Genome Editing in the Chicken: From PGC-Mediated Germline Transmission to Advanced Applications
Abstract
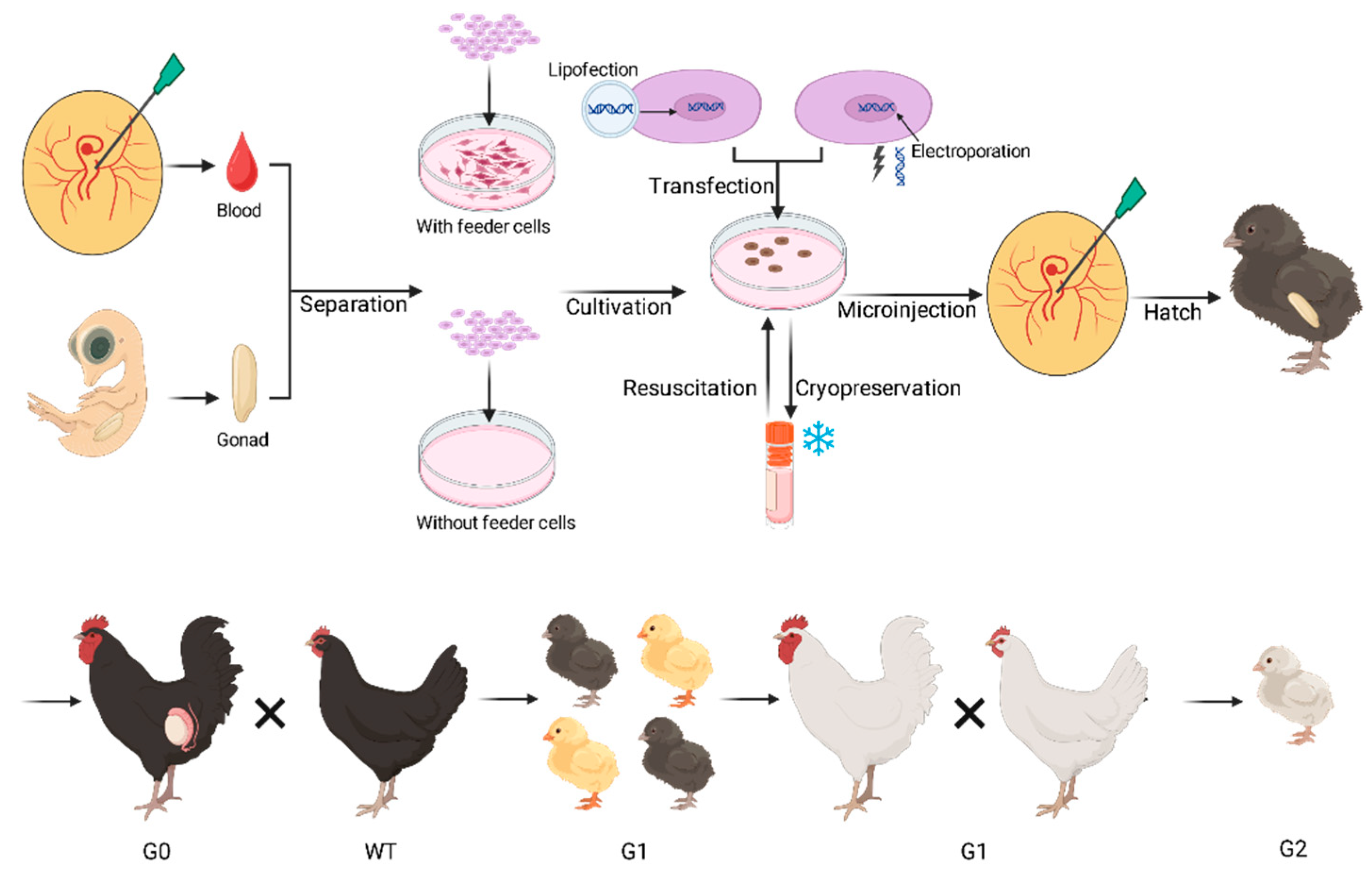
1. Introduction
2. Strategies for Germline Transmission
2.1. Non-PGC-Mediated Strategies
2.1.1. Direct Embryo and Blastoderm Manipulation
2.1.2. EG and ES Cell-Mediated Transmission
2.1.3. Sperm- and Ovary-Mediated Transgenesis
2.2. PGC-Mediated Germline Transmission
2.2.1. Biology and Characterization of PGCs
2.2.2. Sourcing and Isolation of PGCs
2.2.3. In Vitro Cultivation of PGCs
2.2.4. Transfection of PGCs
2.2.5. Cryopreservation of PGCs
3. Genome Editing Methodologies
3.1. Untargeted Genome Editing
3.1.1. Retroviral Vectors
3.1.2. Lentiviral Vectors
3.1.3. Transposon Systems
3.2. Targeted Genome Editing
3.2.1. Nuclease-Based Platforms: ZFNs, TALENs, and CRISPR
3.2.2. Advanced CRISPR-Based Technologies
4. Applications of Genome Editing in Poultry
4.1. Engineering Disease Resistance
4.2. Enhancing Agricultural Production Traits
4.3. Development of Avian Models
4.4. Avian Bioreactors for Pharmaceutical Production
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ichikawa, K.; Matsuzaki, M.; Ezaki, R.; Horiuchi, H. Genome Editing in Chickens. Gene Genome Ed. 2022, 3–4, 100015. [Google Scholar] [CrossRef]
- Han, J.Y.; Park, T.S.; Hong, Y.H.; Jeong, D.K.; Kim, J.N.; Kim, K.D.; Lim, J.M. Production of Germline Chimeras by Transfer of Chicken Gonadal Primordial Germ Cells Maintained in Vitro for an Extended Period. Theriogenology 2002, 58, 1531–1539. [Google Scholar] [CrossRef]
- Park, T.S.; Hong, Y.H.; Kwon, S.C.; Lim, J.M.; Han, J.Y. Birth of Germline Chimeras by Transfer of Chicken Embryonic Germ (EG) Cells into Recipient Embryos. Mol. Reprod. Dev. 2003, 65, 389–395. [Google Scholar] [CrossRef]
- Park, T.S.; Jeong, D.K.; Kim, J.N.; Song, G.H.; Hong, Y.H.; Lim, J.M.; Han, J.Y. Improved Germline Transmission in Chicken Chimeras Produced by Transplantation of Gonadal Primordial Germ Cells into Recipient Embryos1. Biol. Reprod. 2003, 68, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Calos, M. Genome Editing Techniques and Their Therapeutic Applications. Clin. Pharmacol. Ther. 2017, 101, 42–51. [Google Scholar] [CrossRef] [PubMed]
- DeMayo, J.L.; Wang, J.; Liang, D.; Zhang, R.; DeMayo, F.J. Genetically Engineered Mice by Pronuclear DNA Microinjection. Curr. Protoc. Mouse Biol. 2012, 2, 245–262. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Lee, K. Current Approaches and Applications in Avian Genome Editing. Int. J. Mol. Sci. 2020, 21, 3937. [Google Scholar] [CrossRef]
- Sang, H. Transgenic Chickens—Methods and Potential Applications. Trends Biotechnol. 1994, 12, 415–420. [Google Scholar] [CrossRef]
- Marzullo, G. Production of Chick Chimaeras. Nature 1970, 225, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, V.; Hamilton, H.L. A Series of Normal Stages in the Development of the Chick Embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
- Love, J.; Gribbin, C.; Mather, C.; Sang, H. Transgenic Birds by DNA Microinjection. Bio/Technology 1994, 12, 60–63. [Google Scholar] [CrossRef]
- Bosselman, R.A.; Hsu, R.-Y.; Boggs, T.; Hu, S.; Bruszewski, J.; Ou, S.; Kozar, L.; Martin, F.; Green, C.; Jacobsen, F.; et al. Germline Transmission of Exogenous Genes in the Chicken. Science 1989, 243, 533–535. [Google Scholar] [CrossRef]
- Naito, M.; Sasaki, E.; Ohtaki, M.; Sakurai, M. Introduction of Exogenous DNA into Somatic and Germ Cells of Chickens by Microinjection into the Germinal Disc of Fertilized Ova. Mol. Reprod. Dev. 1994, 37, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Kino, K.; Noda, K.; Miyakawa, H.; Ohtsuka, K.; Ono, T.; Mochii, M.; Eguchi, G.; Agata, K. A Low Frequency of Chromosomal Integration of Cloned DNA after Microinjection into the Cytoplasm of Fertilized Eggs of Chickens. Jpn. Poult. Sci. 1998, 35, 356–366. [Google Scholar] [CrossRef]
- Longmuir, K.J.; Haynes, S.M.; Dickinson, M.E.; Murphy, J.C.; Willson, R.C.; Waring, A.J. Optimization of a Peptide/Non-Cationic Lipid Gene Delivery System for Effective Microinjection into Chicken Embryo in Vivo. Mol. Ther. 2001, 4, 66–74. [Google Scholar] [CrossRef]
- Park, T.S.; Han, J.Y. Derivation and Characterization of Pluripotent Embryonic Germ Cells in Chicken. Mol. Reprod. Dev. 2000, 56, 475–482. [Google Scholar] [CrossRef]
- Pain, B.; Clark, M.E.; Shen, M.; Nakazawa, H.; Sakurai, M.; Samarut, J.; Etches, R.J. Long-Term in Vitro Culture and Characterisation of Avian Embryonic Stem Cells with Multiple Morphogenetic Potentialities. Development 1996, 122, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Van De Lavoir, M.-C.; Mather-Love, C.; Leighton, P.; Diamond, J.H.; Heyer, B.S.; Roberts, R.; Zhu, L.; Winters-Digiacinto, P.; Kerchner, A.; Gessaro, T.; et al. High-Grade Transgenic Somatic Chimeras from Chicken Embryonic Stem Cells. Mech. Dev. 2006, 123, 31–41. [Google Scholar] [CrossRef]
- Sato, M. Transgenesis Via Sperm. J. Mamm. Ova Res. 2005, 22, 92–100. [Google Scholar] [CrossRef]
- Lee, Y.M.; Jung, J.G.; Kim, J.N.; Park, T.S.; Kim, T.M.; Shin, S.S.; Kang, D.K.; Lim, J.M.; Han, J.Y. A Testis-Mediated Germline Chimera Production Based on Transfer of Chicken Testicular Cells Directly into Heterologous Testes1. Biol. Reprod. 2006, 75, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Trefil, P.; Micáková, A.; Mucksová, J.; Hejnar, J.; Poplstein, M.; Bakst, M.R.; Kalina, J.; Brillard, J.-P. Restoration of Spermatogenesis and Male Fertility by Transplantation of Dispersed Testicular Cells in the Chicken1. Biol. Reprod. 2006, 75, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Trefil, P.; Aumann, D.; Koslová, A.; Mucksová, J.; Benešová, B.; Kalina, J.; Wurmser, C.; Fries, R.; Elleder, D.; Schusser, B.; et al. Male Fertility Restored by Transplanting Primordial Germ Cells into Testes: A New Way towards Efficient Transgenesis in Chicken. Sci. Rep. 2017, 7, 14246. [Google Scholar] [CrossRef]
- Li, B.; Sun, G.; Sun, H.; Xu, Q.; Gao, B.; Zhou, G.; Zhao, W.; Wu, X.; Bao, W.; Yu, F.; et al. Efficient Generation of Transgenic Chickens Using the Spermatogonial Stem Cells in Vivo and Ex Vivo Transfection. Sci. China Ser. C Life Sci. 2008, 51, 734–742. [Google Scholar] [CrossRef]
- Min, S.; Qing, S.Q.; Hui, Y.Y.; Zhi, F.D.; Rong, Q.Y.; Feng, X.; Chun, L.B. Generation of Antiviral Transgenic Chicken Using Spermatogonial Stem Cell Transfected in Vivo. Afr. J. Biotechnol. 2011, 10, 15678–15683. [Google Scholar] [CrossRef]
- Harel-Markowitz, E.; Gurevich, M.; Shore, L.S.; Katz, A.; Stram, Y.; Shemesh, M. Use of Sperm Plasmid DNA Lipofection Combined with REMI (Restriction Enzyme-Mediated Insertion) for Production of Transgenic Chickens Expressing eGFP (Enhanced Green Fluorescent Protein) or Human Follicle-Stimulating Hormone1. Biol. Reprod. 2009, 80, 1046–1052. [Google Scholar] [CrossRef]
