Gene Therapy Strategies for the Treatment of Bestrophinopathies
Abstract
1. Introduction
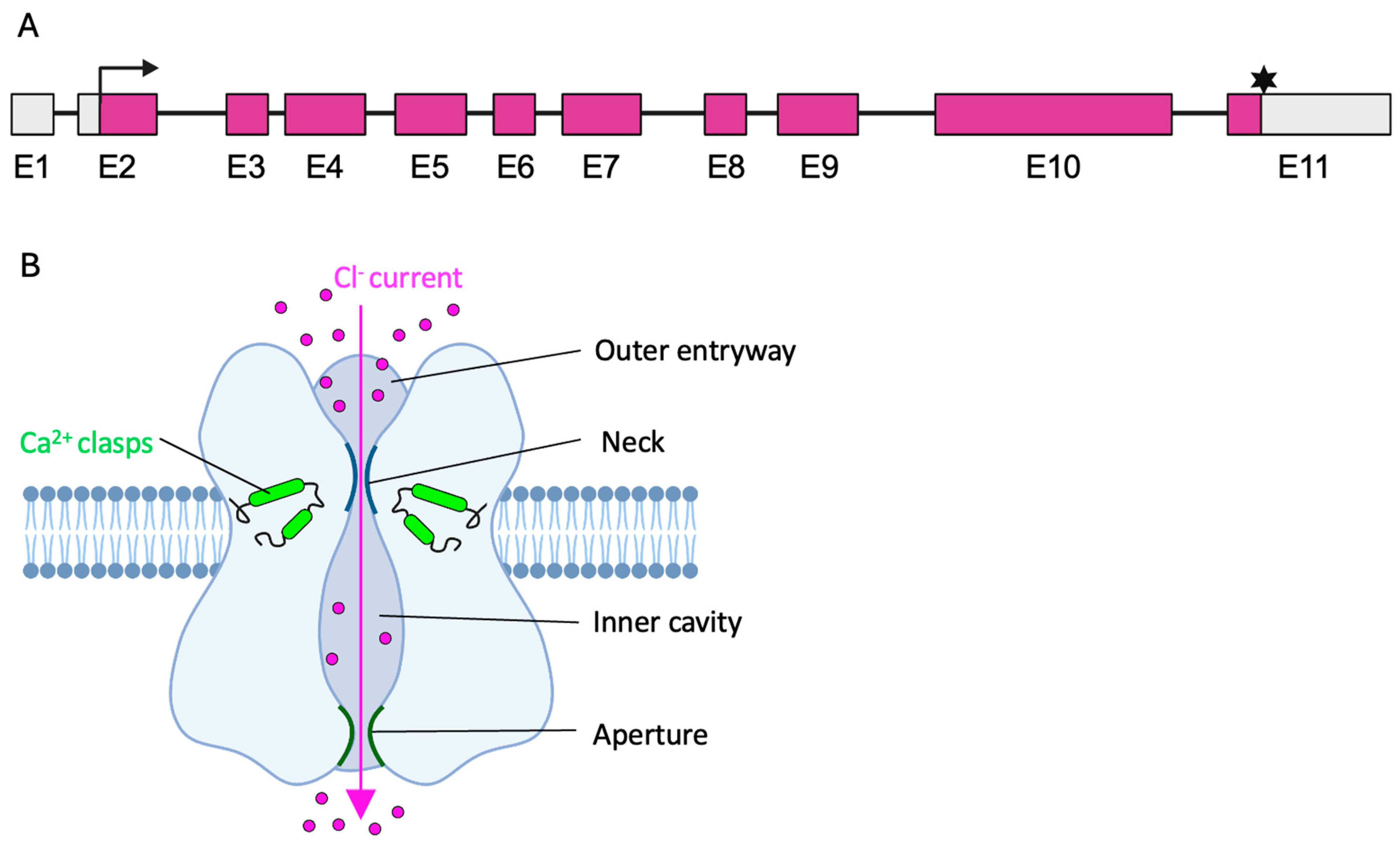
2. The BEST1 Gene and the Bestrophin-1 Channel
3. The Clinical Spectrum of Bestrophinopathies
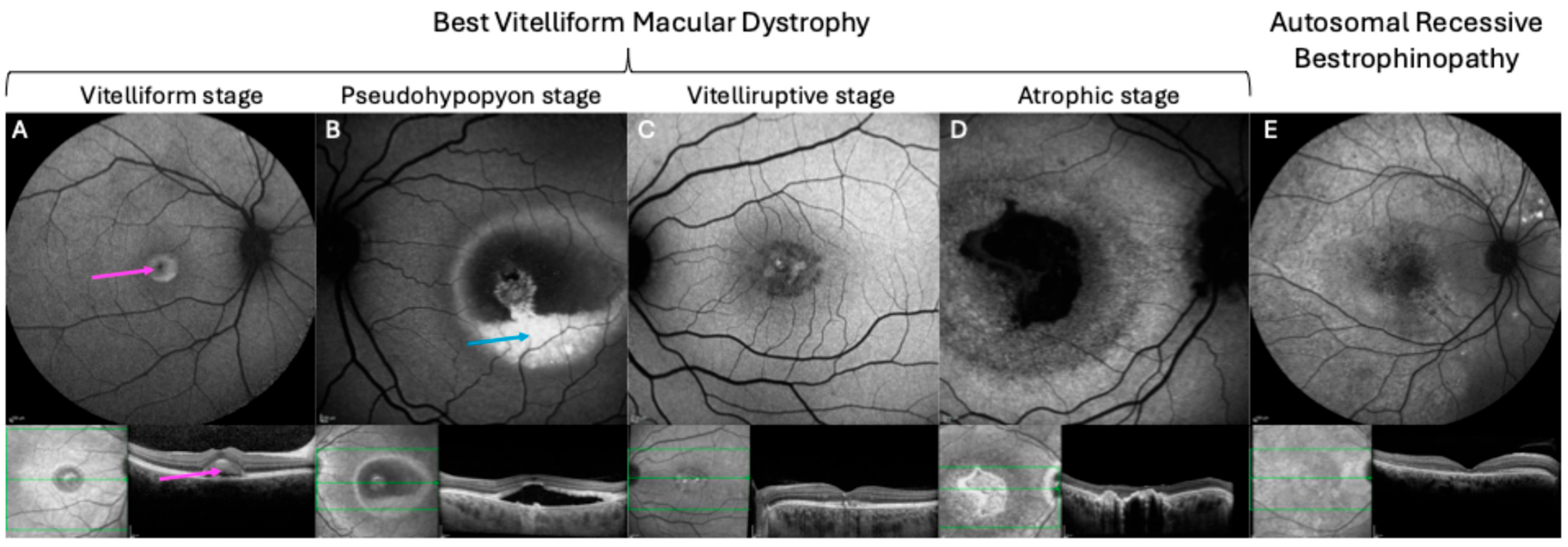
3.1. Best Vitelliform Macular Dystrophy
3.2. Adult Vitelliform Macular Dystrophy
3.3. Autosomal Recessive Bestrophinopathy
3.4. Autosomal Dominant Vitreoretinochoroidopathy
3.5. Retinitis Pigmentosa
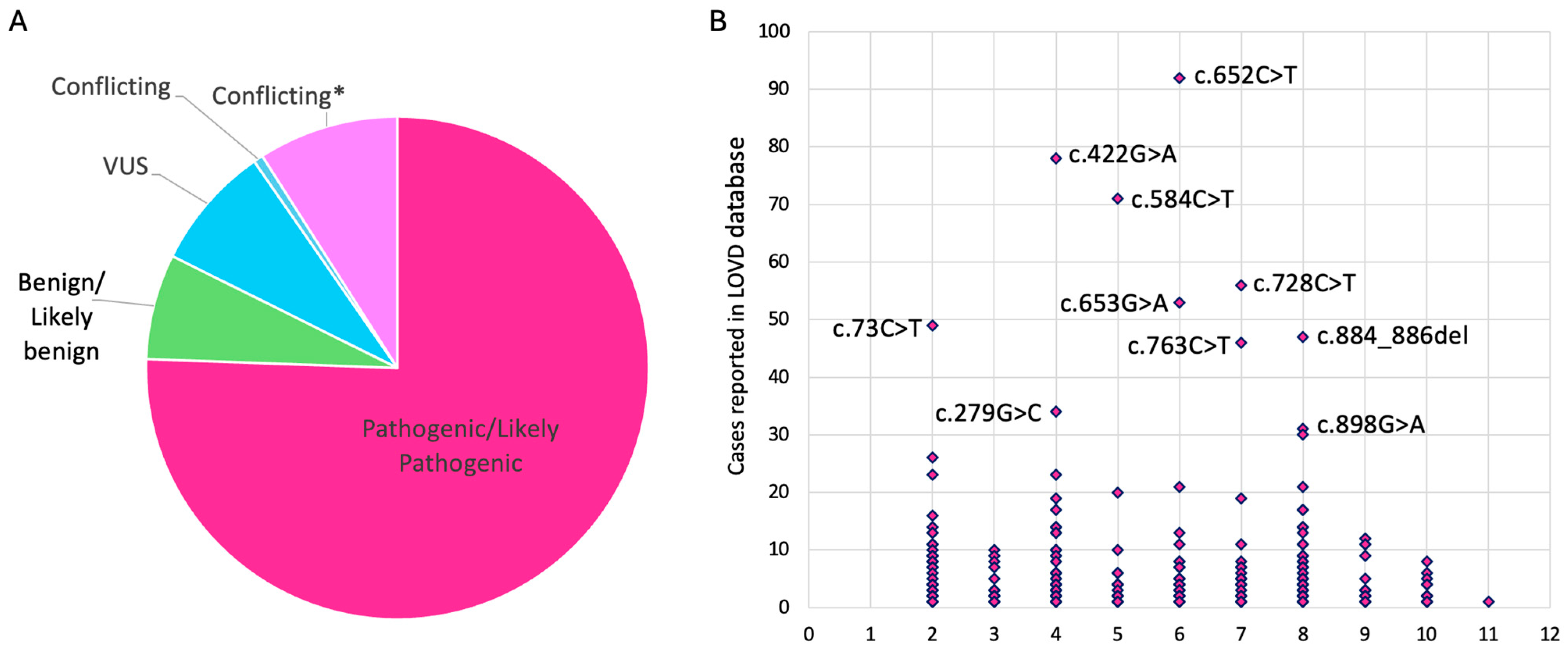
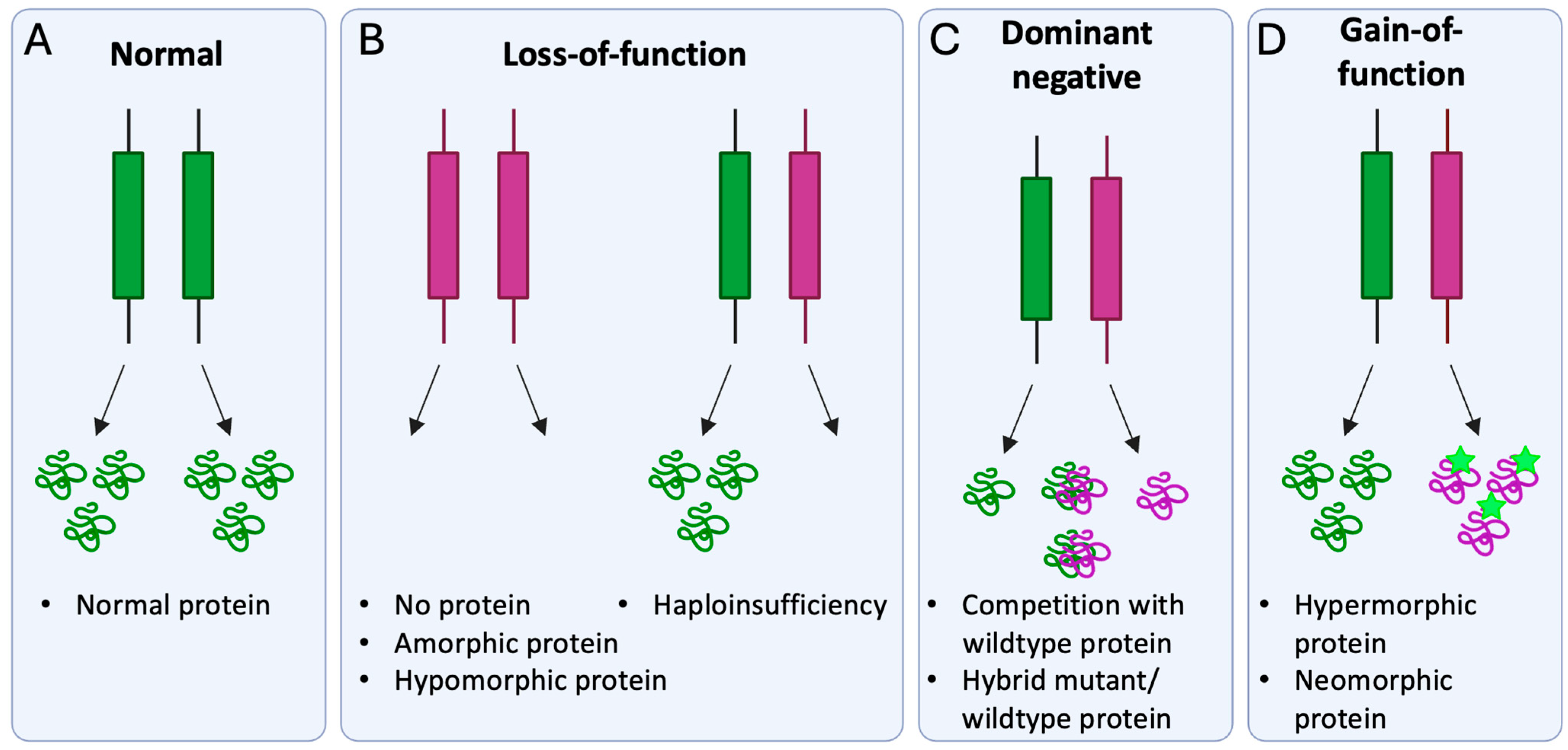
4. Molecular Disease Mechanism of Pathogenic BEST1 Variants
5. Gene Augmentation for the Treatment of Bestrophinopathies
6. CRISPR-Based Treatment Strategies for Pathogenic BEST1 Variants
6.1. Specific Silencing of Disease-Causing Allele
6.1.1. CRISPR/Cas9-Mediated Gene Silencing
6.1.2. Targeting of Allele-Specific Single Nucleotide Polymorphisms
6.2. Ablation-And-Replace Strategy
6.3. Precise Correction of Disease-Causing Variant
6.3.1. Homology-Directed Repair
6.3.2. Homology-Independent Targeted Integration
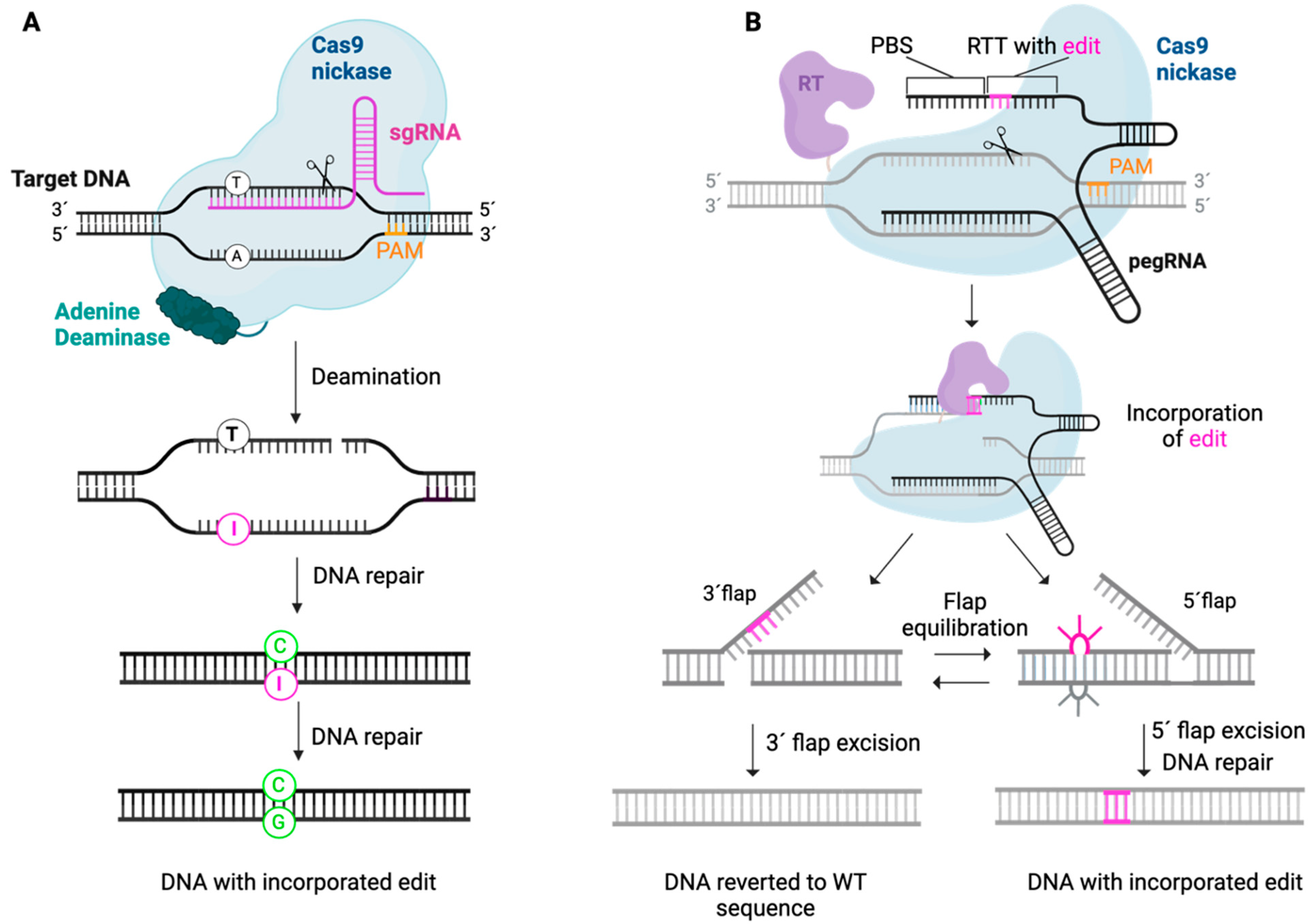
6.3.3. Base Editing
6.3.4. Prime Editing
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Amato, A.; Arrigo, A.; Aragona, E.; Manitto, M.P.; Saladino, A.; Bandello, F.; Battaglia Parodi, M. Gene Therapy in Inherited Retinal Diseases: An Update on Current State of the Art. Front. Med. 2021, 8, 750586. [Google Scholar] [CrossRef]
- Johnson, A.A.; Guziewicz, K.E.; Lee, C.J.; Kalathur, R.C.; Pulido, J.S.; Marmorstein, L.Y.; Marmorstein, A.D. Bestrophin 1 and retinal disease. Prog. Retin. Eye Res. 2017, 58, 45–69. [Google Scholar] [CrossRef]
- Chung, D.C.; Lee, V.; Maguire, A.M. Recent advances in ocular gene therapy. Curr. Opin. Ophthalmol. 2009, 20, 377–381. [Google Scholar] [CrossRef]
- Maguire, A.M.; Russell, S.; Chung, D.C.; Yu, Z.-F.; Tillman, A.; Drack, A.V.; Simonelli, F.; Leroy, B.P.; Reape, K.Z.; High, K.A.; et al. Durability of Voretigene Neparvovec for Biallelic RPE65-Mediated Inherited Retinal Disease: Phase 3 Results at 3 and 4 Years. Ophthalmology 2021, 128, 1460–1468. [Google Scholar] [CrossRef]
- Marmorstein, A.D.; Marmorstein, L.Y.; Rayborn, M.; Wang, X.; Hollyfield, J.G.; Petrukhin, K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2000, 97, 12758–12763. [Google Scholar] [CrossRef]
- Kane Dickson, V.; Pedi, L.; Long, S.B. Structure and insights into the function of a Ca2+-activated Cl− channel. Nature 2014, 516, 213–218. [Google Scholar] [CrossRef]
- Marmorstein, A.D.; Kinnick, T.R.; Stanton, J.B.; Johnson, A.A.; Lynch, R.M.; Marmorstein, L.Y. Bestrophin-1 influences transepithelial electrical properties and Ca2+ signaling in human retinal pigment epithelium. Mol. Vis. 2015, 21, 347–359. [Google Scholar]
