Transcriptional Regulation of WTAP Isoforms by NF-κB Signaling in Human Monocytes
Abstract
1. Introduction
2. Results
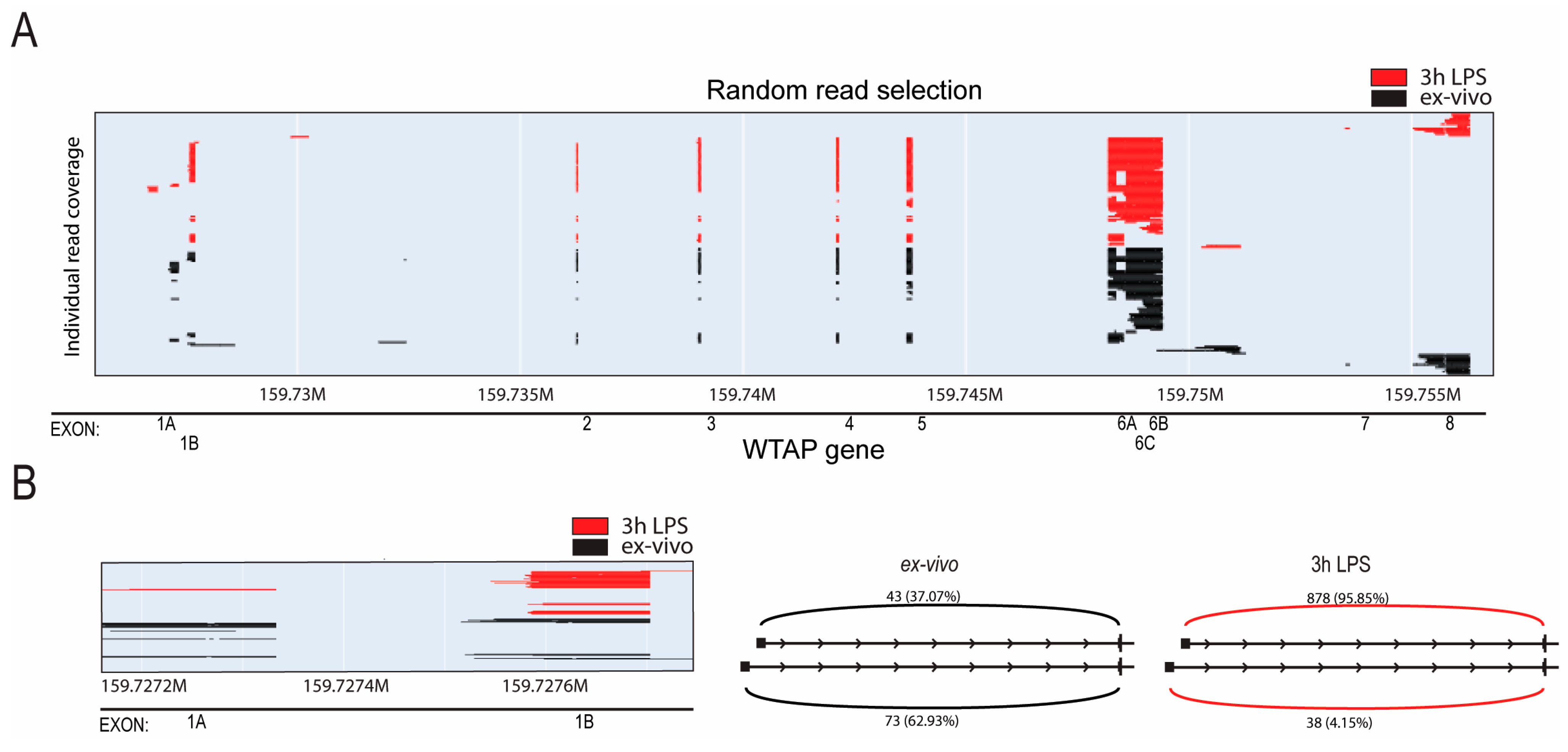
2.1. Alternative Promoter Utilization of WTAP in Human CD14+ Monocyte Activation
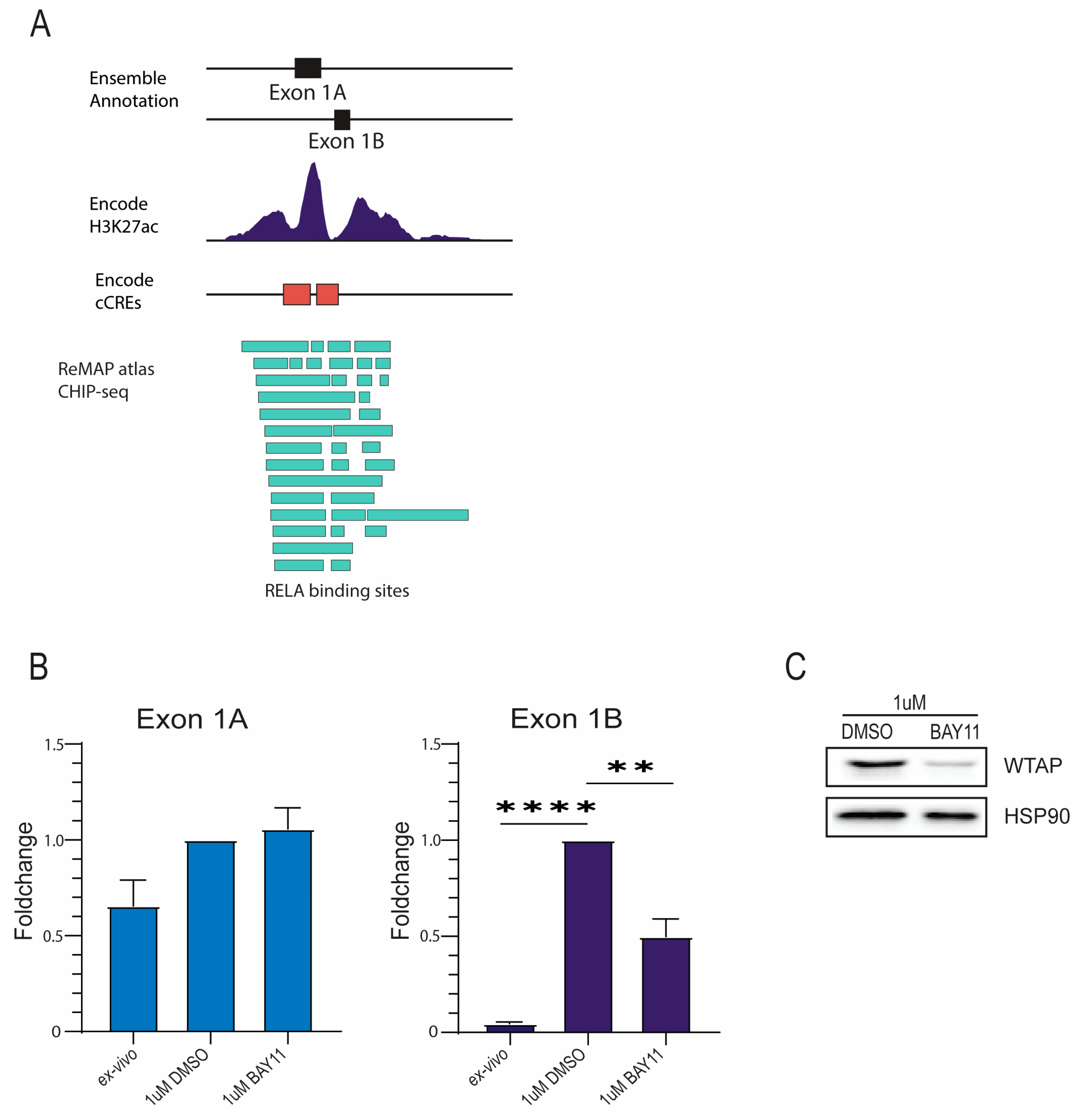
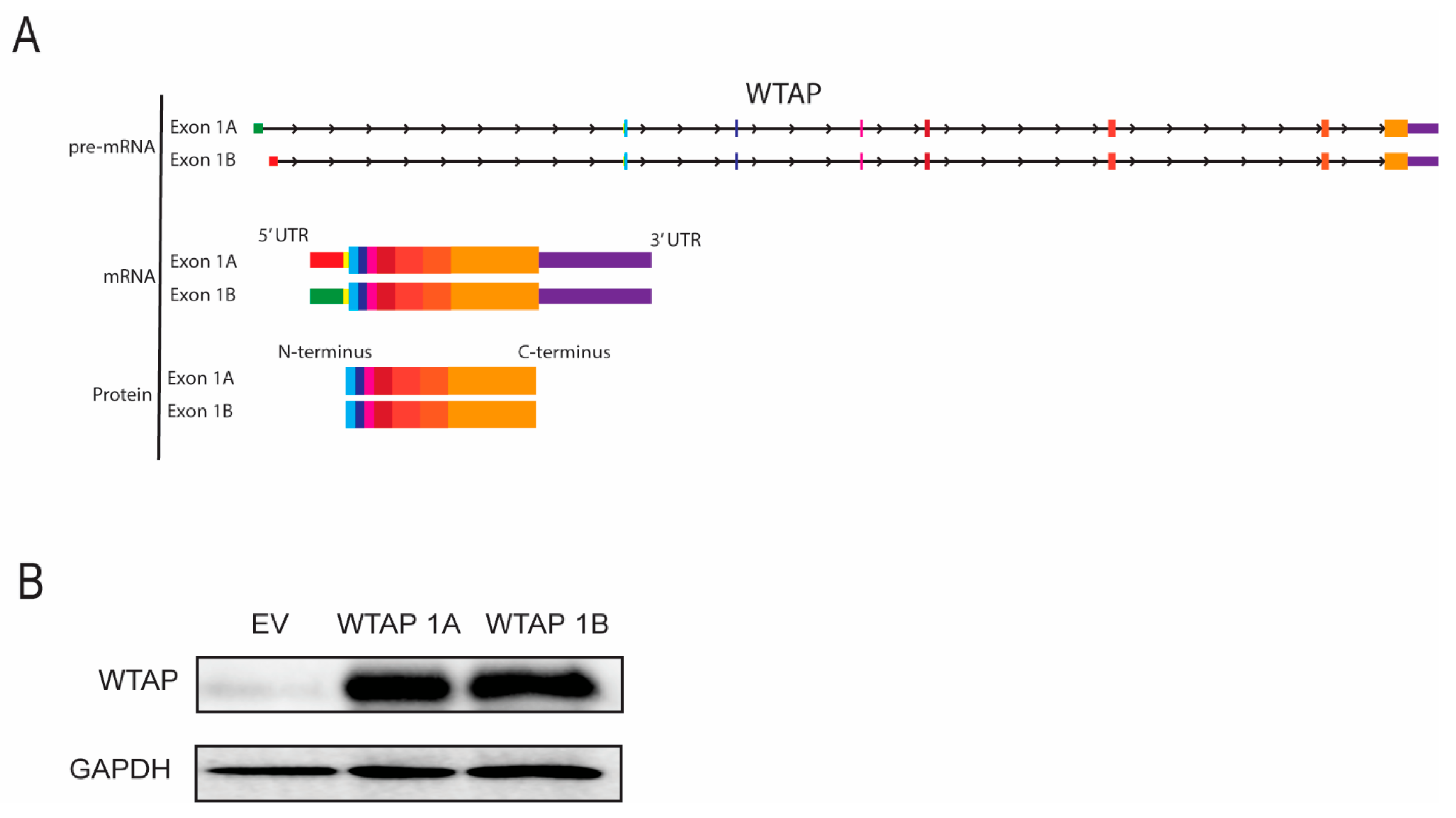
2.2. Transcription Factor RelA Promotes Transcriptional Initiation of Specific WTAP Isoforms in Monocyte Activation
3. Discussion
4. Materials and Methods
4.1. Cell Culturing
4.2. qPCR
4.3. Western Blot
4.4. Nanopore Sequencing
4.5. Next Generation Sequencing
4.6. Transfection of HEK293T Cells with WTAP Isoforms
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teng, Y.; Yi, J.; Chen, J.; Yang, L. N6-Methyladenosine (m6A) Modification in Natural Immune Cell-Mediated Inflammatory Diseases. J. Innate Immun. 2023, 15, 804–821. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Sun, W.; Xia, D.; Wang, Y.; Li, J.; Yang, S. The m6A methyltransferase METTL3 promotes LPS-induced microglia inflammation through TRAF6/NF-κB pathway. NeuroReport 2022, 33, 243–251. [Google Scholar] [CrossRef]
- Yu, R.; Li, Q.; Feng, Z.; Cai, L.; Xu, Q. m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int. J. Mol. Sci. 2019, 20, 1323. [Google Scholar] [CrossRef]
- Tong, J.; Wang, X.; Liu, Y.; Ren, X.; Wang, A.; Chen, Z. Pooled CRISPR screening identifies m6A as a positive regulator of macrophage activation. Sci. Adv. 2021, 7, eabd4742. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Z.; Tang, H.; Shen, Y.; Gong, Z.; Xie, N. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol. 2019, 317, C762–C775. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Lu, H.; Wang, S.; Xu, D. METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-κB signaling pathway. Mediat. Inflamm. 2019, 2019, 3120391. [Google Scholar] [CrossRef]
- Bai, Y.; Jiao, X.; Hu, J.; Xue, W.; Zhou, Z.; Wang, W. WTAP promotes macrophage recruitment and increases VEGF secretion via N6-methyladenosine modification in corneal neovascularization. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166708. [Google Scholar] [CrossRef]
- Li, J.; Wei, L.; Hu, K.; He, Y.; Gong, G.; Liu, Q.; Zhang, Y.; Zhou, K.; Guo, J.; Hua, Y.; et al. Deciphering m6A methylation in monocyte-mediated cardiac fibrosis and monocyte-hitchhiked erythrocyte microvesicle biohybrid therapy. Theranostics 2024, 14, 3486–3508. [Google Scholar] [CrossRef]
- Li, L.J.; Fan, Y.G.; Leng, R.X.; Pan, H.F.; Ye, D.Q. Potential link between m6A modification and systemic lupus erythematosus. Mol. Immunol. 2018, 93, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 2020, 8193895. [Google Scholar] [CrossRef]
- Luo, Q.; Gao, Y.; Zhang, L.; Rao, J.; Guo, Y.; Huang, Z.; Li, J. Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. BioMed Res. Int. 2020, 2020, 5735279. [Google Scholar] [CrossRef] [PubMed]
- Maini, R.N.; Elliott, M.J.; Brennan, F.M.; Feldmann, M. Beneficial effects of tumour necrosis factor-alpha (TNF-α) blockade in rheumatoid arthritis (RA). Clin. Exp. Immunol. 1995, 101, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.B.; Zhang, Y.H.; Lei, S.F. Genome-wide identification of N6-methyladenosine (m6A) SNPs associated with rheumatoid arthritis. Front. Genet. 2018, 9, 299. [Google Scholar] [CrossRef]