- Collares, T.; Campos, V.F.; De Leon, P.M.M.; Cavalcanti, P.V.; Amaral, M.G.; Dellagostin, O.A.; Deschamps, J.C.; Seixas, F.K. Transgene Transmission in Chickens by Sperm-Mediated Gene Transfer after Seminal Plasma Removal and Exogenous DNA Treated with Dimethylsulfoxide or N,N-Dimethylacetamide. J. Biosci. 2011, 36, 613–620. [Google Scholar] [CrossRef]
- Cooper, C.A.; Challagulla, A.; Jenkins, K.A.; Wise, T.G.; O’Neil, T.E.; Morris, K.R.; Tizard, M.L.; Doran, T.J. Generation of Gene Edited Birds in One Generation Using Sperm Transfection Assisted Gene Editing (STAGE). Transgenic Res. 2017, 26, 331–347. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, C.; Du, X.; Gao, F.; Wu, S. Generation of Transgenic Chicken through Ovarian Injection. Anim. Models Exp. Med. 2024, 8, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yan, L.; Guo, H.; Li, L.; Hu, B.; Zhao, Y.; Yong, J.; Hu, Y.; Wang, X.; Wei, Y.; et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 2015, 161, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Magnúsdóttir, E.; Surani, M.A. How to Make a Primordial Germ Cell. Development 2014, 141, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Sasanami, T. Avian Reproduction; Springer Nature: Singapore, 2017; ISBN 978-981-10-3975-1. [Google Scholar]
- Nikolic, A.; Volarevic, V.; Armstrong, L.; Lako, M.; Stojkovic, M. Primordial Germ Cells: Current Knowledge and Perspectives. Stem Cells Int. 2016, 2016, 1741072. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.E.; Lehmann, R. Mechanisms Guiding Primordial Germ Cell Migration: Strategies from Different Organisms. Nat. Rev. Mol. Cell Biol. 2010, 11, 37–49. [Google Scholar] [CrossRef]
- Kang, K.S.; Lee, H.C.; Kim, H.J.; Lee, H.G.; Kim, Y.M.; Lee, H.J.; Park, Y.H.; Yang, S.Y.; Rengaraj, D.; Park, T.S.; et al. Spatial and Temporal Action of Chicken Primordial Germ Cells during Initial Migration. Reproduction 2015, 149, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Mathan; Zaib, G.; Jin, K.; Zuo, Q.; Habib, M.; Zhang, Y.; Li, B. Formation, Application, and Significance of Chicken Primordial Germ Cells: A Review. Animals 2023, 13, 1096. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.G.; Kim, D.K.; Park, T.S.; Lee, S.D.; Lim, J.M.; Han, J.Y. Development of Novel Markers for the Characterization of Chicken Primordial Germ Cells. Stem Cells 2005, 23, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, J.; Glover, J.D.; Taylor, L.; Sang, H.M.; McGrew, M.J. Characterisation and Germline Transmission of Cultured Avian Primordial Germ Cells. PLoS ONE 2010, 5, e15518. [Google Scholar] [CrossRef]
- Motono, M.; Ohashi, T.; Nishijima, K.; Iijima, S. Analysis of Chicken Primordial Germ Cells. Cytotechnology 2008, 57, 199–205. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, J.-W.; Kim, S.W.; Park, J.; Park, T.S. C-X-C Chemokine Receptor Type 4 (CXCR4) Is a Key Receptor for Chicken Primordial Germ Cell Migration. J. Reprod. Dev. 2017, 63, 555–562. [Google Scholar] [CrossRef]
- Petitte, J.N. Avian Germplasm Preservation: Embryonic Stem Cells or Primordial Germ Cells? Poult. Sci. 2006, 85, 237–242. [Google Scholar] [CrossRef]
- Yu, F.; Zhu, Z.; Chen, X.; Huang, J.; Jia, R.; Pan, J. Isolation, Characterization and Germline Chimera Preparation of Primordial Germ Cells from the Chinese Meiling Chicken. Poult. Sci. 2019, 98, 566–572. [Google Scholar] [CrossRef]
- Kang, S.J.; Choi, J.W.; Kim, S.Y.; Park, K.J.; Kim, T.M.; Lee, Y.M.; Kim, H.; Lim, J.M.; Han, J.Y. Reproduction of Wild Birds via Interspecies Germ Cell Transplantation. Biol. Reprod. 2008, 79, 931–937. [Google Scholar] [CrossRef]
- Yasuda, Y.; Tajima, A.; Fujimoto, T.; Kuwana, T. A Method to Obtain Avian Germ-Line Chimaeras Using Isolated Primordial Germ Cells. Reproduction 1992, 96, 521–528. [Google Scholar] [CrossRef]
- Chojnacka-Puchta, L.; Sawicka, D.; Lakota, P.; Plucienniczak, G.; Bednarczyk, M.; Plucienniczak, A. Obtaining Chicken Primordial Germ Cells Used for Gene Transfer: In Vitro and in Vivo Results. J. Appl. Genet. 2015, 56, 493–504. [Google Scholar] [CrossRef]
- Jung, K.M.; Kim, Y.M.; Ono, T.; Han, J.Y. Size-Dependent Isolation of Primordial Germ Cells from Avian Species. Mol. Reprod. Dev. 2017, 84, 508–516. [Google Scholar] [CrossRef]
- Chaipipat, S.; Prukudom, S.; Sritabtim, K.; Kuwana, T.; Piyasanti, Y.; Sinsiri, R.; Piantham, C.; Sangkalerd, S.; Boonsanong, S.; Pitiwong, K.; et al. Primordial Germ Cells Isolated from Individual Embryos of Red Junglefowl and Indigenous Pheasants of Thailand. Theriogenology 2021, 165, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Mozdziak, P.E.; Angerman-Stewart, J.; Rushton, B.; Pardue, S.L.; Petitte, J.N. Isolation of Chicken Primordial Germ Cells Using Fluorescence-Activated Cell Sorting. Poult. Sci. 2005, 84, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Kim, M.A.; Park, T.S.; Kim, D.K.; Park, H.J.; Ono, T.; Lim, J.M.; Han, J.Y. Enriched Gonadal Migration of Donor-derived Gonadal Primordial Germ Cells by Immunomagnetic Cell Sorting in Birds. Mol. Reprod. Dev. 2004, 68, 81–87. [Google Scholar] [CrossRef]
- Karagenç, L.; Cinnamon, Y.; Ginsburg, M.; Petitte, J.N. Origin of Primordial Germ Cells in the Prestreak Chick Embryo. Dev. Genet. 1996, 19, 290–301. [Google Scholar] [CrossRef]
- Chang, I.; Jeong, D.K.; Hong, Y.H.; Park, T.S.; Moon, Y.K.; Ohno, T.; Han, J.Y. Production of Germline Chimeric Chickens by Transfer of Cultured Primordial Germ Cells. Cell Biol. Int. 1997, 21, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Van De Lavoir, M.-C.; Diamond, J.H.; Leighton, P.A.; Mather-Love, C.; Heyer, B.S.; Bradshaw, R.; Kerchner, A.; Hooi, L.T.; Gessaro, T.M.; Swanberg, S.E.; et al. Germline Transmission of Genetically Modified Primordial Germ Cells. Nature 2006, 441, 766–769. [Google Scholar] [CrossRef]
- Miyahara, D.; Mori, T.; Makino, R.; Nakamura, Y.; Oishi, I.; Ono, T.; Nirasawa, K.; Tagami, T.; Kagami, H. Culture Conditions for Maintain Propagation, Long-Term Survival and Germline Transmission of Chicken Primordial Germ Cell-Like Cells. J. Poult. Sci. 2014, 51, 87–95. [Google Scholar] [CrossRef]
- Ji, M.; Guan, W.; Gao, Y.; Li, L.; Bai, C.; Ma, Y.; Li, X. Cultivation and Biological Characterization of Chicken Primordial Germ Cells. Braz. Arch. Biol. Technol. 2016, 59, e16150374. [Google Scholar] [CrossRef]
- Xie, L.; Lu, Z.; Chen, D.; Yang, M.; Liao, Y.; Mao, W.; Mo, L.; Sun, J.; Yang, W.; Xu, H.; et al. Derivation of Chicken Primordial Germ Cells Using an Indirect Co-Culture System. Theriogenology 2019, 123, 83–89. [Google Scholar] [CrossRef]
- Wang, L.; Chen, M.J.; Chen, D.Y.; Peng, S.F.; Zhou, X.L.; Liao, Y.Y.; Yang, X.G.; Xu, H.Y.; Lu, S.S.; Zhang, M.; et al. Derivation and Characterization of Primordial Germ Cells from Guangxi Yellow-Feather Chickens. Poult. Sci. 2017, 96, 1419–1425. [Google Scholar] [CrossRef]