- Owji, A.P.; Wang, J.; Kittredge, A.; Clark, Z.; Zhang, Y.; Hendrickson, W.A.; Yang, T. Structures and gating mechanisms of human bestrophin anion channels. Nat. Commun. 2022, 13, 3836. [Google Scholar] [CrossRef]
- Ji, C.; Kittredge, A.; Hopiavuori, A.; Ward, N.; Chen, S.; Fukuda, Y.; Zhang, Y.; Yang, T. Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations. Commun. Biol. 2019, 2, 240. [Google Scholar] [CrossRef]
- Nachtigal, A.L.; Milenkovic, A.; Brandl, C.; Schulz, H.L.; Duerr, L.M.J.; Lang, G.E.; Reiff, C.; Herrmann, P.; Kellner, U.; Weber, B.H.F. Mutation-Dependent Pathomechanisms Determine the Phenotype in the Bestrophinopathies. Int. J. Mol. Sci. 2020, 21, 1597. [Google Scholar] [CrossRef]
- Casalino, G.; Khan, K.N.; Armengol, M.; Wright, G.; Pontikos, N.; Georgiou, M.; Webster, A.R.; Robson, A.G.; Grewal, P.S.; Michaelides, M. Autosomal Recessive Bestrophinopathy: Clinical Features, Natural History, and Genetic Findings in Preparation for Clinical Trials. Ophthalmology 2021, 128, 706–718. [Google Scholar] [CrossRef]
- Strauß, O.; Müller, C.; Reichhart, N.; Tamm, E.R.; Gomez, N.M. The role of bestrophin-1 in intracellular Ca2+ signaling. Adv. Exp. Med. Biol. 2014, 801, 113–119. [Google Scholar] [CrossRef]
- Gómez, N.M.; Tamm, E.R.; Strauβ, O. Role of bestrophin-1 in store-operated calcium entry in retinal pigment epithelium. Pflugers Arch. 2013, 465, 481–495. [Google Scholar] [CrossRef]
- Fischmeister, R.; Hartzell, H.C. Volume sensitivity of the bestrophin family of chloride channels. J. Physiol. 2005, 562, 477–491. [Google Scholar] [CrossRef]
- Rahman, N.; Georgiou, M.; Khan, K.N.; Michaelides, M. Macular dystrophies: Clinical and imaging features, molecular genetics and therapeutic options. Br. J. Ophthalmol. 2020, 104, 451–460. [Google Scholar] [CrossRef]
- Krämer, F.; White, K.; Pauleikhoff, D.; Gehrig, A.; Passmore, L.; Rivera, A.; Rudolph, G.; Kellner, U.; Andrassi, M.; Lorenz, B.; et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur. J. Hum. Genet. 2000, 8, 286–292. [Google Scholar] [CrossRef]
- Burgess, R.; Millar, I.D.; Leroy, B.P.; Urquhart, J.E.; Fearon, I.M.; De Baere, E.; Brown, P.D.; Robson, A.G.; Wright, G.A.; Kestelyn, P.; et al. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am. J. Hum. Genet. 2008, 82, 19–31. [Google Scholar] [CrossRef]
- Davidson, A.E.; Millar, I.D.; Urquhart, J.E.; Burgess-Mullan, R.; Shweikh, Y.; Parry, N.; O’Sullivan, J.; Maher, G.J.; McKibbin, M.; Downes, S.M.; et al. Missense Mutations in a Retinal Pigment Epithelium Protein, Bestrophin-1, Cause Retinitis Pigmentosa. Am. J. Hum. Genet. 2009, 85, 581–592. [Google Scholar] [CrossRef]
- Nordström, S. Hereditary macular degeneration—A population survey in the country of Vsterbotten, Sweden. Hereditas 1974, 78, 41–62. [Google Scholar] [CrossRef]
- Bitner, H.; Schatz, P.; Mizrahi-Meissonnier, L.; Sharon, D.; Rosenberg, T. Frequency, genotype, and clinical spectrum of best vitelliform macular dystrophy: Data from a national center in Denmark. Am. J. Ophthalmol. 2012, 154, 403–412.e404. [Google Scholar] [CrossRef]
