Decreased Expression of a Phosphoribosylanthranilate Transferase-Encoding Gene, OsPAT1, Causes Lesion Mimics in Rice
Abstract
1. Introduction
2. Results
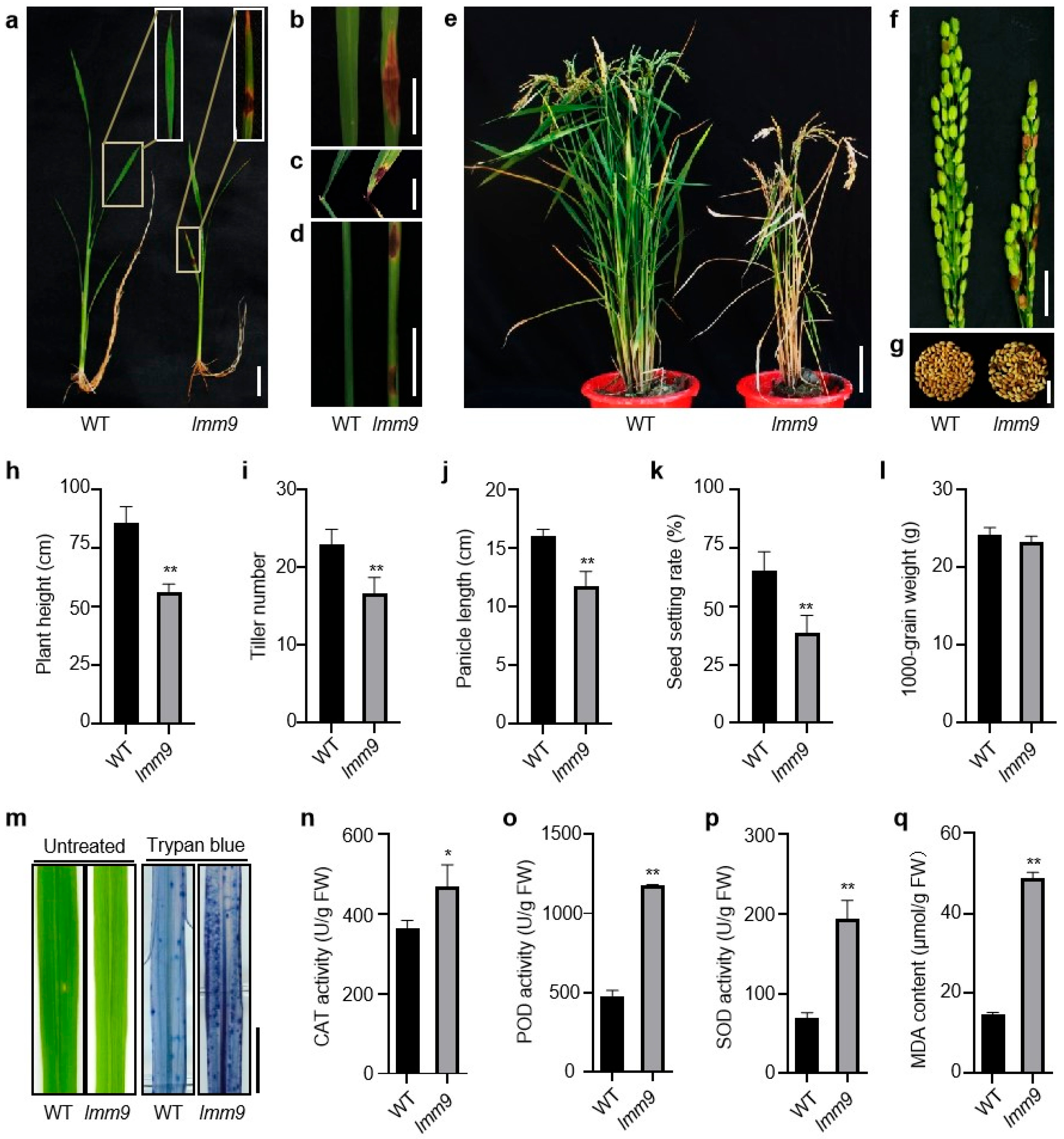
2.1. Discovery and Phenotypic Characterization of the lmm9 Mutant
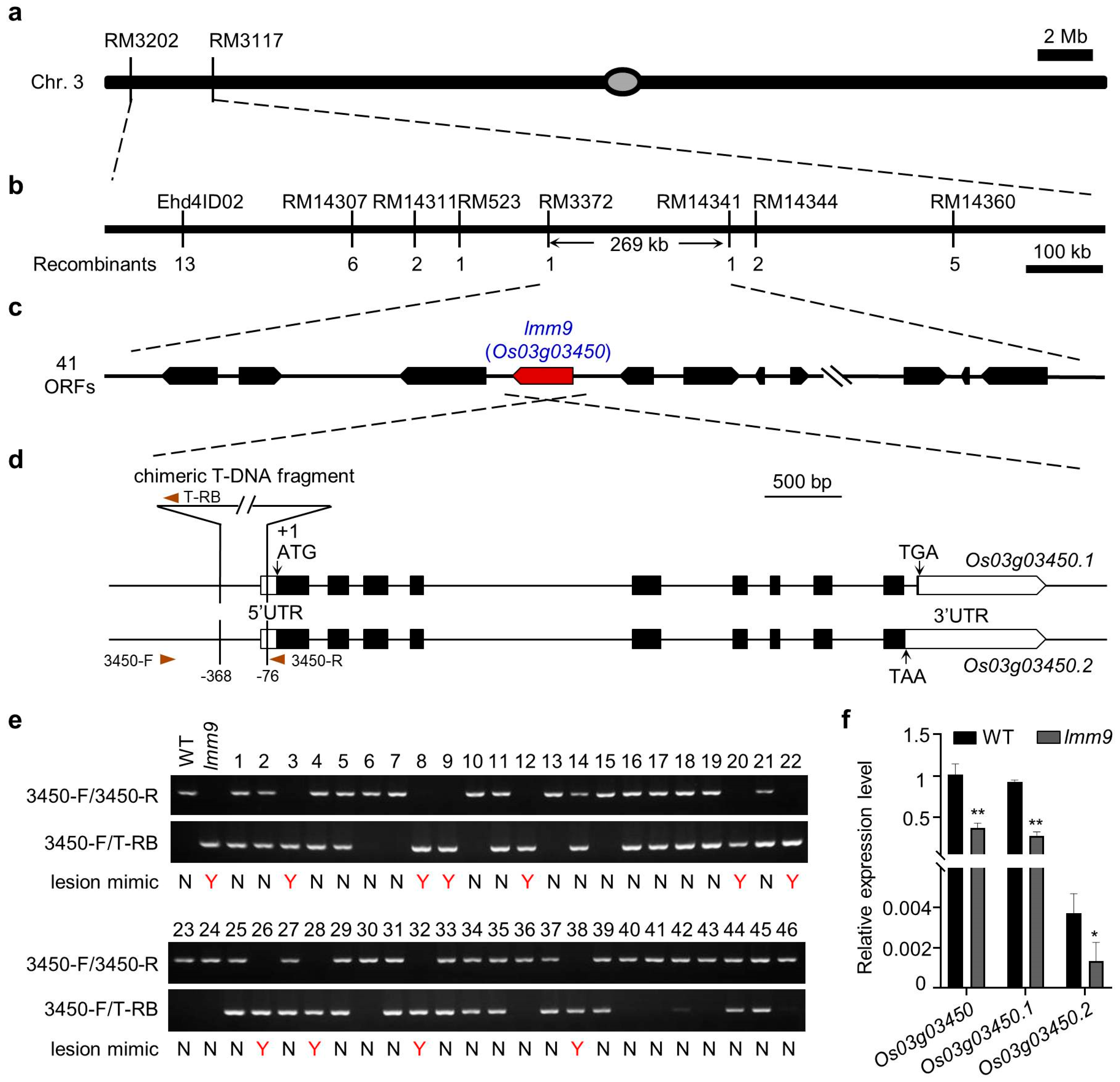
2.2. Localization and Identification of lmm9 Gene
2.3. Downregulation of Os03g03450 Led to the lmm9 Mutant Phenotype
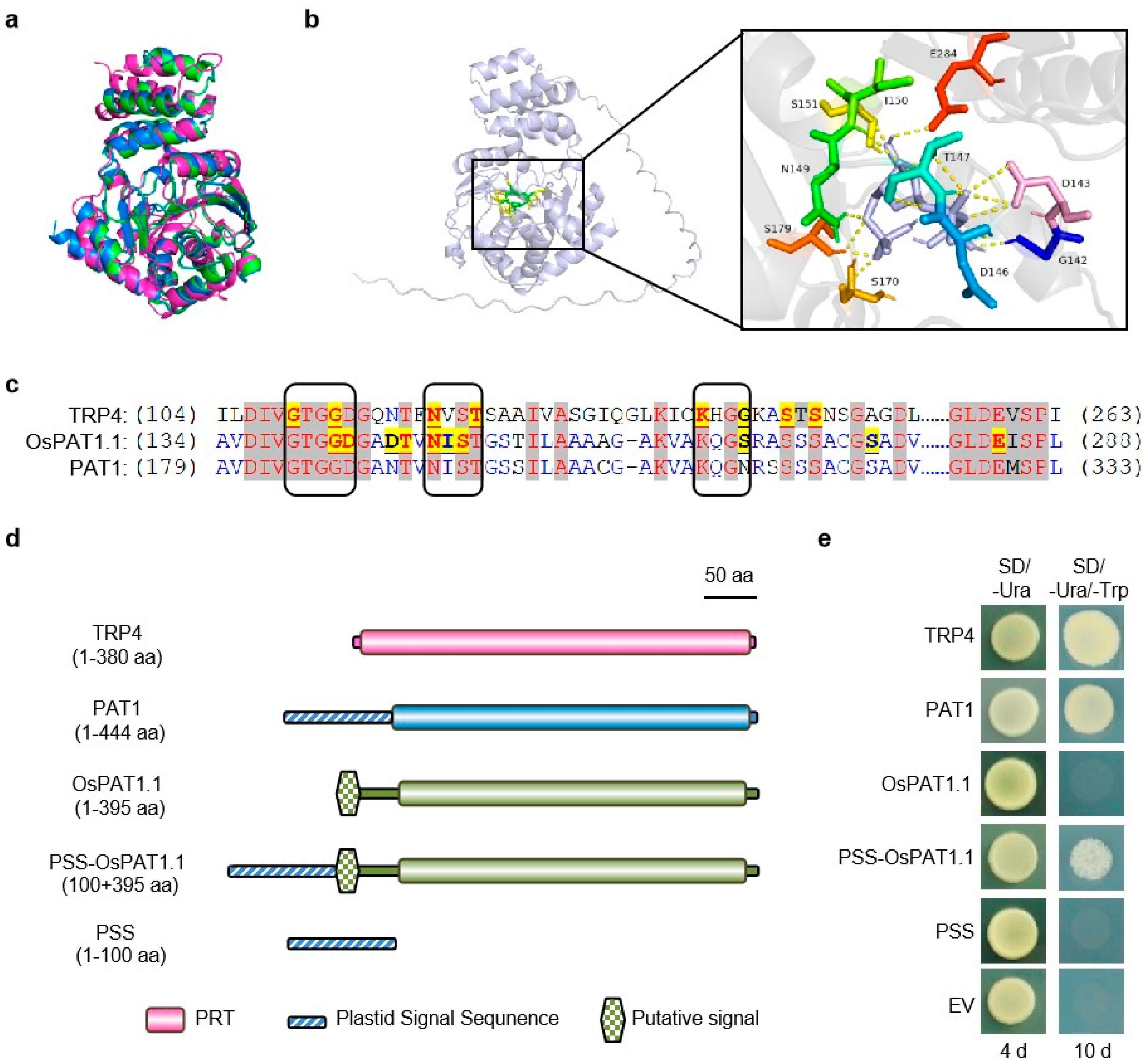
2.4. Os03g03450 Encoded a Conserved and Functional Enzyme–OsPAT1
2.5. Expression Pattern and Protein Subcellular Localization of OsPAT1
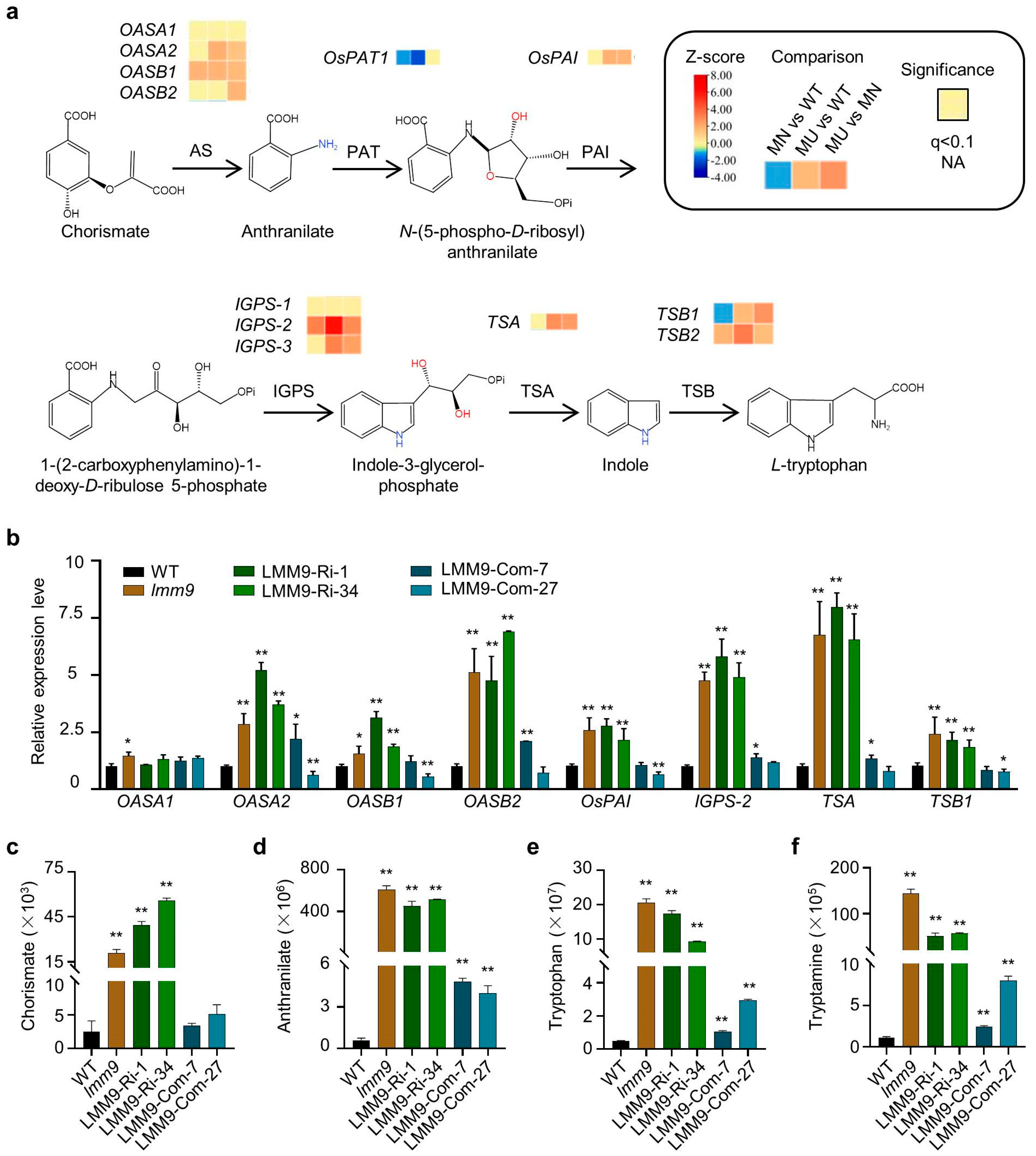
2.6. The Expression of Many Tryptophan Synthesis Pathway Genes Was Induced in the lmm9 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Trypan Blue Staining
4.3. Physiological Measurements
4.4. Mapping of the lmm9 Gene
4.5. Vector Construction and Transformation
4.6. Protein Sequence and Phylogenetic Analysis
4.7. Subcellular Location of OsPAT1
4.8. Protein Structure Prediction
4.9. Molecular Docking
4.10. RNA-Seq Analysis
4.11. qRT-PCR Analysis
4.12. Metabolites Analysis
4.13. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LMMs | Lesion mimic mutants |
| HR | Hypersensitive response |
| PCD | Programmed cell death |
| ROS | reactive oxygen species |
| UROD | Uroporphyrinogen decarboxylase |
| PEPP | Phosphoribosyl group from phosphoribosyl-α1-pyrophosphate |
| Ppi | Pyrophosphate |
| PRA | N-(5′-phosphoribosyl)anthranilate |
| AnPRT | Anthranilate phosphoribosyltransferase |
| PRT | Phosphoribosylanthranilate phosphoribosyltransferase |