- Szczerba, A.; Kuwana, T.; Paradowska, M.; Bednarczyk, M. In Vitro Culture of Chicken Circulating and Gonadal Primordial Germ Cells on a Somatic Feeder Layer of Avian Origin. Animals 2020, 10, 1769. [Google Scholar] [CrossRef]
- Whyte, J.; Glover, J.D.; Woodcock, M.; Brzeszczynska, J.; Taylor, L.; Sherman, A.; Kaiser, P.; McGrew, M.J. FGF, Insulin, and SMAD Signaling Cooperate for Avian Primordial Germ Cell Self-Renewal. Stem Cell Rep. 2015, 5, 1171–1182. [Google Scholar] [CrossRef]
- Lázár, B.; Molnár, M.; Sztán, N.; Végi, B.; Drobnyák, Á.; Tóth, R.; Tokodyné Szabadi, N.; McGrew, M.J.; Gócza, E.; Patakiné Várkonyi, E. Successful Cryopreservation and Regeneration of a Partridge Colored Hungarian Native Chicken Breed Using Primordial Germ Cells. Poult. Sci. 2021, 100, 101207. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, K.; Niu, B.; Wang, Z.; Jie, Y.; Fan, Z.; Li, J.; Sun, C.; Hou, Z.-C.; Shao, L.-W. Hyperthermia Suppresses the Biological Characteristics and Migration of Chicken Primordial Germ Cells. Front. Genome Ed. 2025, 6, 1512108. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Grochowska, E.; Lázár, B.; Várkonyi, E.; Bednarczyk, M.; Stadnicka, K. The Effect of Short- and Long-Term Cryopreservation on Chicken Primordial Germ Cells. Genes 2024, 15, 624. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.-J.; Zheng, D.; Liu, G.; Ren, W.; Wu, G.; Peng, Y.; Wu, J.; Jin, K.; Zuo, Q.; Li, G.; et al. Comparative Study of PGCs Cultivation Systems HiS and FAcs: A Transcriptomic and Cellular Biology Perspective. Poult. Sci. 2024, 103, 104058. [Google Scholar] [CrossRef]
- Zhang, Z.; Elsayed, A.K.; Shi, Q.; Zhang, Y.; Zuo, Q.; Li, D.; Lian, C.; Tang, B.; Xiao, T.; Xu, Q.; et al. Crucial Genes and Pathways in Chicken Germ Stem Cell Differentiation. J. Biol. Chem. 2015, 290, 13605–13621. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, S.; Kim, T.M.; Kim, Y.M.; Seo, H.W.; Park, T.S.; Jeong, J.-W.; Song, G.; Han, J.Y. Basic Fibroblast Growth Factor Activates MEK/ERK Cell Signaling Pathway and Stimulates the Proliferation of Chicken Primordial Germ Cells. PLoS ONE 2010, 5, e12968. [Google Scholar] [CrossRef]
- Liu, X.; Wu, J.; Peng, Y.; Qian, H.; Lv, X.; Li, F.; Jin, K.; Niu, Y.; Song, J.; Han, W.; et al. Chicken Primordial Germ Cells Do Not Proliferate in Insulin-Lacking Media. Int. J. Mol. Sci. 2025, 26, 3122. [Google Scholar] [CrossRef]
- Liu, X.; Wu, J.; Peng, Y.; Liu, G.; Jin, K.; Niu, Y.; Song, J.; Han, W.; Chen, G.; Li, B.; et al. Functional Equivalence of Insulin and IGF-1 in the In Vitro Culture of Chicken Primordial Germ Cells. Genes 2025, 16, 481. [Google Scholar] [CrossRef]
- Liu, X.; Ye, L.; Ding, Y.; Gong, W.; Qian, H.; Jin, K.; Niu, Y.; Zuo, Q.; Song, J.; Han, W.; et al. Role of PI3K/AKT Signaling Pathway Involved in Self-Renewing and Maintaining Biological Properties of Chicken Primordial Germ Cells. Poult. Sci. 2024, 103, 104140. [Google Scholar] [CrossRef] [PubMed]
- Yakhkeshi, S.; Rahimi, S.; Sharafi, M.; Hassani, S.-N.; Taleahmad, S.; Shahverdi, A.; Baharvand, H. In Vitro Improvement of Quail Primordial Germ Cell Expansion through Activation of TGF-Beta Signaling Pathway. J. Cell Biochem. 2018, 119, 4309–4319. [Google Scholar] [CrossRef] [PubMed]
- Rassouli, H.; Sayadmanesh, A.; Rezaeiani, S.; Ghezelayagh, Z.; Gharaati, M.R.; Yaser, T. An Easy and Fast Method for Production of Chinese Hamster Ovary Cell Line Expressing and Secreting Human Recombinant Activin A. Cell J. 2020, 22, 140. [Google Scholar] [CrossRef]
- Gong, W.; Zhao, J.; Yao, Z.; Zhang, Y.; Niu, Y.; Jin, K.; Li, B.; Zuo, Q. The Establishment and Optimization of a Chicken Primordial Germ Cell Induction Model Using Small-Molecule Compounds. Animals 2024, 14, 302. [Google Scholar] [CrossRef]
- Ma, Y.; Meng, L.; Wei, J.; Wu, W.; Zhang, Y.; Wang, X.; Guo, X.; Wang, F.; Mao, Y.; Zhu, G. Suppression of FOXO3 by BMP Signaling Contribute to the Different Primordial Germ Cell Proliferation between Layers and Broilers. Biol. Reprod. 2025, 112, ioaf037. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Jin, K.; Wang, M.; Zhang, Y.; Chen, G.; Li, B. BMP4 Activates the Wnt-Lin28A-Blimp1-Wnt Pathway to Promote Primordial Germ Cell Formation via Altering H3K4me2. J. Cell Sci. 2021, 134, jcs249375. [Google Scholar] [CrossRef]
- Zuo, Q.; Jin, J.; Jin, K.; Sun, C.; Song, J.; Zhang, Y.; Chen, G.; Li, B. Distinct Roles of Retinoic Acid and BMP4 Pathways in the Formation of Chicken Primordial Germ Cells and Spermatogonial Stem Cells. Food Funct. 2019, 10, 7152–7163. [Google Scholar] [CrossRef]
- Lee, H.C.; Lim, S.; Han, J.Y. Wnt/β-Catenin Signaling Pathway Activation Is Required for Proliferation of Chicken Primordial Germ Cells in Vitro. Sci. Rep. 2016, 6, 34510. [Google Scholar] [CrossRef] [PubMed]
- Dehdilani, N.; Yousefi Taemeh, S.; Rival-Gervier, S.; Montillet, G.; Kress, C.; Jean, C.; Goshayeshi, L.; Dehghani, H.; Pain, B. Enhanced Cultivation of Chicken Primordial Germ Cells. Sci. Rep. 2023, 13, 12323. [Google Scholar] [CrossRef]
- Xin-Yan, T.; Wei-Dong, Z.; Yu-Ling, M.; Hong-Yun, L.; Cai-Qiao, Z. Isolation, Culture and Characterization of Chicken Primordial Germ Cells. Chin. J. Agric. Biotechnol. 2006, 3, 183–188. [Google Scholar] [CrossRef]
- Ezaki, R.; Hirose, F.; Furusawa, S.; Horiuchi, H. An Improved Protocol for Stable and Efficient Culturing of Chicken Primordial Germ Cells Using Small-Molecule Inhibitors. Cytotechnology 2020, 72, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Pérez Sáez, J.M.; Bussmann, L.E.; Barañao, J.L.; Bussmann, U.A. Improvement of Chicken Primordial Germ Cell Maintenance in Vitro by Blockade of the Aryl Hydrocarbon Receptor Endogenous Activity. Cell. Reprogramming 2016, 18, 154–161. [Google Scholar] [CrossRef]
- Rengaraj, D.; Won, S.; Jung, K.M.; Woo, S.J.; Lee, H.; Kim, Y.M.; Kim, H.; Han, J.Y. Chicken Blastoderms and Primordial Germ Cells Possess a Higher Expression of DNA Repair Genes and Lower Expression of Apoptosis Genes to Preserve Their Genome Stability. Sci. Rep. 2022, 12, 49. [Google Scholar] [CrossRef]
- Zuo, Q.; Jing, J.; Zhou, J.; Zhang, Y.; Wei, W.; Chen, G.; Li, B. Dual Regulatory Actions of LncBMP4 on BMP4 Promote Chicken Primordial Germ Cell Formation. EMBO Rep. 2022, 23, e52491. [Google Scholar] [CrossRef]
- Ding, Y.; Zhi, Q.; Zuo, Q.; Jin, K.; Han, W.; Li, B. Transcriptome-Based Analysis of Key Signaling Pathways Affecting the Formation of Primordial Germ Cell in Chickens. J. Integr. Agric. 2024, 23, 1644–1657. [Google Scholar] [CrossRef]
- Doddamani, D.; Lázár, B.; Ichikawa, K.; Hu, T.; Taylor, L.; Gócza, E.; Várkonyi, E.; McGrew, M.J. Propagation of Goose Primordial Germ Cells in Vitro Relies on FGF and BMP Signalling Pathways. Commun. Biol. 2025, 8, 301. [Google Scholar] [CrossRef]