- Boon, C.J.; Klevering, B.J.; Leroy, B.P.; Hoyng, C.B.; Keunen, J.E.; den Hollander, A.I. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog. Retin. Eye Res. 2009, 28, 187–205. [Google Scholar] [CrossRef]
- Singh Grewal, S.; Smith, J.J.; Carr, A.F. Bestrophinopathies: Perspectives on clinical disease, Bestrophin-1 function and developing therapies. Ther. Adv. Ophthalmol. 2021, 13, 2515841421997191. [Google Scholar] [CrossRef]
- Marmorstein, A.D.; Cross, H.E.; Peachey, N.S. Functional roles of bestrophins in ocular epithelia. Prog. Retin. Eye Res. 2009, 28, 206–226. [Google Scholar] [CrossRef]
- Renner, A.B.; Tillack, H.; Kraus, H.; Kohl, S.; Wissinger, B.; Mohr, N.; Weber, B.H.; Kellner, U.; Foerster, M.H. Morphology and functional characteristics in adult vitelliform macular dystrophy. Retina 2004, 24, 929–939. [Google Scholar] [CrossRef]
- Fishman, G.A.; Baca, W.; Alexander, K.R.; Derlacki, D.J.; Glenn, A.M.; Viana, M. Visual acuity in patients with best vitelliform macular dystrophy. Ophthalmology 1993, 100, 1665–1670. [Google Scholar] [CrossRef]
- Arora, R.; Khan, K.; Kasilian, M.L.; Strauss, R.W.; Holder, G.E.; Robson, A.G.; Thompson, D.A.; Moore, A.T.; Michaelides, M. Unilateral BEST1-Associated Retinopathy. Am. J. Ophthalmol. 2016, 169, 24–32. [Google Scholar] [CrossRef]
- Kaden, T.R.; Tan, A.C.; Feiner, L.; Freund, K.B. UNILATERAL BEST DISEASE: A CASE REPORT. Retin. Cases Brief. Rep. 2017, 11 (Suppl. S1), S191–S196. [Google Scholar] [CrossRef]
- Querques, G.; Regenbogen, M.; Soubrane, G.; Souied, E.H. High-resolution spectral domain optical coherence tomography findings in multifocal vitelliform macular dystrophy. Surv. Ophthalmol. 2009, 54, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Wittström, E.; Ekvall, S.; Schatz, P.; Bondeson, M.L.; Ponjavic, V.; Andréasson, S. Morphological and functional changes in multifocal vitelliform retinopathy and biallelic mutations in BEST1. Ophthalmic Genet. 2011, 32, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Coussa, R.G.; Binkley, E.M.; Wilkinson, M.E.; Andorf, J.L.; Tucker, B.A.; Mullins, R.F.; Sohn, E.H.; Yannuzzi, L.A.; Stone, E.M.; Han, I.C. Predominance of hyperopia in autosomal dominant Best vitelliform macular dystrophy. Br. J. Ophthalmol. 2022, 106, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Menchini, U.; Giacomelli, G.; Cappelli, S.; Giansanti, F.; Romani, A. Photodynamic Therapy in Adult-Onset Vitelliform Macular Dystrophy Misdiagnosed as Choroidal Neovascularization. Arch. Ophthalmol. 2002, 120, 1761–1763. [Google Scholar]
- Shah, M.; Broadgate, S.; Shanks, M.; Clouston, P.; Yu, J.; MacLaren, R.E.; Németh, A.H.; Halford, S.; Downes, S.M. Association of Clinical and Genetic Heterogeneity With BEST1 Sequence Variations. JAMA Ophthalmol. 2020, 138, 544–551. [Google Scholar] [CrossRef]
