Artificial Intelligence-Driven Multi-Omics Approaches in Glioblastoma
Abstract
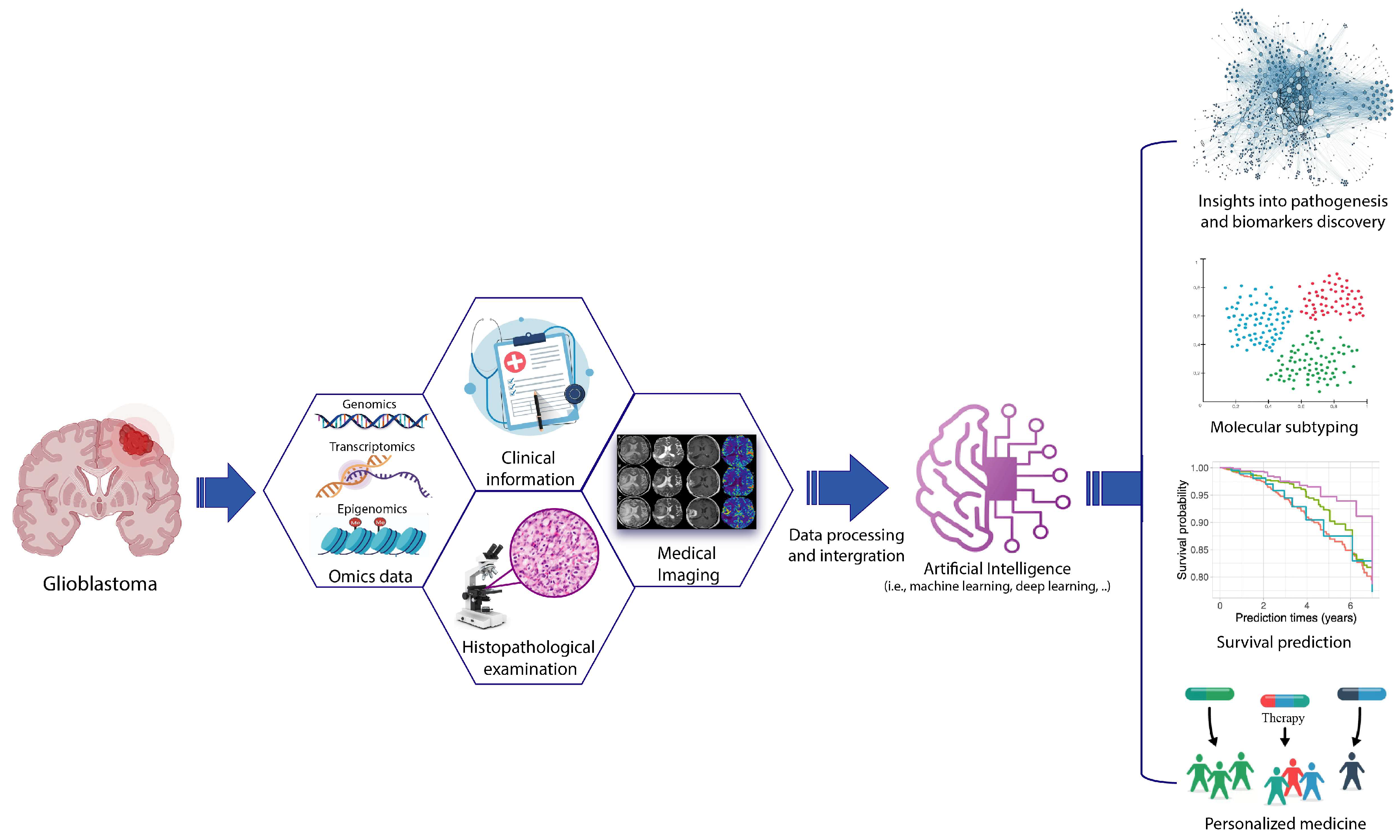
1. Introduction
2. Overview of Machine Learning and Artificial Intelligence Models
3. AI for Omics Data Analysis in GBM
3.1. AI-Assisted Genomic Prediction Models for GBM
3.2. Transcriptome-Based AI Approaches for Advanced GBM Diagnosis and Treatment
3.3. AI Integrates Epigenomic Signatures and Imaging
3.4. AI-Based Multi-Modal Integration in GBM
4. Challenges and Future Perspectives
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Cantrell, J.N.; Waddle, M.R.; Rotman, M.; Peterson, J.L.; Ruiz-Garcia, H.; Heckman, M.G.; Quinones-Hinojosa, A.; Rosenfeld, S.S.; Brown, P.D.; Trifiletti, D.M. Progress Toward Long-Term Survivors of Glioblastoma. Mayo Clin. Proc. 2019, 94, 1278–1286. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Zhang, P.; Xia, Q.; Liu, L.; Li, S.; Dong, L. Current Opinion on Molecular Characterization for GBM Classification in Guiding Clinical Diagnosis, Prognosis, and Therapy. Front. Mol. Biosci. 2020, 7, 562798. [Google Scholar] [CrossRef]
- Migliozzi, S.; Oh, Y.T.; Hasanain, M.; Garofano, L.; D’Angelo, F.; Najac, R.D.; Picca, A.; Bielle, F.; Di Stefano, A.L.; Lerond, J.; et al. Integrative multi-omics networks identify PKCδ and DNA-PK as master kinases of glioblastoma subtypes and guide targeted cancer therapy. Nat. Cancer 2023, 4, 181–202. [Google Scholar] [CrossRef]
- Ravi, V.M.; Will, P.; Kueckelhaus, J.; Sun, N.; Joseph, K.; Salie, H.; Vollmer, L.; Kuliesiute, U.; von Ehr, J.; Benotmane, J.K.; et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell 2022, 40, 639–655.e613. [Google Scholar] [CrossRef]
- Koh, L.; Novera, W.; Lim, S.W.; Chong, Y.K.; Pang, Q.Y.; Low, D.; Ang, B.T.; Tang, C. Integrative multi-omics approach to targeted therapy for glioblastoma. Pharmacol. Res. 2022, 182, 106308. [Google Scholar] [CrossRef]
- Barzegar Behrooz, A.; Latifi-Navid, H.; da Silva Rosa, S.C.; Swiat, M.; Wiechec, E.; Vitorino, C.; Vitorino, R.; Jamalpoor, Z.; Ghavami, S. Integrating Multi-Omics Analysis for Enhanced Diagnosis and Treatment of Glioblastoma: A Comprehensive Data-Driven Approach. Cancers 2023, 15, 3185. [Google Scholar] [CrossRef] [PubMed]
- Anwer, M.S.; Abdel-Rasol, M.A.; El-Sayed, W.M. Emerging therapeutic strategies in glioblastsoma: Drug repurposing, mechanisms of resistance, precision medicine, and technological innovations. Clin. Exp. Med. 2025, 25, 117. [Google Scholar] [CrossRef]
- Valdebenito, J.; Medina, F. Machine learning approaches to study glioblastoma: A review of the last decade of applications. Cancer Rep. 2019, 2, e1226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, M. Deep learning in precision medicine and focus on glioma. Bioeng. Transl. Med. 2023, 8, e10553. [Google Scholar] [CrossRef]
- Pan, X.; Burgman, B.; Wu, E.; Huang, J.H.; Sahni, N.; Yi, S.S. i-Modern: Integrated multi-omics network model identifies potential therapeutic targets in glioma by deep learning with interpretability. Comput. Struct. Biotechnol. J. 2022, 20, 3511–3521. [Google Scholar] [CrossRef]
- Hill, C.S.; Pandit, A.S. Moving towards a unified classification of glioblastomas utilizing artificial intelligence and deep machine learning integration. Front. Oncol. 2023, 13, 1063937. [Google Scholar] [CrossRef]
- Cheung, E.Y.W.; Wu, R.W.K.; Li, A.S.M.; Chu, E.S.M. AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis. Cancers 2023, 15, 5063. [Google Scholar] [CrossRef]
- Sotoudeh, H.; Shafaat, O.; Bernstock, J.D.; Brooks, M.D.; Elsayed, G.A.; Chen, J.A.; Szerip, P.; Chagoya, G.; Gessler, F.; Sotoudeh, E.; et al. Artificial Intelligence in the Management of Glioma: Era of Personalized Medicine. Front. Oncol. 2019, 9, 768. [Google Scholar] [CrossRef]
- Calabrese, E.; Villanueva-Meyer, J.E.; Cha, S. A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci. Rep. 2020, 10, 11852. [Google Scholar] [CrossRef]
- Fawaz, A.; Ferraresi, A.; Isidoro, C. Systems Biology in Cancer Diagnosis Integrating Omics Technologies and Artificial Intelligence to Support Physician Decision Making. J. Pers. Med. 2023, 13, 1590. [Google Scholar] [CrossRef] [PubMed]
- Shafi, S.; Parwani, A.V. Artificial intelligence in diagnostic pathology. Diagn. Pathol. 2023, 18, 109. [Google Scholar] [CrossRef]
- Cui, M.; Zhang, D.Y. Artificial intelligence and computational pathology. Lab. Invest. 2021, 101, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine Learning: Algorithms, Real-World Applications and Research Directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef] [PubMed]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef]