- Yousefi Taemeh, S.; Dehdilani, N.; Goshayeshi, L.; Kress, C.; Rival-Gervier, S.; Montillet, G.; Ebrahimi Vishki, R.; Pain, B.; Dehghani, H. Strain-Specific Variations in the Culture of Chicken Primordial Germ Cells. Sci. Rep. 2025, 15, 11858. [Google Scholar] [CrossRef]
- Kong, L.; Qiu, L.; Guo, Q.; Chen, Y.; Zhang, X.; Chen, B.; Zhang, Y.; Chang, G. Long-term in vitro culture and preliminary establishment of chicken primordial germ cell lines. PLoS ONE 2018, 13, e0196459. [Google Scholar] [CrossRef]
- Hong, Y.H.; Moon, Y.K.; Jeong, D.K.; Han, J.Y. Improved Transfection Efficiency of Chicken Gonadal Primordial Germ Cells for the Production of Transgenic Poultry. Transgenic Res. 1998, 7, 247–252. [Google Scholar] [CrossRef]
- Oishi, I. Improvement of Transfection Efficiency in Cultured Chicken Primordial Germ Cells by Percoll Density Gradient Centrifugation. Biosci. Biotechnol. Biochem. 2010, 74, 2426–2430. [Google Scholar] [CrossRef]
- Zhao, Z.; Zou, X.; Zhu, Y.; He, Y.; Jebessa, E.; Zhang, J.; Ji, J.; Chen, P.; Luo, C. Achieving Optimal Transfection Conditions in Chicken Primordial Germ Cells Under Feeder- and Serum-Free Medium. Animals 2025, 15, 590. [Google Scholar] [CrossRef]
- Sawicka, D.; Chojnacka-Puchta, L. Effective Transfection of Chicken Primordial Germ Cells (PGCs) Using Transposon Vectors and Lipofection. Folia Biol. 2019, 67, 45–52. [Google Scholar] [CrossRef]
- Watanabe, T.; Ochi, Y.; Kajihara, R.; Ichikawa, K.; Ezaki, R.; Matsuzaki, M.; Horiuchi, H. Lipofection with LipofectamineTM 2000 in a Heparin-free Growth Medium Results in High Transfection Efficiency in Chicken Primordial Germ Cells. Biotechnol. J. 2023, 18, 2300328. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Zhu, Z.; Xue, M.; Wang, F.; Ma, Y.; Zhu, G. A Comparative Study of Transfection Techniques for Genetic Modification in Chicken Primordial Germ Cells. Mol. Biotechnol. 2025, 1–13. [Google Scholar] [CrossRef]
- Naito, M.; Tajima, A.; Tagami, T.; Yasuda, Y.; Kuwana, T. Preservation of Chick Primordial Germ Cells in Liquid Nitrogen and Subsequent Production of Viable Offspring. Reproduction 1994, 102, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Usui, F.; Miyahara, D.; Mori, T.; Watanabe, H.; Ono, T.; Takeda, K.; Nirasawa, K.; Kagami, H.; Tagami, T. Viability and Functionality of Primordial Germ Cells after Freeze-Thaw in Chickens. J. Poult. Sci. 2011, 48, 57–63. [Google Scholar] [CrossRef]
- Tajima, A.; Naito, M.; Yasuda, Y.; Kuwana, T. Production of Germ-Line Chimeras by Transfer of Cryopreserved Gonadal Primordial Germ Cells (gPGCs) in Chicken. J. Exp. Zool. 1998, 280, 265–267. [Google Scholar] [CrossRef]
- Moore, D.T.; Purdy, P.H.; Blackburn, H.D. A Method for Cryopreserving Chicken Primordial Germ Cells. Poult. Sci. 2006, 85, 1784–1790. [Google Scholar] [CrossRef]
- Tonus, C.; Connan, D.; Waroux, O.; Vandenhove, B.; Wayet, J.; Gillet, L.; Desmecht, D.; Antoine, N.; Ectors, F.J.; Grobet, L. Cryopreservation of Chicken Primordial Germ Cells by Vitrification and Slow Freezing: A Comparative Study. Theriogenology 2017, 88, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Tonus, C.; Cloquette, K.; Ectors, F.; Piret, J.; Gillet, L.; Antoine, N.; Desmecht, D.; Vanderplasschen, A.; Waroux, O.; Grobet, L. Long Term-Cultured and Cryopreserved Primordial Germ Cells from Various Chicken Breeds Retain High Proliferative Potential and Gonadal Colonisation Competency. Reprod. Fertil. Dev. 2016, 28, 628. [Google Scholar] [CrossRef]
- Hamai, N.; Koide, C.; Tansho, Y.; Ooka, Y.; Hirano, M.; Fatira, E.; Tsudzuki, M.; Nakamura, Y. Development of Cryopreservation Media for the Slow-Freezing of Cultured Primordial Germ Cells in Chicken. J. Reprod. Dev. 2023, 69, 109–117. [Google Scholar] [CrossRef]
- Sid, H.; Schusser, B. Applications of Gene Editing in Chickens: A New Era Is on the Horizon. Front. Genet. 2018, 9, 456. [Google Scholar] [CrossRef]
- Wani, A.K.; Akhtar, N.; Singh, R.; Prakash, A.; Raza, S.H.A.; Cavalu, S.; Chopra, C.; Madkour, M.; Elolimy, A.; Hashem, N.M. Genome Centric Engineering Using ZFNs, TALENs and CRISPR-Cas9 Systems for Trait Improvement and Disease Control in Animals. Vet. Res. Commun. 2023, 47, 1–16. [Google Scholar] [CrossRef]
- Ain, Q.U.; Chung, J.Y.; Kim, Y.-H. Current and Future Delivery Systems for Engineered Nucleases: ZFN, TALEN and RGEN. J. Control. Release 2015, 205, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.; Schambach, A.; Bohne, J.; Galla, M. Retrovirus Vectors: Toward the Plentivirus? Mol. Ther. 2006, 13, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Salter, D.W.; Smith, E.J.; Hughes, S.H.; Wright, S.E.; Crittenden, L.B. Transgenic Chickens: Insertion of Retroviral Genes into the Chicken Germ Line. Virology 1987, 157, 236–240. [Google Scholar] [CrossRef]
- Salter, D.W.; Smith, E.J.; Hughes, S.H.; Wright, S.E.; Fadly, A.M.; Witter, R.L.; Crittenden, L.B. Gene Insertion into the Chicken Germ Line by Retroviruses. Poult. Sci. 1986, 65, 1445–1458. [Google Scholar] [CrossRef]
- Bosselman, R.A.; Hsu, R.Y.; Boggs, T.; Hu, S.; Bruszewski, J.; Ou, S.; Souza, L.; Kozar, L.; Martin, F.; Nicolson, M. Replication-Defective Vectors of Reticuloendotheliosis Virus Transduce Exogenous Genes into Somatic Stem Cells of the Unincubated Chicken Embryo. J. Virol. 1989, 63, 2680–2689. [Google Scholar] [CrossRef]
- Kamihira, M.; Ono, K.; Esaka, K.; Nishijima, K.; Kigaku, R.; Komatsu, H.; Yamashita, T.; Kyogoku, K.; Iijima, S. High-Level Expression of Single-Chain Fv-Fc Fusion Protein in Serum and Egg White of Genetically Manipulated Chickens by Using a Retroviral Vector. J. Virol. 2005, 79, 10864–10874. [Google Scholar] [CrossRef]
- Chapman, S.C.; Lawson, A.; MacArthur, W.C.; Wiese, R.J.; Loechel, R.H.; Burgos-Trinidad, M.; Wakefield, J.K.; Ramabhadran, R.; Mauch, T.J.; Schoenwolf, G.C. Ubiquitous GFP Expression in Transgenic Chickens Using a Lentiviral Vector. Development 2005, 132, 935–940. [Google Scholar] [CrossRef] [PubMed]
- McGrew, M.J.; Sherman, A.; Ellard, F.M.; Lillico, S.G.; Gilhooley, H.J.; Kingsman, A.J.; Mitrophanous, K.A.; Sang, H. Efficient Production of Germline Transgenic Chickens Using Lentiviral Vectors. EMBO Rep. 2004, 5, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.B.; Lois, C. Generation of Tissue-Specific Transgenic Birds with Lentiviral Vectors. Proc. Natl. Acad. Sci. USA 2005, 102, 16443–16447. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Karolak, M.C.; Shin, S.; Lee, K. Generation of Genome-Edited Chicken and Duck Lines by Adenovirus-Mediated in Vivo Genome Editing. Proc. Natl. Acad. Sci. USA 2022, 119, e2214344119. [Google Scholar] [CrossRef]
- Mukae, T.; Yoshii, K.; Watanobe, T.; Tagami, T.; Oishi, I. Production and Characterization of Eggs from Hens with Ovomucoid Gene Mutation. Poult. Sci. 2021, 100, 452–460. [Google Scholar] [CrossRef]