- Gerth, C.; Zawadzki, R.J.; Werner, J.S.; Héon, E. Detailed analysis of retinal function and morphology in a patient with autosomal recessive bestrophinopathy (ARB). Doc. Ophthalmol. 2009, 118, 239–246. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; McAlister, C.; Vandenhoven, C.; Héon, E. BEST1-related autosomal dominant vitreoretinochoroidopathy: A degenerative disease with a range of developmental ocular anomalies. Eye 2011, 25, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, S.J.; Goldberg, M.F.; Orth, D.H.; Fishman, G.A.; Tessler, H.; Mizuno, K. Autosomal dominant vitreoretinochoroidopathy. Arch. Ophthalmol. 1982, 100, 272–278. [Google Scholar] [CrossRef]
- Yardley, J.; Leroy, B.P.; Hart-Holden, N.; Lafaut, B.A.; Loeys, B.; Messiaen, L.M.; Perveen, R.; Reddy, M.A.; Bhattacharya, S.S.; Traboulsi, E.; et al. Mutations of VMD2 splicing regulators cause nanophthalmos and autosomal dominant vitreoretinochoroidopathy (ADVIRC). Investig. Ophthalmol. Vis. Sci. 2004, 45, 3683–3689. [Google Scholar] [CrossRef]
- Burgess, R.; MacLaren, R.E.; Davidson, A.E.; Urquhart, J.E.; Holder, G.E.; Robson, A.G.; Moore, A.T.; Keefe, R.O.; Black, G.C.; Manson, F.D. ADVIRC is caused by distinct mutations in BEST1 that alter pre-mRNA splicing. J. Med. Genet. 2009, 46, 620–625. [Google Scholar] [CrossRef]
- Traboulsi, E. Genetic Diseases of the Eye, 2nd ed.; Oxford University Press: New York, NY, USA, 2012. [Google Scholar]
- Boulanger-Scemama, E.; Sahel, J.A.; Mohand-Said, S.; Antonio, A.; Condroyer, C.; Zeitz, C.; Audo, I. AUTOSOMAL DOMINANT VITREORETINOCHOROIDOPATHY: When Molecular Genetic Testing Helps Clinical Diagnosis. Retina 2019, 39, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Dalvin, L.A.; Abou Chehade, J.E.; Chiang, J.; Fuchs, J.; Iezzi, R.; Marmorstein, A.D. Retinitis pigmentosa associated with a mutation in BEST1. Am. J. Ophthalmol. Case Rep. 2016, 2, 11–17. [Google Scholar] [CrossRef]
- Backwell, L.; Marsh, J.A. Diverse Molecular Mechanisms Underlying Pathogenic Protein Mutations: Beyond the Loss-of-Function Paradigm. Annu. Rev. Genom. Hum. Genet. 2022, 23, 475–498. [Google Scholar] [CrossRef]
- Llavona, P.; Pinelli, M.; Mutarelli, M.; Marwah, V.S.; Schimpf-Linzenbold, S.; Thaler, S.; Yoeruek, E.; Vetter, J.; Kohl, S.; Wissinger, B. Allelic Expression Imbalance in the Human Retinal Transcriptome and Potential Impact on Inherited Retinal Diseases. Genes. 2017, 8, 283. [Google Scholar] [CrossRef]
- Zhao, Q.; Kong, Y.; Kittredge, A.; Li, Y.; Shen, Y.; Zhang, Y.; Tsang, S.H.; Yang, T. Distinct expression requirements and rescue strategies for BEST1 loss- and gain-of-function mutations. Elife 2021, 10, e67622. [Google Scholar] [CrossRef]