- Sedghi, L.; Ijaz, Z.; Noor-A-Rahim, M.; Witheephanich, K.; Pesch, D. Machine Learning in Event-Triggered Control: Recent Advances and Open Issues. IEEE Access 2022, 10, 74671–74690. [Google Scholar] [CrossRef]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69S, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Gao, J.X. Big Data Bioinformatics. Methods 2016, 111, 1–2. [Google Scholar] [CrossRef]
- Lee, J.; Warner, E.; Shaikhouni, S.; Bitzer, M.; Kretzler, M.; Gipson, D.; Pennathur, S.; Bellovich, K.; Bhat, Z.; Gadegbeku, C.; et al. Unsupervised machine learning for identifying important visual features through bag-of-words using histopathology data from chronic kidney disease. Sci. Rep. 2022, 12, 4832. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of Support Vector Machine (SVM) Learning in Cancer Genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, S.; Zhou, X.; Yang, M.; Wu, L.; Liu, B.; Phillips, P.; Wang, S. Comparison of machine learning methods for stationary wavelet entropy-based multiple sclerosis detection: Decision tree, k-nearest neighbors, and support vector machine. SIMULATION 2016, 92, 861–871. [Google Scholar] [CrossRef]
- Murmu, S.; Sinha, D.; Chaurasia, H.; Sharma, S.; Das, R.; Jha, G.K.; Archak, S. A review of artificial intelligence-assisted omics techniques in plant defense: Current trends and future directions. Front. Plant Sci. 2024, 15, 1292054. [Google Scholar] [CrossRef] [PubMed]
- Cios, K.J.; Swiniarski, R.W.; Pedrycz, W.; Kurgan, L.A. Unsupervised Learning: Association Rules. In Data Mining: A Knowledge Discovery Approach; Cios, K.J., Swiniarski, R.W., Pedrycz, W., Kurgan, L.A., Eds.; Springer: Boston, MA, USA, 2007; pp. 289–306. [Google Scholar]
- Vahabi, N.; Michailidis, G. Unsupervised Multi-Omics Data Integration Methods: A Comprehensive Review. Front. Genet. 2022, 13, 854752. [Google Scholar] [CrossRef]
- Ghahramani, Z. Unsupervised Learning. In Advanced Lectures on Machine Learning: ML Summer Schools 2003, Canberra, Australia, February 2–14, 2003, Tübingen, Germany, August 4–16, 2003, Revised Lectures; Bousquet, O., von Luxburg, U., Rätsch, G., Eds.; Springer: Berlin, Heidelberg, 2004; pp. 72–112. [Google Scholar]
- Serra, A.; Tagliaferri, R. Unsupervised Learning: Clustering. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 350–357. [Google Scholar]
- Lopez, C.; Tucker, S.; Salameh, T.; Tucker, C. An unsupervised machine learning method for discovering patient clusters based on genetic signatures. J. Biomed. Inf. 2018, 85, 30–39. [Google Scholar] [CrossRef]
- Mor, U.; Cohen, Y.; Valdes-Mas, R.; Kviatcovsky, D.; Elinav, E.; Avron, H. Dimensionality reduction of longitudinal ’omics data using modern tensor factorizations. PLoS Comput. Biol. 2022, 18, e1010212. [Google Scholar] [CrossRef]
- Picard, M.; Scott-Boyer, M.P.; Bodein, A.; Perin, O.; Droit, A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021, 19, 3735–3746. [Google Scholar] [CrossRef]
- Wang, J.; Liao, N.; Du, X.; Chen, Q.; Wei, B. A semi-supervised approach for the integration of multi-omics data based on transformer multi-head self-attention mechanism and graph convolutional networks. BMC Genom. 2024, 25, 86. [Google Scholar] [CrossRef]
- Eckardt, J.N.; Bornhauser, M.; Wendt, K.; Middeke, J.M. Semi-supervised learning in cancer diagnostics. Front. Oncol. 2022, 12, 960984. [Google Scholar] [CrossRef]
- Zhu, X.; Goldberg, A.B. Introduction to Semi-Supervised Learning; Morgan & Claypool Publishers: San Rafael, CA, USA, 2009. [Google Scholar] [CrossRef]
- Sharma, A.; Lysenko, A.; Jia, S.; Boroevich, K.A.; Tsunoda, T. Advances in AI and machine learning for predictive medicine. J. Hum. Genet. 2024, 10, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Chiranjib, C.; Manojit, B.; Soumen, P.; Sang-Soo, L. From machine learning to deep learning: Advances of the recent data-driven paradigm shift in medicine and healthcare. Curr. Res. Biotechnol. 2024, 7, 100164. [Google Scholar] [CrossRef]
- Sidorova, J.; Lozano, J.J. Review: Deep Learning-Based Survival Analysis of Omics and Clinicopathological Data. Inventions 2024, 9, 59. [Google Scholar] [CrossRef]
- Mohammed, M.A.; Abdulkareem, K.H.; Dinar, A.M.; Zapirain, B.G. Rise of Deep Learning Clinical Applications and Challenges in Omics Data: A Systematic Review. Diagnostics 2023, 13, 664. [Google Scholar] [CrossRef]
- Tran, K.A.; Kondrashova, O.; Bradley, A.; Williams, E.D.; Pearson, J.V.; Waddell, N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021, 13, 152. [Google Scholar] [CrossRef]
