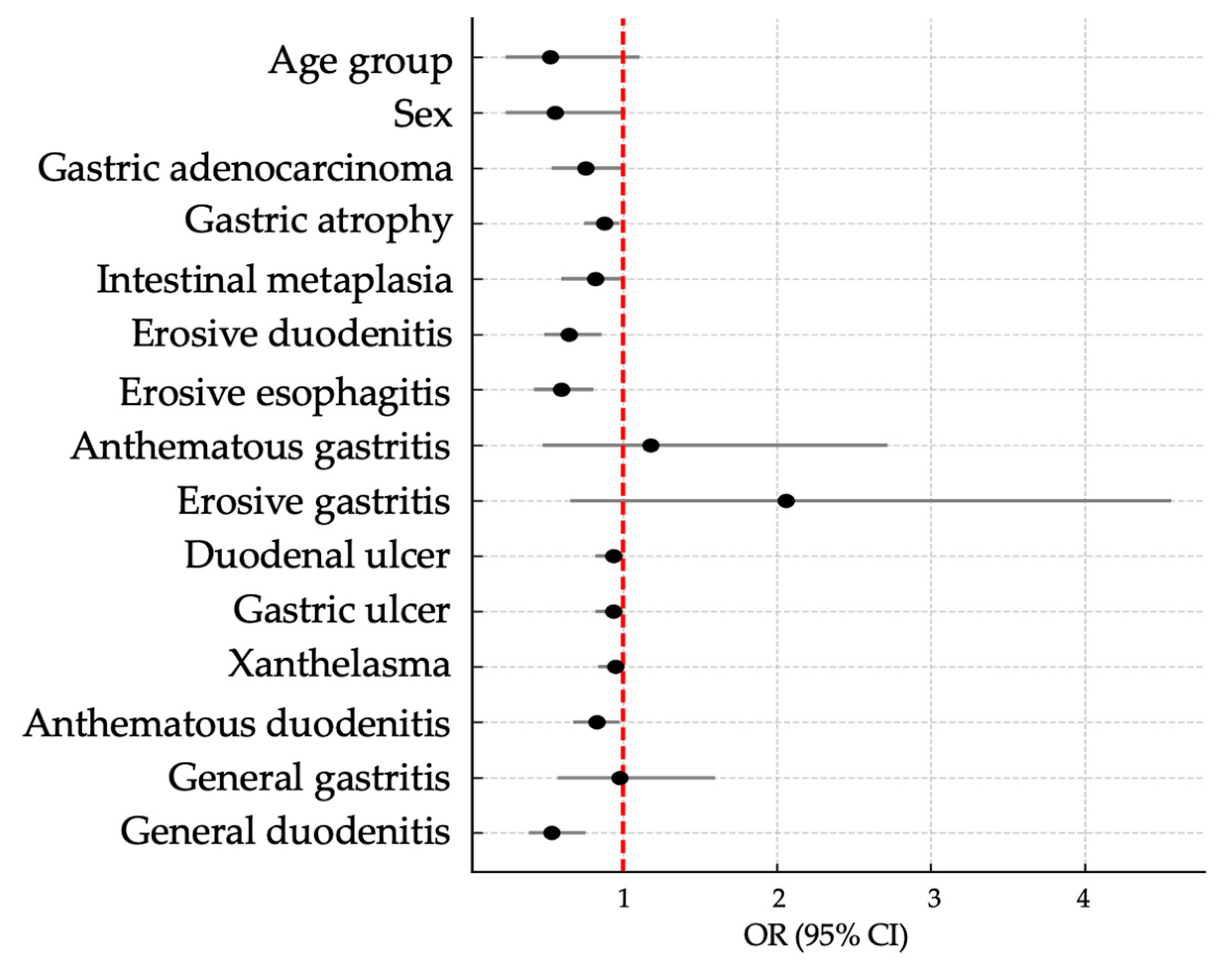
| cagA/vacA/oipA/babA2 | Wald | OR (95% CI) | p * | ||
|---|---|---|---|---|---|
| Absent | Present | ||||
| Age Group | |||||
| 18 to 50 years | 31 (48.4%) | 3 (60.0%) | 2.42 | Ref | |
| 51 years or more | 33 (51.6%) | 2 (40.0%) | 0.53 (0.24–1.11) | 0.120 | |
| Sex | |||||
| Female | 49 (76.6%) | 4 (80.0%) | 2.70 | Ref | |
| Male | 15 (23.4%) | 1 (20.0%) | 0.56 (0.24–1.02) | 0.100 | |
| Severe Diseases | |||||
| Gastric adenocarcinoma | 2 (3.1%) | 0 (0.0%) | 2.67 | 0.76 (0.54–1.00) | 0.102 |
| Gastric atrophy | 4 (6.3%) | 0 (0.0%) | 3.63 | 0.88 (0.75–0.98) | 0.049 |
| Intestinal metaplasia | 2 (3.1%) | 0 (0.0%) | 1.83 | 0.82 (0.60–1.00) | 0.176 |
| Non-severe Diseases | |||||
| Esophagus | |||||
| Erosive esophagitis | 10 (15.6%) | 0 (0.0%) | 9.94 | 0.60 (0.42–0.81) | 0.002 |
| Stomach | |||||
| Anthematous gastritis | 26 (40.6%) | 3 (60.0%) | 0.14 | 1.18 (0.48–2.72) | 0.704 |
| Erosive gastritis | 18 (28.1%) | 3 (60.0%) | 2.26 | 2.06 (0.66–4.56) | 0.133 |
| Gastric ulcer | 1 (1.6%) | 0 (0.0%) | 1.22 | 0.94 (0.82–1.00) | 0.269 |
| General gastritis | 44 (68.8%) | 5 (100.0%) | 0.01 | 0.98 (0.58–1.60) | 0.930 |
| Xanthelasma (gastric) | 1 (1.6%) | 0 (0.0%) | 1.07 | 0.95 (0.84–1.00) | 0.302 |
| Duodenum | |||||
| Anthematous duodenitis | 4 (6.3%) | 0 (0.0%) | 3.81 | 0.83 (0.68–0.98) | 0.049 |
| Duodenal ulcer | 1 (1.6%) | 0 (0.0%) | 1.22 | 0.94 (0.82–1.00) | 0.269 |
| Erosive duodenitis | 8 (12.5%) | 0 (0.0%) | 9.11 | 0.65 (0.49–0.86) | 0.003 |
| General duodenitis | 12 (18.8%) | 0 (0.0%) | 12.50 | 0.54 (0.39–0.76) | <0.001 |
| Gene | Sequence (5′ → 3′) | Amplification Conditions | bp |
|---|---|---|---|
| 16S rRNA | CTGGAGARACTAAGYCCTCC GAGGAATACTCATTGCAAGGCGA | 95 °C, 2 min; 95 °C, 1 min; 60 °C, 1 min; 72 °C, 1 min (40 cycles); 72 °C, 5 min [36] | 150 |