- Yang, T.; Justus, S.; Li, Y.; Tsang, S.H. BEST1: The Best Target for Gene and Cell Therapies. Mol. Ther. 2015, 23, 1805–1809. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Xu, Y.; Kittredge, A.; Ward, N.; Chen, S.; Tsang, S.H.; Yang, T. Patient-specific mutations impair BESTROPHIN1’s essential role in mediating Ca2+-dependent Cl− currents in human RPE. Elife 2017, 6, e29914. [Google Scholar] [CrossRef]
- Guziewicz, K.E.; Cideciyan, A.V.; Beltran, W.A.; Komáromy, A.M.; Dufour, V.L.; Swider, M.; Iwabe, S.; Sumaroka, A.; Kendrick, B.T.; Ruthel, G.; et al. BEST1 gene therapy corrects a diffuse retina-wide microdetachment modulated by light exposure. Proc. Natl. Acad. Sci. USA 2018, 115, E2839–E2848. [Google Scholar] [CrossRef]
- Wood, S.R.; McClements, M.E.; Martinez-Fernandez de la Camara, C.; Patrício, M.I.; Uggenti, C.; Sekaran, S.; Barnard, A.R.; Manson, F.D.; MacLaren, R.E. A Quantitative Chloride Channel Conductance Assay for Efficacy Testing of AAV.BEST1. Hum. Gene Ther. Methods 2019, 30, 44–52. [Google Scholar] [CrossRef]
- Sinha, D.; Steyer, B.; Shahi, P.K.; Mueller, K.P.; Valiauga, R.; Edwards, K.L.; Bacig, C.; Steltzer, S.S.; Srinivasan, S.; Abdeen, A.; et al. Human iPSC Modeling Reveals Mutation-Specific Responses to Gene Therapy in a Genotypically Diverse Dominant Maculopathy. Am. J. Hum. Genet. 2020, 107, 278–292. [Google Scholar] [CrossRef]
- Ji, C.; Li, Y.; Kittredge, A.; Hopiavuori, A.; Ward, N.; Yao, P.; Fukuda, Y.; Zhang, Y.; Tsang, S.H.; Yang, T. Investigation and Restoration of BEST1 Activity in Patient-derived RPEs with Dominant Mutations. Sci. Rep. 2019, 9, 19026. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef]
- Pierce, E.A.; Aleman, T.S.; Jayasundera, K.T.; Ashimatey, B.S.; Kim, K.; Rashid, A.; Jaskolka, M.C.; Myers, R.L.; Lam, B.L.; Bailey, S.T.; et al. Gene Editing for CEP290-Associated Retinal Degeneration. N. Engl. J. Med. 2024, 390, 1972–1984. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143. [Google Scholar] [CrossRef]
- Salman, A.; Song, W.K.; Storm, T.; McClements, M.E.; MacLaren, R.E. CRISPR targeting of SNPs associated with age-related macular degeneration in ARPE-19 cells: A potential model for manipulating the complement system. Gene Ther. 2025, 32, 132–141. [Google Scholar] [CrossRef]
- Tsai, Y.-T.; Wu, W.-H.; Lee, T.-T.; Wu, W.-P.; Xu, C.L.; Park, K.S.; Cui, X.; Justus, S.; Lin, C.-S.; Jauregui, R.; et al. Clustered Regularly Interspaced Short Palindromic Repeats-Based Genome Surgery for the Treatment of Autosomal Dominant Retinitis Pigmentosa. Ophthalmology 2018, 125, 1421–1430. [Google Scholar] [CrossRef]
- Orlans, H.O.; McClements, M.E.; Barnard, A.R.; Martinez-Fernandez de la Camara, C.; MacLaren, R.E. Mirtron-mediated RNA knockdown/replacement therapy for the treatment of dominant retinitis pigmentosa. Nat. Commun. 2021, 12, 4934. [Google Scholar] [CrossRef]