- Alharbi, W.S.; Rashid, M. A review of deep learning applications in human genomics using next-generation sequencing data. Hum. Genom. 2022, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Abouzid, M.; Kaczmarek, E. Deep Learning Approaches in Histopathology. Cancers 2022, 14, 5264. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, Y.; Liao, X.; Shi, W.; Li, K.; Zou, Q.; Peng, S. Deep learning in omics: A survey and guideline. Brief. Funct. Genom. 2019, 18, 41–57. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Ahmed, Z.; Wan, S.; Zhang, F.; Zhong, W. Artificial intelligence for omics data analysis. BMC Methods 2024, 1, 4. [Google Scholar] [CrossRef]
- Yetgin, A. Revolutionizing multi-omics analysis with artificial intelligence and data processing. Quant. Biol. 2025, 13, e70002. [Google Scholar] [CrossRef]
- Ng, S.; Masarone, S.; Watson, D.; Barnes, M.R. The benefits and pitfalls of machine learning for biomarker discovery. Cell Tissue Res. 2023, 394, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Kang, K.; Song, Y.; Kim, T.J. Application of Artificial Intelligence in Pathology: Trends and Challenges. Diagnostics 2022, 12, 2794. [Google Scholar] [CrossRef]
- Brown, E.D.; Pelcher, I.; Leon, S.; Karkare, A.N.; Barbero, J.A.; Ward, M.; Schulder, M. Artificial intelligence applications in the screening and classification of glioblastoma. J. Neurosurg. Sci. 2025, 69, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Poursaeed, R.; Mohammadzadeh, M.; Safaei, A.A. Survival prediction of glioblastoma patients using machine learning and deep learning: A systematic review. BMC Cancer 2024, 24, 1581. [Google Scholar] [CrossRef]
- Tambi, R.; Zehra, B.; Vijayakumar, A.; Satsangi, D.; Uddin, M.; Berdiev, B.K. Artificial intelligence and omics in malignant gliomas. Physiol. Genom. 2024, 56, 876–895. [Google Scholar] [CrossRef]
- El Hachimy, I.; Kabelma, D.; Echcharef, C.; Hassani, M.; Benamar, N.; Hajji, N. A comprehensive survey on the use of deep learning techniques in glioblastoma. Artif. Intell. Med. 2024, 154, 102902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dang, R.; Liu, H.; Dai, L.; Liu, H.; Adegboro, A.A.; Zhang, Y.; Li, W.; Peng, K.; Hong, J.; et al. Machine learning-based investigation of regulated cell death for predicting prognosis and immunotherapy response in glioma patients. Sci. Rep. 2024, 14, 4173. [Google Scholar] [CrossRef]
- Hollon, T.; Jiang, C.; Chowdury, A.; Nasir-Moin, M.; Kondepudi, A.; Aabedi, A.; Adapa, A.; Al-Holou, W.; Heth, J.; Sagher, O.; et al. Artificial-intelligence-based molecular classification of diffuse gliomas using rapid, label-free optical imaging. Nat. Med. 2023, 29, 828–832. [Google Scholar] [CrossRef]
- Shawon, S.S.; Afrin, S.; Yeasmin, N.; Azim, M.A.; Alom, Z. Prediction of IDH1 Gene expression in Glioblastoma Using Machine Learning Techniques. In Proceedings of the 2022 25th International Conference on Computer and Information Technology (ICCIT), Cox’s Bazar, Bangladesh, 17–19 December 2022; pp. 336–341. [Google Scholar]
- Nuechterlein, N.; Shapiro, L.G.; Holland, E.C.; Cimino, P.J. Machine learning modeling of genome-wide copy number alteration signatures reliably predicts IDH mutational status in adult diffuse glioma. Acta Neuropathol. Commun. 2021, 9, 191. [Google Scholar] [CrossRef]
- Al Mamlook, R.E.; Nasayreh, A.; Gharaibeh, H.; Shrestha, S. Classification Of Cancer Genome Atlas Glioblastoma Multiform (TCGA-GBM) Using Machine Learning Method. In Proceedings of the 2023 IEEE International Conference on Electro Information Technology (eIT), Romeoville, IL, USA, 18–20 May 2023; pp. 265–270. [Google Scholar]
- Dal Bo, M.; Polano, M.; Ius, T.; Di Cintio, F.; Mondello, A.; Manini, I.; Pegolo, E.; Cesselli, D.; Di Loreto, C.; Skrap, M.; et al. Machine learning to improve interpretability of clinical, radiological and panel-based genomic data of glioma grade 4 patients undergoing surgical resection. J. Transl. Med. 2023, 21, 450. [Google Scholar] [CrossRef] [PubMed]
- Felici, A.; Peduzzi, G.; Pellungrini, R.; Campa, D.; Canzian, F. Regression and machine learning approaches identify potential risk factors for glioblastoma multiforme. Brain Commun. 2025, 7, fcaf187. [Google Scholar] [CrossRef]
- He, W.; Huang, W.; Zhang, L.; Wu, X.; Zhang, S.; Zhang, B. Radiogenomics: Bridging the gap between imaging and genomics for precision oncology. MedComm 2024, 5, e722. [Google Scholar] [CrossRef]
- Hu, L.S.; Wang, L.; Hawkins-Daarud, A.; Eschbacher, J.M.; Singleton, K.W.; Jackson, P.R.; Clark-Swanson, K.; Sereduk, C.P.; Peng, S.; Wang, P.; et al. Uncertainty quantification in the radiogenomics modeling of EGFR amplification in glioblastoma. Sci. Rep. 2021, 11, 3932. [Google Scholar] [CrossRef]
- Liang, H.-X.; Wang, Z.-Y.; Li, Y.; Ren, A.-N.; Chen, Z.-F.; Wang, X.-Z.; Wang, X.-M.; Yuan, Z.-G. The application value of support vector machine model based on multimodal MRI in predicting IDH-1mutation and Ki-67 expression in glioma. BMC Med. Imaging 2024, 24, 244. [Google Scholar] [CrossRef]