- Lu, Y.; Lin, C.; Wang, X. PiggyBac Transgenic Strategies in the Developing Chicken Spinal Cord. Nucleic Acids Res. 2009, 37, e141. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Villegas, N.; Nurieva, W.; Amberger, M.; Ivics, Z. Contemporary Transposon Tools: A Review and Guide through Mechanisms and Applications of Sleeping Beauty, piggyBac and Tol2 for Genome Engineering. Int. J. Mol. Sci. 2021, 22, 5084. [Google Scholar] [CrossRef] [PubMed]
- Sherman, A.; Dawson, A.; Mather, C.; Gilhooley, H.; Li, Y.; Mitchell, R.; Finnegan, D.; Sang, H. Transpostion of the Drosophila Element Mariner into the Chicken Germ Line. Nat. Biotechnol. 1998, 16, 1050–1053, Erratum in Nat. Biotechnol. 1999, 17, 81. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kasai, T.; Nakagawa, S.; Tanabe, K.; Watanabe, T.; Kawakami, K.; Takahashi, Y. Stable Integration and Conditional Expression of Electroporated Transgenes in Chicken Embryos. Dev. Biol. 2007, 305, 616–624. [Google Scholar] [CrossRef]
- Tyack, S.G.; Jenkins, K.A.; O’Neil, T.E.; Wise, T.G.; Morris, K.R.; Bruce, M.P.; McLeod, S.; Wade, A.J.; McKay, J.; Moore, R.J.; et al. A New Method for Producing Transgenic Birds via Direct in Vivo Transfection of Primordial Germ Cells. Transgenic Res. 2013, 22, 1257–1264. [Google Scholar] [CrossRef]
- Park, T.S.; Han, J.Y. piggyBac Transposition into Primordial Germ Cells Is an Efficient Tool for Transgenesis in Chickens. Proc. Natl. Acad. Sci. USA 2012, 109, 9337–9341. [Google Scholar] [CrossRef]
- Jordan, B.J.; Vogel, S.; Stark, M.R.; Beckstead, R.B. Expression of Green Fluorescent Protein in the Chicken Using in Vivo Transfection of the piggyBac Transposon. J. Biotechnol. 2014, 173, 86–89. [Google Scholar] [CrossRef]
- LaFountaine, J.S.; Fathe, K.; Smyth, H.D.C. Delivery and Therapeutic Applications of Gene Editing Technologies ZFNs, TALENs, and CRISPR/Cas9. Int. J. Pharm. 2015, 494, 180–194. [Google Scholar] [CrossRef]
- Liu, Q.; Sun, Q.; Yu, J. Gene Editing’s Sharp Edge: Understanding Zinc Finger Nucleases (ZFN), Transcription Activator-Like Effector Nucleases (TALEN) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR). Trans. Mater. Biotechnol. Life Sci. 2024, 3, 170–179. [Google Scholar] [CrossRef]
- Park, T.S.; Lee, H.J.; Kim, K.H.; Kim, J.-S.; Han, J.Y. Targeted Gene Knockout in Chickens Mediated by TALENs. Proc. Natl. Acad. Sci. USA 2014, 111, 12716–12721. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.; Carlson, D.F.; Nandi, S.; Sherman, A.; Fahrenkrug, S.C.; McGrew, M.J. Efficient TALEN-Mediated Gene Targeting of Chicken Primordial Germ Cells. Development 2017, 144, 928–934. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA Maturation by Trans-Encoded Small RNA and Host Factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 Transcriptional Activators for Target Specificity Screening and Paired Nickases for Cooperative Genome Engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Kagami, H.; Tagami, T. Targeted Mutagenesis in Chicken Using CRISPR/Cas9 System. Sci. Rep. 2016, 6, 23980. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, L.; Pedersen, D.; Ching, K.H.; Yi, H.; Collarini, E.J.; Izquierdo, S.; Van De Lavoir, M.-C.; Leighton, P.A. Germline Gene Editing in Chickens by Efficient CRISPR-Mediated Homologous Recombination in Primordial Germ Cells. PLoS ONE 2016, 11, e0154303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zuo, Q.; Li, D.; Zhang, W.; Wang, F.; Ji, Y.; Jin, J.; Lu, Z.; Wang, M.; et al. CRISPR/Cas9 Mediated Chicken Stra8 Gene Knockout and Inhibition of Male Germ Cell Differentiation. PLoS ONE 2017, 12, e0172207. [Google Scholar] [CrossRef]
- Lee, H.J.; Yoon, J.W.; Jung, K.M.; Kim, Y.M.; Park, J.S.; Lee, K.Y.; Park, K.J.; Hwang, Y.S.; Park, Y.H.; Rengaraj, D.; et al. Targeted Gene Insertion into Z Chromosome of Chicken Primordial Germ Cells for Avian Sexing Model Development. FASEB J. 2019, 33, 8519–8529. [Google Scholar] [CrossRef]
- Ezaki, R.; Ichikawa, K.; Matsuzaki, M.; Horiuchi, H. Targeted Knock-in of a Fluorescent Protein Gene into the Chicken Vasa Homolog Locus of Chicken Primordial Germ Cells Using CRIS-PITCh Method. J. Poult. Sci. 2022, 59, 182–190. [Google Scholar] [CrossRef]
- Kinoshita, K.; Tanabe, K.; Nakamura, Y.; Nishijima, K.-I.; Suzuki, T.; Okuzaki, Y.; Mizushima, S.; Wang, M.-S.; Khan, S.U.; Xu, K.; et al. PGC-Based Cryobanking, Regeneration through Germline Chimera Mating, and CRISPR/Cas9-Mediated TYRP1 Modification in Indigenous Chinese Chickens. Commun. Biol. 2024, 7, 1127. [Google Scholar] [CrossRef]
- Zuo, Z.; Babu, K.; Ganguly, C.; Zolekar, A.; Newsom, S.; Rajan, R.; Wang, Y.-C.; Liu, J. Rational Engineering of CRISPR-Cas9 Nuclease to Attenuate Position-Dependent Off-Target Effects. CRISPR J. 2022, 5, 329–340. [Google Scholar] [CrossRef]
- Altgilbers, S.; Dierks, C.; Klein, S.; Weigend, S.; Kues, W.A. Quantitative Analysis of CRISPR/Cas9-Mediated Provirus Deletion in Blue Egg Layer Chicken PGCs by Digital PCR. Sci. Rep. 2022, 12, 15587. [Google Scholar] [CrossRef]
- Ruffolo, J.A.; Nayfach, S.; Gallagher, J.; Bhatnagar, A.; Beazer, J.; Hussain, R.; Russ, J.; Yip, J.; Hill, E.; Pacesa, M.; et al. Design of Highly Functional Genome Editors by Modelling CRISPR–Cas Sequences. Nature 2025, 645, 518–525. [Google Scholar] [CrossRef]
- Chia, B.S.; Seah, Y.F.S.; Wang, B.; Shen, K.; Srivastava, D.; Chew, W.L. Engineering a New Generation of Gene Editors: Integrating Synthetic Biology and AI Innovations. ACS Synth. Biol. 2025, 14, 636–647. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J. A CRISPR View of Development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A Review of the Challenges and Approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef] [PubMed]
- Chapman, B.; Han, J.H.; Lee, H.J.; Ruud, I.; Kim, T.H. Targeted Modulation of Chicken Genes In Vitro Using CRISPRa and CRISPRi Toolkit. Genes 2023, 14, 906. [Google Scholar] [CrossRef] [PubMed]
- Han, J.H.; Lee, H.J.; Kim, T.H. Characterization of Transcriptional Enhancers in the Chicken Genome Using CRISPR-Mediated Activation. Front. Genome Ed. 2023, 5, 1269115. [Google Scholar] [CrossRef]
- Sokka, J.; Yoshihara, M.; Kvist, J.; Laiho, L.; Warren, A.; Stadelmann, C.; Jouhilahti, E.-M.; Kilpinen, H.; Balboa, D.; Katayama, S.; et al. CRISPR Activation Enables High-Fidelity Reprogramming into Human Pluripotent Stem Cells. Stem Cell Rep. 2022, 17, 413–426. [Google Scholar] [CrossRef]
- Urrutia-Cabrera, D.; Hsiang-Chi Liou, R.; Lin, J.; Shi, Y.; Liu, K.; Hung, S.S.C.; Hewitt, A.W.; Wang, P.-Y.; Ching-Bong Wong, R. Combinatorial Approach of Binary Colloidal Crystals and CRISPR Activation to Improve Induced Pluripotent Stem Cell Differentiation into Neurons. ACS Appl. Mater. Interfaces 2022, 14, 8669–8679. [Google Scholar] [CrossRef]
- Leng, K.; Rose, I.V.L.; Kim, H.; Xia, W.; Romero-Fernandez, W.; Rooney, B.; Koontz, M.; Li, E.; Ao, Y.; Wang, S.; et al. CRISPRi Screens in Human iPSC-Derived Astrocytes Elucidate Regulators of Distinct Inflammatory Reactive States. Nat. Neurosci. 2022, 25, 1528–1542. [Google Scholar] [CrossRef]