| PAT1 | Phosphoribosylanthranilate transferase 1 |
| WT | Wide type |
| CAT | Catalase |
| POD | Peroxidase |
| SOD | Superoxide dismutase |
References
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Lorrain, S.; Vailleau, F.; Balague, C.; Roby, D. Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003, 8, 263–271. [Google Scholar] [CrossRef]
- Kang, S.G.; Lee, K.E.; Singh, M.; Kumar, P.; Matin, M.N. Rice lesion mimic mutants (LMM): The current understanding of genetic mutations in the failure of ROS scavenging during lesion formation. Plants 2021, 10, 1598. [Google Scholar] [CrossRef]
- Li, C.; Liu, H.; Wang, J.; Pan, Q.; Wang, Y.; Wu, K.; Jia, P.; Mu, Y.; Tang, H.; Xu, Q.; et al. Characterization and fine mapping of a lesion mimic mutant (Lm5) with enhanced stripe rust and powdery mildew resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 421–438. [Google Scholar] [CrossRef]
- Yu, M.; Fan, Y.; Li, X.; Chen, X.; Yu, S.; Wei, S.; Li, S.; Chang, W.; Qu, C.; Li, J.; et al. LESION MIMIC MUTANT 1 confers basal resistance to Sclerotinia sclerotiorum in rapeseed via a salicylic acid-dependent pathway. J. Exp. Bot. 2023, 74, 5620–5634. [Google Scholar] [CrossRef]
- Rao, Y.; Jiao, R.; Wang, S.; Wu, X.; Ye, H.; Pan, C.; Li, S.; Xin, D.; Zhao, W.; Dai, G.; et al. SPL36 Encodes a Receptor-like Protein Kinase that Regulates Programmed Cell Death and Defense Responses in Rice. Rice 2021, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Sha, G.; Sun, P.; Kong, X.; Han, X.; Sun, Q.; Fouillen, L.; Zhao, J.; Li, Y.; Yang, L.; Wang, Y.; et al. Genome editing of a rice CDP-DAG synthase confers multipathogen resistance. Nature 