- Hu, L.S.; Ning, S.; Eschbacher, J.M.; Baxter, L.C.; Gaw, N.; Ranjbar, S.; Plasencia, J.; Dueck, A.C.; Peng, S.; Smith, K.A.; et al. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro-Oncol. 2017, 19, 128–137. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; D’Angelo, F.; Curtin, L.; Sereduk, C.P.; Leon, G.; Singleton, K.W.; Urcuyo, J.; Hawkins-Daarud, A.; Jackson, P.R.; et al. Quantifying intra-tumoral genetic heterogeneity of glioblastoma toward precision medicine using MRI and a data-inclusive machine learning algorithm. PLoS ONE 2024, 19, e0299267. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Bakas, S.; Pisapia, J.M.; Nasrallah, M.P.; Rozycki, M.; Martinez-Lage, M.; Morrissette, J.J.D.; Dahmane, N.; O’Rourke, D.M.; Davatzikos, C. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncol. 2018, 20, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Nasrallah, M.P.; Zhao, J.; Tsai, C.C.; Meredith, D.; Marostica, E.; Ligon, K.L.; Golden, J.A.; Yu, K.H. Machine learning for cryosection pathology predicts the 2021 WHO classification of glioma. Med 2023, 4, 526–540.e524. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, Y.; Jin, L.; Aibaidula, A.; Lu, J.; Jiao, Z.; Wu, J.; Zhang, H.; Shen, D. Deep Learning of Imaging Phenotype and Genotype for Predicting Overall Survival Time of Glioblastoma Patients. IEEE Trans. Med. Imaging 2020, 39, 2100–2109. [Google Scholar] [CrossRef]
- Young, J.D.; Cai, C.; Lu, X. Unsupervised deep learning reveals prognostically relevant subtypes of glioblastoma. BMC Bioinform. 2017, 18, 381. [Google Scholar] [CrossRef]
- Tang, J.; He, D.; Yang, P.; He, J.; Zhang, Y. Genome-wide expression profiling of glioblastoma using a large combined cohort. Sci. Rep. 2018, 8, 15104. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Morrison, C.; Weterings, E.; Gravbrot, N.; Hammer, M.; Weinand, M.; Sanan, A.; Pandey, R.; Mahadevan, D.; Stea, B. Gene Expression Patterns Associated with Survival in Glioblastoma. Int. J. Mol. Sci. 2024, 25, 3668. [Google Scholar] [CrossRef]
- Nayak, C.; Singh, S.K. Integrated Transcriptome Profiling Identifies Prognostic Hub Genes as Therapeutic Targets of Glioblastoma: Evidenced by Bioinformatics Analysis. ACS Omega 2022, 7, 22531–22550. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wu, Y.; Shi, Q.; Wu, J.; Kong, D.; Wu, X.; He, X.; Liu, T.; Li, S. Systematic characterization of cancer transcriptome at transcript resolution. Nat. Commun. 2022, 13, 6803. [Google Scholar] [CrossRef] [PubMed]
- Supplitt, S.; Karpinski, P.; Sasiadek, M.; Laczmanska, I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int. J. Mol. Sci. 2021, 22, 1422. [Google Scholar] [CrossRef]
- Modelska, A.; Quattrone, A.; Re, A. Molecular portraits: The evolution of the concept of transcriptome-based cancer signatures. Brief. Bioinform. 2015, 16, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Golub, T.R.; Slonim, D.K.; Tamayo, P.; Huard, C.; Gaasenbeek, M.; Mesirov, J.P.; Coller, H.; Loh, M.L.; Downing, J.R.; Caligiuri, M.A.; et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 1999, 286, 531–537. [Google Scholar] [CrossRef]
- Ferlier, T.; Coulouarn, C. Regulation of Gene Expression in Cancer-An Overview. Cells 2022, 11, 4058. [Google Scholar] [CrossRef]
- Creighton, C.J. Gene Expression Profiles in Cancers and Their Therapeutic Implications. Cancer J. 2023, 29, 9–14. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Park, J.; Shim, J.K.; Yoon, S.J.; Kim, S.H.; Chang, J.H.; Kang, S.G. Transcriptome profiling-based identification of prognostic subtypes and multi-omics signatures of glioblastoma. Sci. Rep. 2019, 9, 10555. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Mo, Q.; Schultz, N.; Seshan, V.E.; Olshen, A.B.; Huse, J.; Ladanyi, M.; Sander, C. Integrative subtype discovery in glioblastoma using iCluster. PLoS ONE 2012, 7, e35236. [Google Scholar] [CrossRef] [PubMed]
- Teo, W.Y.; Sekar, K.; Seshachalam, P.; Shen, J.; Chow, W.Y.; Lau, C.C.; Yang, H.; Park, J.; Kang, S.G.; Li, X.; et al. Relevance of a TCGA-derived Glioblastoma Subtype Gene-Classifier among Patient Populations. Sci. Rep. 2019, 9, 7442. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Shi, J. Machine learning and bioinformatics approaches for classification and clinical detection of bevacizumab responsive glioblastoma subtypes based on miRNA expression. Sci. Rep. 2022, 12, 8685. [Google Scholar] [CrossRef]
- Steponaitis, G.; Kucinskas, V.; Golubickaite, I.; Skauminas, K.; Saudargiene, A. Glioblastoma Molecular Classification Tool Based on mRNA Analysis: From Wet-Lab to Subtype. Int. J. Mol. Sci. 2022, 23, 5875. [Google Scholar] [CrossRef]