- Leung, R.K.-K.; Cheng, Q.-X.; Wu, Z.-L.; Khan, G.; Liu, Y.; Xia, H.-Y.; Wang, J. CRISPR-Cas12-Based Nucleic Acids Detection Systems. Methods 2022, 203, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, S.; Ma, Y.; Jiang, Y.; Li, Y.; Shi, J.; Deng, G.; Tian, G.; Kong, H.; Wang, X. On-Site and Visual Detection of the H5 Subtype Avian Influenza Virus Based on RT-RPA and CRISPR/Cas12a. Viruses 2024, 16, 753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, G.; Ding, X.; Zhang, K.; Sun, W.; Li, Q.; Yi, Y.; Wang, J.; Pang, X.; Chen, L. A Rapid Simultaneous Detection of Duck Hepatitis a Virus 3 and Novel Duck Reovirus Based on RPA CRISPR Cas12a/Cas13a. Int. J. Biol. Macromol. 2024, 274, 133246. [Google Scholar] [CrossRef]
- Wu, Y.; Zhan, J.; Shan, Z.; Li, Y.; Liu, Y.; Li, Y.; Wang, Y.; Liu, Z.; Wen, X.; Wang, X. CRISPR-Cas13a-Based Detection Method for Avian Influenza Virus. Front. Microbiol. 2023, 14, 1288951. [Google Scholar] [CrossRef]
- Chen, Q.; Su, C.; Li, S.; Zhang, Z.; Yang, Y.; Yang, Y.; Tao, D.; Xie, S.; Gong, P.; Feng, Y. A Sensitive and Rapid Visual Method of Chicken Sexing Based on LAMP-CRISPR/Cas12a System. Br. Poult. Sci. 2025, 66, 531–538. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Saito, D.; Suzuki, T.; Takemoto, T. An Inducible Germ Cell Ablation Chicken Model for High-Grade Germline Chimeras. Development 2023, 150, dev202079. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Challagulla, A.; Schat, K.A.; Doran, T.J. In Vitro Inhibition of Influenza Virus Using CRISPR/Cas13a in Chicken Cells. Methods Protoc. 2021, 4, 40. [Google Scholar] [CrossRef]
- Asmamaw Mengstie, M.; Teshome Azezew, M.; Asmamaw Dejenie, T.; Teshome, A.A.; Tadele Admasu, F.; Behaile Teklemariam, A.; Tilahun Mulu, A.; Mekonnen Agidew, M.; Adugna, D.G.; Geremew, H.; et al. Recent Advancements in Reducing the Off-Target Effect of CRISPR-Cas9 Genome Editing. Biol. Targets Ther. 2024, 18, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base Editing in Crops: Current Advances, Limitations and Future Implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar] [CrossRef]
- Nidhi, S.; Anand, U.; Oleksak, P.; Tripathi, P.; Lal, J.A.; Thomas, G.; Kuca, K.; Tripathi, V. Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int. J. Mol. Sci. 2021, 22, 3327. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, J.; Harrison, P.T. Prime Editing—An Update on the Field. Gene Ther. 2021, 28, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Lee, H.J.; Choi, H.J.; Han, S.T.; Lee, K.H.; Park, K.J.; Park, J.S.; Jung, K.M.; Kim, Y.M.; Han, H.J.; et al. Highly Elevated Base Excision Repair Pathway in Primordial Germ Cells Causes Low Base Editing Activity in Chickens. FASEB J. 2020, 34, 15907–15921. [Google Scholar] [CrossRef]
- Xu, T.; Zhong, J.; Huang, Z.; Yu, L.; Zheng, J.; Xie, L.; Sun, L.; Liu, X.; Lu, Y. Optimization of the Base Editor BE4max in Chicken Somatic Cells. Poult. Sci. 2022, 101, 102174. [Google Scholar] [CrossRef]
- Atsuta, Y.; Suzuki, K.; Iikawa, H.; Yaguchi, H.; Saito, D. Prime Editing in Chicken Fibroblasts and Primordial Germ Cells. Dev. Growth Differ. 2022, 64, 548–557. [Google Scholar] [CrossRef]
- Smith, J.; Smith, N.; Yu, L.; Paton, I.R.; Gutowska, M.W.; Forrest, H.L.; Danner, A.F.; Seiler, J.P.; Digard, P.; Webster, R.G.; et al. A Comparative Analysis of Host Responses to Avian Influenza Infection in Ducks and Chickens Highlights a Role for the Interferon-Induced Transmembrane Proteins in Viral Resistance. BMC Genom. 2015, 16, 574. [Google Scholar] [CrossRef]
- Lee, C.-W.; Saif, Y.M. Avian Influenza Virus. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Spackman, E.; Suarez, D. Avian Influenza Virus; Humana Press: Totowa, NJ, USA, 2008; Volume 8. [Google Scholar]
- Lyall, J.; Irvine, R.M.; Sherman, A.; McKinley, T.J.; Núñez, A.; Purdie, A.; Outtrim, L.; Brown, I.H.; Rolleston-Smith, G.; Sang, H.; et al. Suppression of Avian Influenza Transmission in Genetically Modified Chickens. Science 2011, 331, 223–226. [Google Scholar] [CrossRef]
- Jun, H.-R.; Pham, C.D.; Lim, S.-I.; Lee, S.-C.; Kim, Y.-S.; Park, S.; Kwon, M.-H. An RNA-Hydrolyzing Recombinant Antibody Exhibits an Antiviral Activity against Classical Swine Fever Virus. Biochem. Biophys. Res. Commun. 2010, 395, 484–489. [Google Scholar] [CrossRef]
- June Byun, S.; Yuk, S.; Jang, Y.-J.; Choi, H.; Jeon, M.-H.; Erdene-Ochir, T.; Kwon, J.-H.; Noh, J.-Y.; Kim, J.S.; Yoo, J.G.; et al. Transgenic Chickens Expressing the 3D8 Single Chain Variable Fragment Protein Suppress Avian Influenza Transmission. Sci. Rep. 2017, 7, 5938. [Google Scholar] [CrossRef]
- Idoko-Akoh, A.; Goldhill, D.H.; Sheppard, C.M.; Bialy, D.; Quantrill, J.L.; Sukhova, K.; Brown, J.C.; Richardson, S.; Campbell, C.; Taylor, L.; et al. Creating Resistance to Avian Influenza Infection through Genome Editing of the ANP32 Gene Family. Nat. Commun. 2023, 14, 6136. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, K.Y.; Jung, K.M.; Park, K.J.; Lee, K.O.; Suh, J.-Y.; Yao, Y.; Nair, V.; Han, J.Y. Precise Gene Editing of Chicken Na+/H+ Exchange Type 1 (chNHE1) Confers Resistance to Avian Leukosis Virus Subgroup J (ALV-J). Dev. Comp. Immunol. 2017, 77, 340–349. [Google Scholar] [CrossRef]
- Hellmich, R.; Sid, H.; Lengyel, K.; Flisikowski, K.; Schlickenrieder, A.; Bartsch, D.; Thoma, T.; Bertzbach, L.D.; Kaufer, B.B.; Nair, V.; et al. Acquiring Resistance Against a Retroviral Infection via CRISPR/Cas9 Targeted Genome Editing in a Commercial Chicken Line. Front. Genome Ed. 2020, 2, 3. [Google Scholar] [CrossRef]
- Koslová, A.; Trefil, P.; Mucksová, J.; Reinišová, M.; Plachý, J.; Kalina, J.; Kučerová, D.; Geryk, J.; Krchlíková, V.; Lejčková, B.; et al. Precise CRISPR/Cas9 Editing of the NHE1 Gene Renders Chickens Resistant to the J Subgroup of Avian Leukosis Virus. Proc. Natl. Acad. Sci. USA 2020, 117, 2108–2112. [Google Scholar] [CrossRef]
- Luo, J.; Teng, M.; Zai, X.; Tang, N.; Zhang, Y.; Mandviwala, A.; Reddy, V.R.A.P.; Baigent, S.; Yao, Y.; Nair, V. Efficient Mutagenesis of Marek’s Disease Virus-Encoded microRNAs Using a CRISPR/Cas9-Based Gene Editing System. Viruses 2020, 12, 466. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, W.; Tang, N.; Moffat, K.; Nair, V.; Yao, Y. Targeted Deletion of Glycoprotein B Gene by CRISPR/Cas9 Nuclease Inhibits Gallid Herpesvirus Type 3 in Dually Infected Marek’s Disease Virus-Transformed Lymphoblastoid Cell Line MSB-1. J. Virol. 2022, 96, e02027-21. [Google Scholar] [CrossRef]
- Challagulla, A.; Jenkins, K.A.; O’Neil, T.E.; Shi, S.; Morris, K.R.; Wise, T.G.; Paradkar, P.N.; Tizard, M.L.; Doran, T.J.; Schat, K.A. In Vivo Inhibition of Marek’s Disease Virus in Transgenic Chickens Expressing Cas9 and gRNA against ICP4. Microorganisms 2021, 9, 164. [Google Scholar] [CrossRef]