2023, 618, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Guo, Y.; Sun, H.; Dai, J.; Peng, X.; Wu, X.; Yun, H.; Zhang, L.; Qian, Y.; Li, X.; et al. Lesion mimic mutant 8 balances disease resistance and growth in rice. Front. Plant Sci. 2023, 14, 1189926. [Google Scholar] [CrossRef]
- Zhang, A.; Jiang, H.; Chu, H.; Cao, L.; Chen, J. Rice lesion mimic gene cloning and association analysis for disease resistance. Curr. Issues Mol. Biol. 2022, 44, 2350–2361. [Google Scholar] [CrossRef]
- Hu, G.; Yalpani, N.; Briggs, S.P.; Johal, G.S. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 1998, 10, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Yang, S.; Wang, D.; Tang, K.; Feng, X.X.; Feng, X.Z. Genetic mapping of a light-dependent lesion mimic mutant reveals the function of coproporphyrinogen III oxidase homolog in soybean. Front. Plant Sci. 2020, 11, 557, Corrigendum in Front. Plant Sci. 2020, 11, 893. [Google Scholar]
- Zhao, Y.; Xu, W.; Wang, L.; Han, S.; Zhang, Y.; Liu, Q.; Liu, B.; Zhao, X. A maize necrotic leaf mutant caused by defect of coproporphyrinogen III oxidase in the porphyrin pathway. Genes 2022, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.R.; Qu, S.; Bordeos, A.; Yang, C.; Baraoidan, M.; Yan, H.; Xie, Q.; Nahm, B.H.; Leung, H.; Wang, G.L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004, 16, 2795–2808. [Google Scholar] [CrossRef]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.L.; et al. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef]