- Lu, C.H.; Wei, S.T.; Liu, J.J.; Chang, Y.J.; Lin, Y.F.; Yu, C.S.; Chang, S.L. Recognition of a Novel Gene Signature for Human Glioblastoma. Int. J. Mol. Sci. 2022, 23, 4157. [Google Scholar] [CrossRef]
- Pasquini, L.; Napolitano, A.; Lucignani, M.; Tagliente, E.; Dellepiane, F.; Rossi-Espagnet, M.C.; Vidiri, A.; Villani, V.; Ranazzi, G.; Stoppacciaro, A.; et al. Comparison of Machine Learning Classifiers to Predict Patient Survival and Genetics of High-Grade Glioma: Towards a Standardized Model for Clinical Implementation. JMIR Prepr. 2021. [Google Scholar] [CrossRef]
- Kalya, M.; Kel, A.; Leha, A.; Altynbekova, K.; Wingender, E.; Beissbarth, T. Machine Learning based Survival Group Prediction in Glioblastoma. Preprints 2022. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Guan, G.; Zhao, W.; Zhuang, M. A Risk Classification System With Five-Gene for Survival Prediction of Glioblastoma Patients. Front. Neurol. 2019, 10, 745. [Google Scholar] [CrossRef]
- Bao, Z.S.; Li, M.Y.; Wang, J.Y.; Zhang, C.B.; Wang, H.J.; Yan, W.; Liu, Y.W.; Zhang, W.; Chen, L.; Jiang, T. Prognostic value of a nine-gene signature in glioma patients based on mRNA expression profiling. CNS Neurosci. Ther. 2014, 20, 112–118. [Google Scholar] [CrossRef]
- Chen, D.; Liu, Z.; Wang, J.; Yang, C.; Pan, C.; Tang, Y.; Zhang, P.; Liu, N.; Li, G.; Li, Y.; et al. Integrative genomic analysis facilitates precision strategies for glioblastoma treatment. iScience 2022, 25, 105276. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, H.; Hu, H.; Cai, Z.; Lu, C.; Liang, Q.; Qian, J.; Wang, C.; Jiang, L. A Novel Six-mRNA Signature Predicts Survival of Patients With Glioblastoma Multiforme. Front. Genet. 2021, 12, 634116. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Cai, J.; Yuan, Y.; Shi, Y.; Wu, H.; Liu, Q.; Yao, Y.; Chen, L.; Dang, W.; Zhang, X.; et al. A four-gene signature-derived risk score for glioblastoma: Prospects for prognostic and response predictive analyses. Cancer Biol. Med. 2019, 16, 595–605. [Google Scholar] [CrossRef]
- Yin, W.; Tang, G.; Zhou, Q.; Cao, Y.; Li, H.; Fu, X.; Wu, Z.; Jiang, X. Expression Profile Analysis Identifies a Novel Five-Gene Signature to Improve Prognosis Prediction of Glioblastoma. Front. Genet. 2019, 10, 419. [Google Scholar] [CrossRef]
- Zhao, Z.; Le, N.Q.K.; Chua, M.C.H. AI-driven multi-modal framework for prognostic modeling in glioblastoma: Enhancing clinical decision support. Comput. Med. Imaging Graph. 2025, 124, 102628. [Google Scholar] [CrossRef]
- Zhou, J.; Duan, S.; Zhu, Z.; Wang, H.; Tian, C.; Yang, H.; Chen, S.; Ye, M.; Zhang, X.; Zhang, B. Identification of intrinsic imaging subtypes using clustering analysis based on dynamic contrast-enhanced magnetic resonance imaging radiomics features for gliomas: Preliminary associations with gene expression profiles. Quant. Imaging Med. Surg. 2025, 15, 4734–4747. [Google Scholar] [CrossRef]
- Fatai, A.A.; Gamieldien, J. A 35-gene signature discriminates between rapidly- and slowly-progressing glioblastoma multiforme and predicts survival in known subtypes of the cancer. BMC Cancer 2018, 18, 377. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.G.; Xue, X.Y.; Wang, L.; Lin, W.; Zhang, X. Deep learning identified glioblastoma subtypes based on internal genomic expression ranks. BMC Cancer 2022, 22, 86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Huang, C.; Cao, H.; Lin, J.; Gong, X.; Li, J.; Chen, Y.; Tian, Z.; Fang, Z.; Huang, J. A Novel Prognostic Signature of Transcription Factors for the Prediction in Patients With GBM. Front. Genet. 2019, 10, 906. [Google Scholar] [CrossRef]
- Handayani, L.; Chegodaev, D.; Steven, R.; Satou, K. Identification of Key Genes Associated with Overall Survival in Glioblastoma Multiforme Using TCGA RNA-Seq Expression Data. Genes 2025, 16, 755. [Google Scholar] [CrossRef]
- Wong, K.K.; Rostomily, R.; Wong, S.T.C. Prognostic Gene Discovery in Glioblastoma Patients using Deep Learning. Cancers 2019, 11, 53. [Google Scholar] [CrossRef]
- Thomas, M.P.H.; Ajaib, S.; Tanner, G.; Bulpitt, A.J.; Stead, L.F. GBMPurity: A machine learning tool for estimating glioblastoma tumor purity from bulk RNA-sequencing data. Neuro-Oncol. 2025, 27, 1458–1473. [Google Scholar] [CrossRef]
- Jiang, Q.; Yang, X.; Deng, T.; Yan, J.; Guo, F.; Mo, L.; An, S.; Huang, Q. Comprehensive machine learning-based integration develops a novel prognostic model for glioblastoma. Mol. Ther. Oncol. 2024, 32, 200838. [Google Scholar] [CrossRef]
- Li, N.; Ward, M.; Bashir, M.; Cao, Y.; Datta, A.; Li, Z.; Zhang, S. Machine learning reveals novel targets for both glioblastoma and osteosarcoma. Heliyon 2025, 11, e42997. [Google Scholar] [CrossRef]