- Ullah, A.; Khan, M.; Akram, M.; Abdullah, M.; Sayka, S.; Iqbal, I.; Siddique, F.; Arain, M.A.; Hassan, F.-U.; Shafi, M. CRISPR Cas-9 for Treatment and Control of Avian Viral Diseases: Challenges and Opportunities. World’s Poult. Sci. J. 2024, 80, 745–765. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Lee, K. Myostatin Gene Role in Regulating Traits of Poultry Species for Potential Industrial Applications. J. Anim. Sci. Biotechnol. 2024, 15, 82. [Google Scholar] [CrossRef] [PubMed]
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of Skeletal Muscle Mass in Mice by a New TGF-p Superfamily Member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Xu, K.; Han, C.X.; Zhou, H.; Ding, J.M.; Xu, Z.; Yang, L.Y.; He, C.; Akinyemi, F.; Zheng, Y.M.; Qin, C. Effective MSTN Gene Knockout by AdV-Delivered CRISPR/Cas9 in Postnatal Chick Leg Muscle. Int. J. Mol. Sci. 2020, 21, 2584. [Google Scholar] [CrossRef]
- Lee, J.; Kim, D.-H.; Lee, K. Muscle Hyperplasia in Japanese Quail by Single Amino Acid Deletion in MSTN Propeptide. Int. J. Mol. Sci. 2020, 21, 1504. [Google Scholar] [CrossRef]
- Bhattacharya, T.K.; Shukla, R.; Chatterjee, R.N.; Bhanja, S.K. Comparative Analysis of Silencing Expression of Myostatin (MSTN) and Its Two Receptors (ACVR2A and ACVR2B) Genes Affecting Growth Traits in Knock down Chicken. Sci. Rep. 2019, 9, 7789. [Google Scholar] [CrossRef]
- Park, T.S.; Park, J.; Lee, J.H.; Park, J.-W.; Park, B.-C. Disruption of G0/G1 Switch Gene 2 (G0S2) Reduced Abdominal Fat Deposition and Altered Fatty Acid Composition in Chicken. FASEB J. 2019, 33, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Dhanapala, P.; Withanage-Dona, D.; Tang, M.; Doran, T.; Suphioglu, C. Hypoallergenic Variant of the Major Egg White Allergen Gal d 1 Produced by Disruption of Cysteine Bridges. Nutrients 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, K.; Rudra, M.; Berghof, T.V.L.; Leitão, A.; Frankl-Vilches, C.; Dittrich, F.; Duda, D.; Klinger, R.; Schleibinger, S.; Sid, H.; et al. Unveiling the Critical Role of Androgen Receptor Signaling in Avian Sexual Development. Nat. Commun. 2024, 15, 8970. [Google Scholar] [CrossRef]
- Ioannidis, J.; Taylor, G.; Zhao, D.; Liu, L.; Idoko-Akoh, A.; Gong, D.; Lovell-Badge, R.; Guioli, S.; McGrew, M.J.; Clinton, M. Primary Sex Determination in Birds Depends on DMRT1 Dosage, but Gonadal Sex Does Not Determine Adult Secondary Sex Characteristics. Proc. Natl. Acad. Sci. USA 2021, 118, e2020909118. [Google Scholar] [CrossRef] [PubMed]
- Fallahshahroudi, A.; Yousefi Taemeh, S.; Rodríguez-Montes, L.; Trost, N.; Frank, D.; Lafrenz, P.; Koubek, J.; Tellez, G.; Ballantyne, M.; Idoko-Akoh, A.; et al. A Male-Essential miRNA Is Key for Avian Sex Chromosome Dosage Compensation. Nature 2025, 645, 148–157. [Google Scholar] [CrossRef]
- Schusser, B.; Collarini, E.J.; Pedersen, D.; Yi, H.; Ching, K.; Izquierdo, S.; Thoma, T.; Lettmann, S.; Kaspers, B.; Etches, R.J.; et al. Expression of Heavy Chain-only Antibodies Can Support B-cell Development in Light Chain Knockout Chickens. Eur. J. Immunol. 2016, 46, 2137–2148. [Google Scholar] [CrossRef]
- Schusser, B.; Collarini, E.J.; Yi, H.; Izquierdo, S.M.; Fesler, J.; Pedersen, D.; Klasing, K.C.; Kaspers, B.; Harriman, W.D.; Van De Lavoir, M.-C.; et al. Immunoglobulin Knockout Chickens via Efficient Homologous Recombination in Primordial Germ Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20170–20175. [Google Scholar] [CrossRef]
- Rieblinger, B.; Sid, H.; Duda, D.; Bozoglu, T.; Klinger, R.; Schlickenrieder, A.; Lengyel, K.; Flisikowski, K.; Flisikowska, T.; Simm, N.; et al. Cas9-Expressing Chickens and Pigs as Resources for Genome Editing in Livestock. Proc. Natl. Acad. Sci. USA 2021, 118, e2022562118. [Google Scholar] [CrossRef]
- Ho Ching Chan, B.; Hardy, H.; Requena, T.; Findlay, A.; Ioannidis, J.; Meunier, D.; Toms, M.; Moosajee, M.; Raper, A.; McGrew, M.J.; et al. A Stable NTN1 Fluorescent Reporter Chicken Reveals Cell Specific Molecular Signatures during Optic Fissure Closure. Sci. Rep. 2025, 15, 10096. [Google Scholar] [CrossRef] [PubMed]
- Hagihara, Y.; Okuzaki, Y.; Matsubayashi, K.; Kaneoka, H.; Suzuki, T.; Iijima, S.; Nishijima, K. Primordial Germ Cell-specific Expression of ᴇGFP in Transgenic Chickens. Genesis 2020, 58, e23388. [Google Scholar] [CrossRef]
- Doddamani, D.; Carlson, D.F.; McTeir, L.; Taylor, L.; Nandi, S.; Davey, M.G.; McGrew, M.J.; Glover, J.D. PRDM14 Is Essential for Vertebrate Gastrulation and Safeguards Avian Germ Cell Identity. Dev. Biol. 2025, 521, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Matsui, S.; Watanabe, D. Transgenic Songbirds with Suppressed or Enhanced Activity of CREB Transcription Factor. Proc. Natl. Acad. Sci. USA 2015, 112, 7599–7604. [Google Scholar] [CrossRef]
- Liu, W.; Kohn, J.; Szwed, S.K.; Pariser, E.; Sepe, S.; Haripal, B.; Oshimori, N.; Marsala, M.; Miyanohara, A.; Lee, R. Human Mutant Huntingtin Disrupts Vocal Learning in Transgenic Songbirds. Nat. Neurosci. 2015, 18, 1617–1622. [Google Scholar] [CrossRef] [PubMed]
- Molnár, M.; Lázár, B.; Sztán, N.; Végi, B.; Drobnyák, Á.; Tóth, R.; Liptói, K.; Marosán, M.; Gócza, E.; Nandi, S.; et al. Investigation of the Guinea Fowl and Domestic Fowl Hybrids as Potential Surrogate Hosts for Avian Cryopreservation Programmes. Sci. Rep. 2019, 9, 14284. [Google Scholar] [CrossRef]
- Ballantyne, M.; Taylor, L.; Hu, T.; Meunier, D.; Nandi, S.; Sherman, A.; Flack, B.; Henshall, J.M.; Hawken, R.J.; McGrew, M.J. Avian Primordial Germ Cells Are Bipotent for Male or Female Gametogenesis. Front. Cell Dev. Biol. 2021, 9, 726827. [Google Scholar] [CrossRef]
- Ballantyne, M.; Woodcock, M.; Doddamani, D.; Hu, T.; Taylor, L.; Hawken, R.J.; McGrew, M.J. Direct Allele Introgression into Pure Chicken Breeds Using Sire Dam Surrogate (SDS) Mating. Nat. Commun. 2021, 12, 659. [Google Scholar] [CrossRef]
- Meng, S.; Miao, A.; Wu, S.; Du, X.; Gao, F. Genetically Modified Chickens as Bioreactors for Protein-Based Drugs. Front. Genome Ed. 2025, 6, 1522837. [Google Scholar] [CrossRef] [PubMed]
- Woodfint, R.M.; Hamlin, E.; Lee, K. Avian Bioreactor Systems: A Review. Mol. Biotechnol. 2018, 60, 975–983. [Google Scholar] [CrossRef]
- Kwon, M.S.; Koo, B.C.; Choi, B.R.; Park, Y.; Lee, Y.M.; Suh, H.S.; Park, Y.S.; Lee, H.T.; Kim, J.; Roh, J.Y.; et al. Generation of Transgenic Chickens That Produce Bioactive Human Granulocyte-colony Stimulating Factor. Mol. Reprod. Dev. 2008, 75, 1120–1126. [Google Scholar] [CrossRef]
- Lee, S.H.; Gupta, M.K.; Ho, Y.T.; Kim, T.; Lee, H.T. Transgenic Chickens Expressing Human Urokinase-Type Plasminogen Activator. Poult. Sci. 2013, 92, 2396–2403. [Google Scholar] [CrossRef]