- You, Q.; Zhai, K.; Yang, D.; Yang, W.; Wu, J.; Liu, J.; Pan, W.; Wang, J.; Zhu, X.; Jian, Y.; et al. An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 2016, 20, 758–769. [Google Scholar] [CrossRef]
- Tang, J.; Zhu, X.; Wang, Y.; Liu, L.; Xu, B.; Li, F.; Fang, J.; Chu, C. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J. 2011, 66, 996–1007. [Google Scholar] [CrossRef]
- Vega-Sanchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Zhao, J.; Li, X.; Xiao, J.; Wang, S. A pair of orthologs of a leucine-rich repeat receptor kinase-like disease resistance gene family regulates rice response to raised temperature. BMC Plant Biol. 2011, 11, 160. [Google Scholar] [CrossRef]
- Li, T.; Chen, X.; Zhong, X.; Zhao, Y.; Liu, X.; Zhou, S.; Cheng, S.; Zhou, D.X. Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 2013, 25, 4725–4736. [Google Scholar] [CrossRef]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef]
- Ueno, M.; Shibata, H.; Kihara, J.; Honda, Y.; Arase, S. Increased tryptophan decarboxylase and monoamine oxidase activities induce Sekiguchi lesion formation in rice infected with Magnaporthe grisea. Plant J. 2003, 36, 215–228. [Google Scholar] [CrossRef]
- Fujiwara, T.; Maisonneuve, S.; Isshiki, M.; Mizutani, M.; Chen, L.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 2010, 285, 11308–11313. [Google Scholar] [CrossRef]
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2021, 105, 942–956. [Google Scholar] [CrossRef]
- Xia, S.; Ruan, B.; Rao, Y.; Cui, Y.; Zhang, Q.; Zeng, D.; Qian, Q.; Ren, D. The ell1 mutation disrupts tryptophan metabolism and induces cell death. Plant Signal. Behav. 2021, 16, 1905336. [Google Scholar] [CrossRef]