- Sanjurjo-Soriano, C.; Erkilic, N.; Baux, D.; Mamaeva, D.; Hamel, C.P.; Meunier, I.; Roux, A.F.; Kalatzis, V. Genome Editing in Patient iPSCs Corrects the Most Prevalent USH2A Mutations and Reveals Intriguing Mutant mRNA Expression Profiles. Mol. Ther. Methods Clin. Dev. 2020, 17, 156–173. [Google Scholar] [CrossRef]
- Siles, L.; Ruiz-Nogales, S.; Navinés-Ferrer, A.; Méndez-Vendrell, P.; Pomares, E. Efficient correction of ABCA4 variants by CRISPR-Cas9 in hiPSCs derived from Stargardt disease patients. Mol. Ther. Nucleic Acids 2023, 32, 64–79. [Google Scholar] [CrossRef]
- Botto, C.; Rucli, M.; Tekinsoy, M.D.; Pulman, J.; Sahel, J.A.; Dalkara, D. Early and late stage gene therapy interventions for inherited retinal degenerations. Prog. Retin. Eye Res. 2022, 86, 100975. [Google Scholar] [CrossRef]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016, 19, 1–9. [Google Scholar] [CrossRef]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z.; et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.K.; Song, B.; Hwang, G.H.; Bae, S. Current trends in gene recovery mediated by the CRISPR-Cas system. Exp. Mol. Med. 2020, 52, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Jia, R.; Zhao, X.; Zhang, F.; Chen, S.; Yu, S.; Liu, X.; Dou, H.; Feng, X.; Zhang, J.; et al. In vivo genome editing via CRISPR/Cas9-mediated homology-independent targeted integration for Bietti crystalline corneoretinal dystrophy treatment. Nat. Commun. 2024, 15, 3773. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef]
- Hansen, S.; McClements, M.E.; Corydon, T.J.; MacLaren, R.E. Future Perspectives of Prime Editing for the Treatment of Inherited Retinal Diseases. Cells 2023, 12, 440. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Chen, Y.; Zhi, S.; Liu, W.; Wen, J.; Hu, S.; Cao, T.; Sun, H.; Li, Y.; Huang, L.; Liu, Y.; et al. Development of Highly Efficient Dual-AAV Split Adenosine Base Editor for In Vivo Gene Therapy. Small Methods 2020, 4, 2000309. [Google Scholar] [CrossRef]
- Fry, L.E.; McClements, M.E.; MacLaren, R.E. Analysis of Pathogenic Variants Correctable With CRISPR Base Editing Among Patients With Recessive Inherited Retinal Degeneration. JAMA Ophthalmol. 2021, 139, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Kaukonen, M.; McClements, M.E.; MacLaren, R.E. CRISPR DNA Base Editing Strategies for Treating Retinitis Pigmentosa Caused by Mutations in Rhodopsin. Genes 2022, 13, 1327. [Google Scholar] [CrossRef]
- Suh, S.; Choi, E.H.; Leinonen, H.; Foik, A.T.; Newby, G.A.; Yeh, W.H.; Dong, Z.; Kiser, P.D.; Lyon, D.C.; Liu, D.R.; et al. Restoration of visual function in adult mice with an inherited retinal disease via adenine base editing. Nat. Biomed. Eng. 2021, 5, 169–178. [Google Scholar] [CrossRef]