- Song, Z.; Wang, F.; Yang, C.; Guo, Y.; Li, J.; Huang, R.; Ling, H.; Cheng, G.; Chen, Z.; Zhu, Z.; et al. Meta-analysis of multi-center transcriptomic profiles and machine learning reveal phospholipase Cβ4 as a Wnt/Ca2+ signaling mediator in glioblastoma immunotherapy. Front. Immunol. 2025, 16, 1610683. [Google Scholar] [CrossRef]
- Kint, S.; Younes, S.T.; Bao, S.; Long, G.; Wouters, D.; Stephenson, E.; Lou, X.; Zhong, M.; Zhang, D.; Su, G.; et al. Spatial epigenomic niches underlie glioblastoma cell state plasticity. bioRxiv 2025. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, N.; Wu, W.; Zhou, R.; Li, S.; Wang, Z.; Dai, Z.; Zhang, L.; Liu, Z.; Zhang, J.; et al. Machine learning-based tumor-infiltrating immune cell-associated lncRNAs for predicting prognosis and immunotherapy response in patients with glioblastoma. Brief. Bioinform. 2022, 23, bbac386. [Google Scholar] [CrossRef]
- Li, C.; Liu, W.; Liu, C.; Luo, Q.; Luo, K.; Wei, C.; Li, X.; Qin, J.; Zheng, C.; Lan, C.; et al. Integrating machine learning and bioinformatics analysis to m6A regulator-mediated methylation modification models for predicting glioblastoma patients’ prognosis and immunotherapy response. Aging 2023, 15, 4051–4070. [Google Scholar] [CrossRef]
- Shaw, R.; Basu, M.; Karmakar, S.; Ghosh, M.K. MGMT in TMZ-based glioma therapy: Multifaceted insights and clinical trial perspectives. Biochim. Biophys. Acta Mol. Cell Res. 2024, 1871, 119673. [Google Scholar] [CrossRef]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Wang, Y. Radiomics in glioma: Emerging trends and challenges. Ann. Clin. Transl. Neurol. 2025, 12, 460–477. [Google Scholar] [CrossRef]
- Xi, Y.B.; Guo, F.; Xu, Z.L.; Li, C.; Wei, W.; Tian, P.; Liu, T.T.; Liu, L.; Chen, G.; Ye, J.; et al. Radiomics signature: A potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J. Magn. Reson. Imaging 2018, 47, 1380–1387. [Google Scholar] [CrossRef]
- Hajianfar, G.; Shiri, I.; Maleki, H.; Oveisi, N.; Haghparast, A.; Abdollahi, H.; Oveisi, M. Noninvasive O(6) Methylguanine-DNA Methyltransferase Status Prediction in Glioblastoma Multiforme Cancer Using Magnetic Resonance Imaging Radiomics Features: Univariate and Multivariate Radiogenomics Analysis. World Neurosurg. 2019, 132, e140–e161. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Do, D.T.; Chiu, F.Y.; Yapp, E.K.Y.; Yeh, H.Y.; Chen, C.Y. XGBoost Improves Classification of MGMT Promoter Methylation Status in IDH1 Wildtype Glioblastoma. J. Pers. Med. 2020, 10, 128. [Google Scholar] [CrossRef]
- Do, D.T.; Yang, M.R.; Lam, L.H.T.; Le, N.Q.K.; Wu, Y.W. Improving MGMT methylation status prediction of glioblastoma through optimizing radiomics features using genetic algorithm-based machine learning approach. Sci. Rep. 2022, 12, 13412. [Google Scholar] [CrossRef]
- Yogananda, C.G.B.; Shah, B.R.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Mickey, B.; Patel, T.R.; Fei, B.; et al. MRI-Based Deep-Learning Method for Determining Glioma MGMT Promoter Methylation Status. AJNR Am. J. Neuroradiol. 2021, 42, 845–852. [Google Scholar] [CrossRef]
- Chen, X.; Zeng, M.; Tong, Y.; Zhang, T.; Fu, Y.; Li, H.; Zhang, Z.; Cheng, Z.; Xu, X.; Yang, R.; et al. Automatic Prediction of MGMT Status in Glioblastoma via Deep Learning-Based MR Image Analysis. Biomed. Res. Int. 2020, 2020, 9258649. [Google Scholar] [CrossRef]
- Calabrese, E.; Rudie, J.D.; Rauschecker, A.M.; Villanueva-Meyer, J.E.; Clarke, J.L.; Solomon, D.A.; Cha, S. Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma. Neurooncol Adv. 2022, 4, vdac060. [Google Scholar] [CrossRef]
- Mikkelsen, V.E.; Dai, H.Y.; Stensjoen, A.L.; Berntsen, E.M.; Salvesen, O.; Solheim, O.; Torp, S.H. MGMT Promoter Methylation Status Is Not Related to Histological or Radiological Features in IDH Wild-type Glioblastomas. J. Neuropathol. Exp. Neurol. 2020, 79, 855–862. [Google Scholar] [CrossRef]
- Saeed, N.; Ridzuan, M.; Alasmawi, H.; Sobirov, I.; Yaqub, M. MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models. Med. Image Anal. 2023, 90, 102989. [Google Scholar] [CrossRef]
- Faghani, S.; Khosravi, B.; Moassefi, M.; Conte, G.M.; Erickson, B.J. A Comparison of Three Different Deep Learning-Based Models to Predict the MGMT Promoter Methylation Status in Glioblastoma Using Brain MRI. J. Digit. Imaging 2023, 36, 837–846. [Google Scholar] [CrossRef]
- Goodman, R.M.; Bouroncle, B.A.; Miller, F.; North, C. Unusual chromosomal findings in a case of myelofibrosis. J. Hered. 1968, 59, 348–350. [Google Scholar] [CrossRef]
- Saeed, N. Is it Possible to Predict MGMT Promoter Methylation from Brain Tumor MRI Scans using Deep Learning Models? arXiv 2022, arXiv:2201.06086. [Google Scholar] [CrossRef]