- Li, W.; Cheng, W.; Jiang, H.; Fang, C.; Peng, L.; Tao, L.; Zhan, Y.; Huang, X.; Ma, B.; Chen, X.; et al. Mutation of rice EARLY LEAF LESION AND SENESCENCE 1 (ELS1), which encodes an anthranilate synthase alpha-subunit, induces ROS accumulation and cell death through activating the tryptophan synthesis pathway in rice. Plant J. 2024, 120, 2723–2737. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, J.; Lins, T.; Martin, W.; Eilert, U. Anthranilate synthase from Ruta graveolens. Duplicated AS alpha genes encode tryptophan-sensitive and tryptophan-insensitive isoenzymes specific to amino acid and alkaloid biosynthesis. Plant Physiol. 1996, 111, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Tozawa, Y.; Hasegawa, H.; Terakawa, T.; Wakasa, K. Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1. Plant Physiol. 2001, 126, 1493–1506. [Google Scholar] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Crawford, I.P. Synthesis of tryptophan from chorismate: Comparative aspects. Methods Enzymol. 1987, 142, 293–300. [Google Scholar]
- Furter, R.; Paravicini, G.; Aebi, M.; Braus, G.; Prantl, F.; Niederberger, P.; Hutter, R. The TRP4 gene of Saccharomyces cerevisiae: Isolation and structural analysis. Nucleic Acids Res. 1986, 14, 6357–6373. [Google Scholar] [CrossRef]
- Rose, A.B.; Casselman, A.L.; Last, R.L. A Phosphoribosylanthranilate Transferase Gene Is Defective in Blue Fluorescent Arabidopsis thaliana Tryptophan Mutants. Plant Physiol. 1992, 100, 582–592. [Google Scholar] [CrossRef]
- Last, R.L.; Fink, G.R. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 1988, 240, 305–310. [Google Scholar] [CrossRef]