- Muller, A.; Sullivan, J.; Schwarzer, W.; Wang, M.; Park-Windhol, C.; Hasler, P.W.; Janeschitz-Kriegl, L.; Duman, M.; Klingler, B.; Matsell, J.; et al. High-efficiency base editing in the retina in primates and human tissues. Nat. Med. 2025, 31, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Jang, H.; Jo, D.H.; Cho, C.S.; Shin, J.H.; Seo, J.H.; Yu, G.; Gopalappa, R.; Kim, D.; Cho, S.R.; Kim, J.H.; et al. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat. Biomed. Eng. 2022, 6, 181–194. [Google Scholar] [CrossRef]
- Qin, H.; Zhang, W.; Zhang, S.; Feng, Y.; Xu, W.; Qi, J.; Zhang, Q.; Xu, C.; Liu, S.; Zhang, J.; et al. Vision rescue via unconstrained in vivo prime editing in degenerating neural retinas. J. Exp. Med. 2023, 220, e20220776. [Google Scholar] [CrossRef]
- Testa, F.; Maguire, A.M.; Rossi, S.; Pierce, E.A.; Melillo, P.; Marshall, K.; Banfi, S.; Surace, E.M.; Sun, J.; Acerra, C.; et al. Three-year follow-up after unilateral subretinal delivery of adeno-associated virus in patients with Leber congenital Amaurosis type 2. Ophthalmology 2013, 120, 1283–1291. [Google Scholar] [CrossRef]
- Hauswirth, W.W.; Aleman, T.S.; Kaushal, S.; Cideciyan, A.V.; Schwartz, S.B.; Wang, L.; Conlon, T.J.; Boye, S.L.; Flotte, T.R.; Byrne, B.J.; et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: Short-term results of a phase I trial. Hum. Gene Ther. 2008, 19, 979–990. [Google Scholar] [CrossRef]
- Fu, Y.; He, X.; Ma, L.; Gao, X.D.; Liu, P.; Shi, H.; Chai, P.; Ge, S.; Jia, R.; Liu, D.R.; et al. In vivo prime editing rescues photoreceptor degeneration in nonsense mutant retinitis pigmentosa. Nat. Commun. 2025, 16, 2394. [Google Scholar] [CrossRef]
- Arsenijevic, Y.; Berger, A.; Udry, F.; Kostic, C. Lentiviral Vectors for Ocular Gene Therapy. Pharmaceutics 2022, 14, 1605. [Google Scholar] [CrossRef]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265.e216. [Google Scholar] [CrossRef]
- Haldrup, J.; Andersen, S.; Labial, A.R.L.; Wolff, J.H.; Frandsen, F.P.; Skov, T.W.; Rovsing, A.B.; Nielsen, I.; Jakobsen, T.S.; Askou, A.L.; et al. Engineered lentivirus-derived nanoparticles (LVNPs) for delivery of CRISPR/Cas ribonucleoprotein complexes supporting base editing, prime editing and in vivo gene modification. Nucleic Acids Res. 2023, 51, 10059–10074. [Google Scholar] [CrossRef] [PubMed]
- Hołubowicz, R.; Du, S.W.; Felgner, J.; Smidak, R.; Choi, E.H.; Palczewska, G.; Menezes, C.R.; Dong, Z.; Gao, F.; Medani, O.; et al. Safer and efficient base editing and prime editing via ribonucleoproteins delivered through optimized lipid-nanoparticle formulations. Nat. Biomed. Eng. 2025, 9, 57–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Zhan, W.; Gallagher, T.L.; Gao, G. Recombinant adeno-associated virus as a delivery platform for ocular gene therapy: A comprehensive review. Mol. Ther. 2024, 32, 4185–4207. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Wu, Z. Ocular delivery of CRISPR/Cas genome editing components for treatment of eye diseases. Adv. Drug Deliv. Rev. 2021, 168, 181–195. [Google Scholar] [CrossRef]