- Saxena, S.; Jena, B.; Mohapatra, B.; Gupta, N.; Kalra, M.; Scartozzi, M.; Saba, L.; Suri, J.S. Fused deep learning paradigm for the prediction of o6-methylguanine-DNA methyltransferase genotype in glioblastoma patients: A neuro-oncological investigation. Comput. Biol. Med. 2023, 153, 106492. [Google Scholar] [CrossRef]
- Ma, Y.; Xi, Z. Integrated Analysis of Multiomics Data Identified Molecular Subtypes and Oxidative Stress-Related Prognostic Biomarkers in Glioblastoma Multiforme. Oxid. Med. Cell Longev. 2022, 2022, 9993319. [Google Scholar] [CrossRef]
- Binder, H.; Schmidt, M.; Hopp, L.; Davitavyan, S.; Arakelyan, A.; Loeffler-Wirth, H. Integrated Multi-Omics Maps of Lower-Grade Gliomas. Cancers 2022, 14, 2797. [Google Scholar] [CrossRef]
- Wu, C.; Zhou, F.; Ren, J.; Li, X.; Jiang, Y.; Ma, S. A Selective Review of Multi-Level Omics Data Integration Using Variable Selection. High-Throughput 2019, 8, 4. [Google Scholar] [CrossRef]
- Meng, C.; Zeleznik, O.A.; Thallinger, G.G.; Kuster, B.; Gholami, A.M.; Culhane, A.C. Dimension reduction techniques for the integrative analysis of multi-omics data. Brief. Bioinform. 2016, 17, 628–641. [Google Scholar] [CrossRef]
- Rappoport, N.; Shamir, R. Multi-omic and multi-view clustering algorithms: Review and cancer benchmark. Nucleic Acids Res. 2019, 47, 1044. [Google Scholar] [CrossRef]
- Chauvel, C.; Novoloaca, A.; Veyre, P.; Reynier, F.; Becker, J. Evaluation of integrative clustering methods for the analysis of multi-omics data. Brief. Bioinform. 2020, 21, 541–552. [Google Scholar] [CrossRef]
- Yan, J.; Risacher, S.L.; Shen, L.; Saykin, A.J. Network approaches to systems biology analysis of complex disease: Integrative methods for multi-omics data. Brief. Bioinform. 2017, 19, 1370–1381. [Google Scholar] [CrossRef]
- Zarayeneh, N.; Ko, E.; Oh, J.H.; Suh, S.; Liu, C.; Gao, J.; Kim, D.; Kang, M. Integration of multi-omics data for integrative gene regulatory network inference. Int. J. Data Min. Bioinform. 2017, 18, 223–239. [Google Scholar] [CrossRef]
- Wani, N.; Raza, K. Integrative Approaches to Reconstruct Regulatory Networks From Multi-Omics Data: A Review of State-of-the-Art Methods. Preprints 2018. [Google Scholar] [CrossRef]
- Lee, B.; Zhang, S.; Poleksic, A.; Xie, L. Heterogeneous Multi-Layered Network Model for Omics Data Integration and Analysis. Front. Genet. 2020, 10, 1381. [Google Scholar] [CrossRef]
- Morabito, A.; De Simone, G.; Pastorelli, R.; Brunelli, L.; Ferrario, M. Algorithms and tools for data-driven omics integration to achieve multilayer biological insights: A narrative review. J. Transl. Med. 2025, 23, 425. [Google Scholar] [CrossRef]
- Lu, J.; Cowperthwaite, M.C.; Burnett, M.G.; Shpak, M. Molecular Predictors of Long-Term Survival in Glioblastoma Multiforme Patients. PLoS ONE 2016, 11, e0154313. [Google Scholar] [CrossRef]
- Ensenyat-Mendez, M.; Iniguez-Munoz, S.; Sese, B.; Marzese, D.M. iGlioSub: An integrative transcriptomic and epigenomic classifier for glioblastoma molecular subtypes. BioData Min. 2021, 14, 42. [Google Scholar] [CrossRef]
- Fathi Kazerooni, A.; Saxena, S.; Toorens, E.; Tu, D.; Bashyam, V.; Akbari, H.; Mamourian, E.; Sako, C.; Koumenis, C.; Verginadis, I.; et al. Clinical measures, radiomics, and genomics offer synergistic value in AI-based prediction of overall survival in patients with glioblastoma. Sci. Rep. 2022, 12, 8784. [Google Scholar] [CrossRef]
- Yan, J.; Sun, Q.; Tan, X.; Liang, C.; Bai, H.; Duan, W.; Mu, T.; Guo, Y.; Qiu, Y.; Wang, W.; et al. Image-based deep learning identifies glioblastoma risk groups with genomic and transcriptomic heterogeneity: A multi-center study. Eur. Radiol. 2023, 33, 904–914. [Google Scholar] [CrossRef]
- Munquad, S.; Das, A.B. DeepAutoGlioma: A deep learning autoencoder-based multi-omics data integration and classification tools for glioma subtyping. BioData Min. 2023, 16, 32. [Google Scholar] [CrossRef]
- Munquad, S.; Si, T.; Mallik, S.; Das, A.B.; Zhao, Z. A Deep Learning-Based Framework for Supporting Clinical Diagnosis of Glioblastoma Subtypes. Front. Genet. 2022, 13, 855420. [Google Scholar] [CrossRef] [PubMed]
- Șerban, M.; Toader, C.; Covache-Busuioc, R.-A. Precision Neuro-Oncology in Glioblastoma: AI-Guided CRISPR Editing and Real-Time Multi-Omics for Genomic Brain Surgery. Int. J. Mol. Sci. 2025, 26, 7364. [Google Scholar] [CrossRef]
- Ding, Y.; Xu, Z.; Hu, W.; Deng, P.; Ma, M.; Wu, J. Comprehensive multi-omics and machine learning framework for glioma subtyping and precision therapeutics. Sci. Rep. 2025, 15, 24874. [Google Scholar] [CrossRef]
- Xu, C.; Yang, J.; Xiong, H.; Cui, X.; Zhang, Y.; Gao, M.; He, L.; Fang, Q.; Han, C.; Liu, W.; et al. Machine learning and multi-omics analysis reveal key regulators of proneural–mesenchymal transition in glioblastoma. Sci. Rep. 2025, 15, 19731. [Google Scholar] [CrossRef]