- Cohen, J.D.; Strader, L.C. An auxin research odyssey: 1989–2023. Plant Cell 2024, 36, 1410–1428. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Sarkar, A.K. Serotonin: A frontline player in plant growth and stress responses. Physiol. Plant 2023, 175, e13968. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Wang, L.; Li, B.; Jin, C.; Lin, X. Melatonin: A master regulator of plant development and stress responses. J. Integr. Plant Biol. 2021, 63, 126–145. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, M.; Kuang, Z.; Yue, J.; Xue, L.; Zhu, M.; Zhu, Z.; Khan, M.H.; Niu, L. Crystal structures of anthranilate phosphoribosyltransferase from Saccharomyces cerevisiae. Acta Crystallogr. F Struct. Biol. Commun. 2021, 77, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Last, R.L. The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiol. 1996, 110, 51–59. [Google Scholar] [CrossRef]
- Janero, D.R. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radic. Biol. Med. 1990, 9, 515–540. [Google Scholar] [CrossRef]
- Regni, L.; Del Pino, A.M.; Mousavi, S.; Palmerini, C.A.; Baldoni, L.; Mariotti, R.; Mairech, H.; Gardi, T.; D’Amato, R.; Proietti, P. Behavior of four olive cultivars during salt stress. Front. Plant Sci. 2019, 10, 867. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, C.; Mu, J.Q.; Zhang, H.; Yao, W.; Ding, X.; Ding, J.; Chang, Y. All-in-one sequencing: An improved library preparation method for cost-effective and high-throughput next-generation sequencing. Plant Methods 2020, 16, 74. [Google Scholar] [CrossRef]