| Gene Therapy Strategy | Delivery | Clinical Condition | Inheritance | Disease Model | Outcome | Ref |
|---|---|---|---|---|---|---|
| Gene augmentation | Baculovirus vector | ARB | Autosomal recessive | iPSC-RPE cells | Rescue of Ca2+-dependent Cl− current (whole-cell patch clamp recording) | [46] |
| Gene augmentation | AAV2 vector | ARB (canine model) | Autosomal recessive | Canine BEST1 disease model | Revision of subretinal detachment and microdetachment. Correction of PR/RPE interface | [47] |
| Gene augmentation | Lentiviral vector | ARB | Autosomal recessive | iPSC-RPE cells | Increased levels of BEST1 protein. Restoration of BEST1 calcium-activated chloride channel activity. Improvement of RPE function (Rhodopsin degradation) | [49] |
| Gene augmentation | Lentiviral vector | Best disease | Autosomal dominant | iPSC-RPE cells | Increased levels of BEST1 protein. Restoration of BEST1 calcium-activated chloride channel activity. Improvement of RPE function (Rhodopsin degradation) | [49] |
| CRISPR/Cas9-mediated knock out of mutant allele | Lentiviral vector | Best disease | Autosomal dominant | iPSC-RPE cells | Increased levels of BEST1 protein. Restoration of BEST1 calcium-activated chloride channel activity | [49] |
| Gene augmentation | AAV2 vector/ Baculovirus vector | BVMD | Autosomal dominant (loss-of-function) | iPSC-RPE | Rescue of Ca2+-dependent Cl− current (whole-cell patch clamp recording) | [50] |
| CRISPR/dCas9-mediated knock down of both alleles + gene augmentation | (i) Baculavirus vector expressing dCas9- KRAB-MeCP2) (ii) Baculovirus vector expressing wildtype BEST1 | Autosomal dominant (gain-of-function) | hPSC-RPE H1-iCas9 cells * | Rescue of Ca2+ dependent Cl− current (whole-cell patch clamp recording) | [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haldrup, S.B.; McClements, M.E.; Cehajic-Kapetanovic, J.; Corydon, T.J.; MacLaren, R.E. Gene Therapy Strategies for the Treatment of Bestrophinopathies. Int. J. Mol. Sci. 2025, 26, 9421. https://doi.org/10.3390/ijms26199421
Haldrup SB, McClements ME, Cehajic-Kapetanovic J, Corydon TJ, MacLaren RE. Gene Therapy Strategies for the Treatment of Bestrophinopathies. International Journal of Molecular Sciences. 2025; 26(19):9421. https://doi.org/10.3390/ijms26199421
Chicago/Turabian StyleHaldrup, Silja B., Michelle E. McClements, Jasmina Cehajic-Kapetanovic, Thomas J. Corydon, and Robert E. MacLaren. 2025. "Gene Therapy Strategies for the Treatment of Bestrophinopathies" International Journal of Molecular Sciences 26, no. 19: 9421. https://doi.org/10.3390/ijms26199421
APA StyleHaldrup, S. B., McClements, M. E., Cehajic-Kapetanovic, J., Corydon, T. J., & MacLaren, R. E. (2025). Gene Therapy Strategies for the Treatment of Bestrophinopathies. International Journal of Molecular Sciences, 26(19), 9421. https://doi.org/10.3390/ijms26199421