| cagA | ATGACTAACGAAACTATTGATC CAGGATTTTTGATCGCTTTATT | 94 °C, 1 min; 53 °C, 1 min; 72 °C, 1 min (40 cycles) [37] | 232 |
| vacA | ATGGAAATACAACAAACACAC CCTGARACCGTTCCTACAGC | 94 °C, 45 s; 54 °C, 45 s; 72 °C, 45 s (35 cycles) [39] | 286 |
| oipA | GTTTTTGATGCATGGGATTT GTGCATCTCTTATGGCTTT | 94 °C, 1 min; 56 °C, 1 min; 72 °C, 1 min (35 cycles) [38] | 401 |
| babA2 | CCAAACGAAACAAAAAGCGT GCTTGTGTAAAAGCCGTCGT | 94 °C, 2 min; 94 °C, 45 s; 48 °C, 30 s; 72 °C, 60 s (35 cycles); 72 °C, 10 min [37] | 271 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maciel, D.N.; Santos-Dutra, H.C.d.O.; Rocha, V.L.; Rasmussen, L.T.; Barbosa, M.S. Impact of Helicobacter pylori Virulence Genotypes cagA, vacA, oipA, and babA2 on Severity of Gastropathies in Brazilian Patients. Int. J. Mol. Sci. 2025, 26, 9471. https://doi.org/10.3390/ijms26199471
Maciel DN, Santos-Dutra HCdO, Rocha VL, Rasmussen LT, Barbosa MS. Impact of Helicobacter pylori Virulence Genotypes cagA, vacA, oipA, and babA2 on Severity of Gastropathies in Brazilian Patients. International Journal of Molecular Sciences. 2025; 26(19):9471. https://doi.org/10.3390/ijms26199471
Chicago/Turabian StyleMaciel, Diogo Nery, Hellen Christina de Oliveira Santos-Dutra, Viviane Lopes Rocha, Lucas Trevizani Rasmussen, and Mônica Santiago Barbosa. 2025. "Impact of Helicobacter pylori Virulence Genotypes cagA, vacA, oipA, and babA2 on Severity of Gastropathies in Brazilian Patients" International Journal of Molecular Sciences 26, no. 19: 9471. https://doi.org/10.3390/ijms26199471
APA StyleMaciel, D. N., Santos-Dutra, H. C. d. O., Rocha, V. L., Rasmussen, L. T., & Barbosa, M. S. (2025). Impact of Helicobacter pylori Virulence Genotypes cagA, vacA, oipA, and babA2 on Severity of Gastropathies in Brazilian Patients. International Journal of Molecular Sciences, 26(19), 9471. https://doi.org/10.3390/ijms26199471