- Shi, W.; Zheng, Y.; Luo, S.; Li, X.; Zhang, Y.; Meng, X.; Huang, C.; Li, J. METTL3 Promotes Activation and Inflammation of FLSs Through the NF-κB Signaling Pathway in Rheumatoid Arthritis. Front. Med. 2021, 8, 607585. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Chen, R.; Ling, T.; Liu, B.; Huang, J.; Cheng, Y.; Lin, Y.; Chen, H.; Xie, X.; Xia, G.; et al. WTAP, transcriptionally regulated by p65, promotes inflammation through m6A modification and phase separation. Front. Immunol. 2023, 14, 1123456. [Google Scholar]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.W.; Schoenleber, R.; Jesmok, G.; Best, J.; Moore, S.A.; Collins, T.; Gerritsen, M.E. Novel Inhibitors of Cytokine-induced IκB Phosphorylation and Endothelial Cell Adhesion Molecule Expression Show Anti-inflammatory Effects in Vivo. J. Biol. Chem. 1997, 272, 21096–21103. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Sun, H.; Gao, Z.; Zhu, Z.; Yuan, K. Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential. Biomolecules 2022, 12, 1244. [Google Scholar] [CrossRef]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.A.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef]
- Weng, L.; Qiu, K.; Gao, W.; Shi, C.; Shu, F. LncRNA PCGEM1 accelerates non-small cell lung cancer progression via sponging miR-433-3p to upregulate WTAP. BMC Pulm. Med. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chang, Y.; Zhu, L.; Shen, C.; Qian, J.; Chang, R. LINC00839/miR-144-3p/WTAP (WT1 Associated protein) axis is involved in regulating hepatocellular carcinoma progression. Bioengineered 2021, 12, 10849–10861. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Liu, M.; Zhang, Y.; Xie, L.; Shi, Z.; Wang, G. SNHG10/miR-141-3p/WTAP axis promotes osteosarcoma proliferation and migration. J. Biochem. Mol. Toxicol. 2022, 36, e23031. [Google Scholar] [CrossRef] [PubMed]
- Li, J.R.; Liu, L.; Luo, H.; Chen, Z.G.; Wang, J.H.; Li, N.F. Long Noncoding RNA DUXAP8 Promotes Pancreatic Carcinoma Cell Migration and Invasion Via Pathway by miR-448/WTAP/Fak Signaling Axis. Pancreas 2021, 50, 317–326. [Google Scholar] [CrossRef]
- Huang, Q.; Mo, J.; Liao, Z.; Chen, X.; Zhang, B. The RNA m6A writer WTAP in diseases: Structure, roles, and mechanisms. Cell Death Dis. 2022, 13, 852. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, J.; Ye, Y.; Liu, K.; Zeng, L.; Huang, J.; Pan, L.; Li, M.; Bai, R.; Zhuang, L.; et al. N6-methyladenosine–Mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res. 2021, 81, 5268–5283. [Google Scholar] [CrossRef]
- Ou, B.; Liu, Y.; Yang, X.; Xu, X.; Yan, Y.; Zhang, J. C5aR1-positive neutrophils promote breast cancer glycolysis through WTAP-dependent m6A methylation of ENO1. Cell Death Dis. 2021, 12, 737. [Google Scholar] [CrossRef]
- Cho, S.; Lee, G.; Pickering, B.F.; Jang, C.; Park, J.H.; He, L.; Mathur, L.; Kim, S.S.; Jung, S.; Tang, H.W.; et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Mol. Cell 2021, 81, 2064–2075.e8. [Google Scholar] [CrossRef]
- Xiao, H.; Zhang, Y.; Sun, L.; Zhao, Z.; Liu, W.; Luo, B. EBV downregulates the m6A “writer” WTAP in EBV-associated gastric carcinoma. Virus Res. 2021, 304, 198510. [Google Scholar] [CrossRef]