- Kawabe, Y.; Hayashida, Y.; Numata, K.; Harada, S.; Hayashida, Y.; Ito, A.; Kamihira, M. Oral Immunotherapy for Pollen Allergy Using T-Cell Epitope-Containing Egg White Derived from Genetically Manipulated Chickens. PLoS ONE 2012, 7, e48512. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Lee, H.G.; Moon, J.K.; Lee, H.J.; Yoon, J.W.; Yun, B.N.R.; Kang, S.; Kim, J.; Kim, H.; Han, J.Y.; et al. Deposition of Bioactive Human Epidermal Growth Factor in the Egg White of Transgenic Hens Using an Oviduct-specific Minisynthetic Promoter. FASEB J. 2015, 29, 2386–2396. [Google Scholar] [CrossRef]
- Kwon, M.S.; Koo, B.C.; Kim, D.; Nam, Y.H.; Cui, X.-S.; Kim, N.-H.; Kim, T. Generation of Transgenic Chickens Expressing the Human Erythropoietin (hEPO) Gene in an Oviduct-Specific Manner: Production of Transgenic Chicken Eggs Containing Human Erythropoietin in Egg Whites. PLoS ONE 2018, 13, e0194721. [Google Scholar] [CrossRef]
- Liu, T.; Wu, H.; Cao, D.; Li, Q.; Zhang, Y.; Li, N.; Hu, X. Oviduct-Specific Expression of Human Neutrophil Defensin 4 in Lentivirally Generated Transgenic Chickens. PLoS ONE 2015, 10, e0127922. [Google Scholar] [CrossRef]
- Oishi, I.; Yoshii, K.; Miyahara, D.; Tagami, T. Efficient Production of Human Interferon Beta in the White of Eggs from Ovalbumin Gene–Targeted Hens. Sci. Rep. 2018, 8, 10203. [Google Scholar] [CrossRef]
- Yoo, E.; Choi, H.J.; Han, J.Y. Enhanced Activation of Signaling Pathway by Recombinant Human Adiponectin from Genome-Edited Chickens. J. Biotechnol. 2024, 395, 95–99. [Google Scholar] [CrossRef]
- Kim, Y.M.; Park, J.S.; Choi, H.J.; Jung, K.M.; Lee, K.Y.; Shim, J.H.; Park, K.J.; Han, J.Y. Efficient Production of Recombinant Human Adiponectin in Egg White Using Genome Edited Chickens. Front. Nutr. 2023, 9, 1068558. [Google Scholar] [CrossRef] [PubMed]

| Pathway | Key Ligands | Key Effectors | Principal Functions in PGCs |
|---|---|---|---|
| FGF/MAPK | FGF2 | p-ERK1/2 | Mitogenesis, Proliferation |
| PI3K/AKT/mTOR | Insulin, IGF-1, FGF2 | p-AKT, p-mTOR | Survival, Anti-apoptosis, Metabolism, Proliferation |
| Activin/SMAD | Activin A | p-SMAD2/3 | Maintenance of Self-Renewal |
| BMP/SMAD | BMP4 | p-SMAD1/5/8 | Germline Specification, Proliferation (non-clonal) |
| WNT | Wnt proteins | β-catenin | Modulation of Self-Renewal, Germline Specification |
| Method | Protocol Specifics | Efficiency | References |
|---|---|---|---|
| Electroporation | With DMSO | 80% | [84] |
| Electroporation | Without DMSO | 26.5% | [84] |
| Electroporation | Percoll density gradients purified | 16.6% | [85] |
| Electroporation | Unpurified | 2.3% | [85] |
| Electroporation | Percoll density gradient purified | 75.8% | [44] |
| Lipofection | Ammonium chloride-potassium | 35.2% | [44] |
| Electroporation | Lonza system | 71.13% | [86] |
| Lipofection | Lipofectamine™ 3000 | 1.38% | [86] |
| Lipofection | Transposon vector | 67% | [87] |
| Lipofection | Heparin-free PGC medium | 64.48% | [88] |
| Lipofection | Opti-MEM | 19.56% | [88] |
| Lipofection | KO-DMEM | 23.98% | [88] |
| Lipofection | GFP plasmid | <5% | [89] |
| Electroporation | GFP plasmid | 80% | [89] |
| Adenovirus | ADV-GFP plasmid | 83% | [89] |
| Mediator | Method | Target Gene | Purpose | References |
|---|---|---|---|---|
| Embryo cells | lentiviral vector | influenza A virus polymerase | Resist to IVA | [161] |
| Embryo cells | lentiviral vector | 3D8 scFv | Resist to IVA | [162,163] |
| PGCs | CRISPR/Cas9 | ANP32A | Resist to IVA | [164] |
| PGCs | CRISPR/Cas9 | chNHE1 W38 | Resist to ALV-J | [166] |
| PGCs | CRISPR/Cas9 | chNHE1 W38 | Resist to ALV-J | [167] |
| PGCs | CRISPR/Cas9, Tol2 Transposon | ICP4 | Resist to MDV | [170] |
| Mediator | Method | Target Gene | Purpose | References |
|---|---|---|---|---|
| Skeletal muscle | Adenovirus vector, CRISPR/Cas9 | MSTN | Muscle production | [174] |
| Embryo cells | Adenovirus vector, CRISPR/Cas9 | MSTN | Muscle production | [175] |
| Sperm | Lentiviral vector | MSTN, ACVR2A, ACVR2B | Muscle production | [176] |
| PGCs | CRISPR/Cas9 | G0S2 | Reduction in fat deposition | [177] |
| PGCs | CRISPR/Cas9 | OVA, OVM | Egg production | [125] |
| PGCs | CRISPR/Cas9 | AR | Sex determination | [179] |
| PGCs | CRISPR/Cas9 | DMRT1 | Sex determination | [180] |
| PGCs | CRISPR/Cas9 | miR-2954 | Sex differentiation | [181] |
| Mediator | Method | Target Gene | Purpose | References |
|---|---|---|---|---|
| PGCs | phiC31 integrase | Ig H | Model for disease analysis | [182] |
| PGCs | phiC31 integrase | Ig L | Model for disease analysis | [183] |
| PGCs | phiC31 integrase | SpCas9 | Model for genome editing | [184] |
| PGCs | CRISPR/Cas9 | NTN1 | Model for disease analysis | [185] |
| PGCs | CRISPR/Cas9 | PRDM14 | Model for gene functional analysis | [186] |
| PGCs | TALEN, PiggyBac transposons | PRDM14 | Model for gene functional analysis | [187] |
| Embryo cells | Lentiviral vectors | CREB | Model for disease analysis | [188] |
| Embryo cells | Lentiviral vectors | HTT | Model for disease analysis | [189] |
| PGCs | TALEN | DDX4 | Model for sterility | [120] |
| PGCs | CRISPR/Cas9 | DAZL, DOW, FRZ | Model for sterility | [192] |
| PGCs | CRISPR/Cas9 | DDX4 | Model for sterility | [148] |
| Mediator | Method | Target Proteins | Purpose | References |
|---|---|---|---|---|
| Embryo cells | Retrovirus vector | scFv-Fc | Bioreactor | [104] |
| Embryo cells | Retrovirus vector | hG-CSF | Bioreactor | [195] |
| Embryo cells | Retrovirus vector | huPA | Bioreactor | [196] |
| Embryo cells | Retrovirus vector | 7Crp | Bioreactor | [197] |
| PGCs | PiggyBac transposons | EGF | Bioreactor | [198] |
| Embryo cells | Lentiviral vectors | hEPO | Bioreactor | [199] |
| Embryo cells | Lentiviral vectors | HNP4 | Bioreactor | [200] |
| PGCs | CRISPR/Cas9 | hIFN-β | Bioreactor | [201] |
| PGCs | CRISPR/Cas9 | ADPN | Bioreactor | [203] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Shi, N.; Yao, H.; Li, J.; Wang, Y.; Zhang, J. Genome Editing in the Chicken: From PGC-Mediated Germline Transmission to Advanced Applications. Int. J. Mol. Sci. 2025, 26, 9426. https://doi.org/10.3390/ijms26199426
He J, Shi N, Yao H, Li J, Wang Y, Zhang J. Genome Editing in the Chicken: From PGC-Mediated Germline Transmission to Advanced Applications. International Journal of Molecular Sciences. 2025; 26(19):9426. https://doi.org/10.3390/ijms26199426
Chicago/Turabian StyleHe, Jiliang, Ningkun Shi, Hongqin Yao, Juan Li, Yajun Wang, and Jiannan Zhang. 2025. "Genome Editing in the Chicken: From PGC-Mediated Germline Transmission to Advanced Applications" International Journal of Molecular Sciences 26, no. 19: 9426. https://doi.org/10.3390/ijms26199426
APA StyleHe, J., Shi, N., Yao, H., Li, J., Wang, Y., & Zhang, J. (2025). Genome Editing in the Chicken: From PGC-Mediated Germline Transmission to Advanced Applications. International Journal of Molecular Sciences, 26(19), 9426. https://doi.org/10.3390/ijms26199426