- Liu, H.; Ding, Y.; Zhou, Y.; Jin, W.; Xie, K.; Chen, L.L. CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 2017, 10, 530–532. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Tang, W.; Liang, K.; Zhao, C.; Yu, F.; Qiu, F. A candidate association study of transcription factors in maize revealed the ZmPLATZ15-ZmEREB200 module as a key regulator of waterlogging tolerance at the seedling stage. Plant Physiol. Biochem. 2025, 222, 109664. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Zhang, Q.; Xu, Y.; Zhang, B.; Li, H.; Li, Y.; Zhang, H.; Yuan, W. Decreased Expression of a Phosphoribosylanthranilate Transferase-Encoding Gene, OsPAT1, Causes Lesion Mimics in Rice. Int. J. Mol. Sci. 2025, 26, 9428. https://doi.org/10.3390/ijms26199428
Ren J, Zhang Q, Xu Y, Zhang B, Li H, Li Y, Zhang H, Yuan W. Decreased Expression of a Phosphoribosylanthranilate Transferase-Encoding Gene, OsPAT1, Causes Lesion Mimics in Rice. International Journal of Molecular Sciences. 2025; 26(19):9428. https://doi.org/10.3390/ijms26199428
Chicago/Turabian StyleRen, Jun, Qingwen Zhang, Yafei Xu, Biaoming Zhang, Haitao Li, Yan Li, Haitao Zhang, and Wenya Yuan. 2025. "Decreased Expression of a Phosphoribosylanthranilate Transferase-Encoding Gene, OsPAT1, Causes Lesion Mimics in Rice" International Journal of Molecular Sciences 26, no. 19: 9428. https://doi.org/10.3390/ijms26199428
APA StyleRen, J., Zhang, Q., Xu, Y., Zhang, B., Li, H., Li, Y., Zhang, H., & Yuan, W. (2025). Decreased Expression of a Phosphoribosylanthranilate Transferase-Encoding Gene, OsPAT1, Causes Lesion Mimics in Rice. International Journal of Molecular Sciences, 26(19), 9428. https://doi.org/10.3390/ijms26199428