- Tang, A.D.; Soulette, C.M.; van Baren, M.J.; Hart, K.; Hrabeta-Robinson, E.; Wu, C.J.; Brooks, A.N. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 2020, 11, 1438. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Picavet, L.W.; Abu-Tawil, H.I.; Sijbers, L.J.P.M.; Calis, J.J.A.; ter Haar, N.; Bodelón, A.; Vastert, S.J.; van Loosdregt, J. Transcriptional Regulation of WTAP Isoforms by NF-κB Signaling in Human Monocytes. Int. J. Mol. Sci. 2025, 26, 9364. https://doi.org/10.3390/ijms26199364
Picavet LW, Abu-Tawil HI, Sijbers LJPM, Calis JJA, ter Haar N, Bodelón A, Vastert SJ, van Loosdregt J. Transcriptional Regulation of WTAP Isoforms by NF-κB Signaling in Human Monocytes. International Journal of Molecular Sciences. 2025; 26(19):9364. https://doi.org/10.3390/ijms26199364
Chicago/Turabian StylePicavet, Lucas W., Hisham I. Abu-Tawil, Lyanne J. P. M. Sijbers, Jorg J. A. Calis, Nienke ter Haar, Alejandra Bodelón, Sebastiaan J. Vastert, and Jorg van Loosdregt. 2025. "Transcriptional Regulation of WTAP Isoforms by NF-κB Signaling in Human Monocytes" International Journal of Molecular Sciences 26, no. 19: 9364. https://doi.org/10.3390/ijms26199364
APA StylePicavet, L. W., Abu-Tawil, H. I., Sijbers, L. J. P. M., Calis, J. J. A., ter Haar, N., Bodelón, A., Vastert, S. J., & van Loosdregt, J. (2025). Transcriptional Regulation of WTAP Isoforms by NF-κB Signaling in Human Monocytes. International Journal of Molecular Sciences, 26(19), 9364. https://doi.org/10.3390/ijms26199364