- Tasci, E.; Chappidi, S.; Zhuge, Y.; Zhang, L.; Cooley Zgela, T.; Sproull, M.; Mackey, M.; Camphausen, K.; Krauze, A.V. GLIO-Select: Machine Learning-Based Feature Selection and Weighting of Tissue and Serum Proteomic and Metabolomic Data Uncovers Sex Differences in Glioblastoma. Int. J. Mol. Sci. 2025, 26, 4339. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; He, Z.; Guo, J.; Bu, D.; Han, D.; Qu, X.; Li, Q.; Cheng, S.; Han, A.; Guo, J. Leveraging TME features and multi-omics data with an advanced deep learning framework for improved Cancer survival prediction. Sci. Rep. 2025, 15, 14282. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Liu, Y.; Mo, Z.; Liu, X.; Sun, H.; Li, J. Integrated Multi-Omics and Whole Slide Images for Survival Prediction in Glioblastoma Using Multiple Instance Learning and Co-Attention. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15–19 July 2024; pp. 1–4. [Google Scholar]
- Ye, L.; Ye, C.; Li, P.; Wang, Y.; Ma, W. Inferring the genetic relationships between unsupervised deep learning–derived imaging phenotypes and glioblastoma through multi-omics approaches. Brief. Bioinform. 2025, 26, bbaf037. [Google Scholar] [CrossRef]
- Oh, J.H.; Choi, W.; Ko, E.; Kang, M.; Tannenbaum, A.; Deasy, J.O. PathCNN: Interpretable convolutional neural networks for survival prediction and pathway analysis applied to glioblastoma. Bioinformatics 2021, 37, i443–i450. [Google Scholar] [CrossRef]
- Hao, Y.; Jing, X.-Y.; Sun, Q. Cancer survival prediction by learning comprehensive deep feature representation for multiple types of genetic data. BMC Bioinform. 2023, 24, 267. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, H.; Li, F. Investigating the relevance of major signaling pathways in cancer survival using a biologically meaningful deep learning model. BMC Bioinform. 2021, 22, 47. [Google Scholar] [CrossRef]
- Camps, J.; Jiménez-Franco, A.; García-Pablo, R.; Joven, J.; Arenas, M. Artificial intelligence-driven integration of multi-omics and radiomics: A new hope for precision cancer diagnosis and prognosis. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2025, 1871, 167841. [Google Scholar] [CrossRef]
- Lai, J.; Shen, Z.; Yuan, L. Bio-ATT-CNN: A Novel Method for Identification of Glioblastoma. In Intelligent Computing Theories and Application; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2022; pp. 767–776. [Google Scholar]
- Chaddad, A.; Daniel, P.; Sabri, S.; Desrosiers, C.; Abdulkarim, B. Integration of Radiomic and Multi-omic Analyses Predicts Survival of Newly Diagnosed IDH1 Wildtype Glioblastoma. Cancers 2019, 11, 1148. [Google Scholar] [CrossRef]
- Zheng, Y.; Carrillo-Perez, F.; Pizurica, M.; Heiland, D.H.; Gevaert, O. Spatial cellular architecture predicts prognosis in glioblastoma. Nat. Commun. 2023, 14, 4122. [Google Scholar] [CrossRef]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.J.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Nandoe Tewarie, R.; Lycklama, G.J.; De Witt Hamer, P.C.; Eijgelaar, R.S.; et al. Combined molecular subtyping, grading, and segmentation of glioma using multi-task deep learning. Neuro-Oncol. 2023, 25, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Arteaga-Arteaga, H.B.; Candamil-Cortés, M.S.; Breaux, B.; Guillen-Rondon, P.; Orozco-Arias, S.; Tabares-Soto, R. Machine learning applications on intratumoral heterogeneity in glioblastoma using single-cell RNA sequencing data. Brief. Funct. Genom. 2023, 22, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.H.B.; Leon, A.J.; Hui, W.; Lee, S.C.-E.; Batruch, I.; Faust, K.; Klekner, A.; Hutóczki, G.; Koritzinsky, M.; Richer, M.; et al. Topographic mapping of the glioblastoma proteome reveals a triple-axis model of intra-tumoral heterogeneity. Nat. Commun. 2022, 13, 116. [Google Scholar] [CrossRef]
- Hoffmann, C. MOSAIC: Intra-tumoral heterogeneity characterization through large-scale spatial and cell-resolved multi-omics profiling. bioRxiv 2025. [Google Scholar] [CrossRef]
- Ouhmouk, M.; Baichoo, S.; Abik, M. Challenges in AI-driven multi-omics data analysis for Oncology: Addressing dimensionality, sparsity, transparency and ethical considerations. Inform. Med. Unlocked 2025, 57, 101679. [Google Scholar] [CrossRef]
- Conte, L.; Caruso, G.; Philip, A.K.; Cucci, F.; De Nunzio, G.; Cascio, D.; Caffo, M. Artificial Intelligence-Assisted Drug and Biomarker Discovery for Glioblastoma: A Scoping Review of the Literature. Cancers 2025, 17, 571. [Google Scholar] [CrossRef]
- Acuña-Villaorduña, A.; Baranda, J.C.; Boehmer, J.; Fashoyin-Aje, L.; Gore, S.D. Equitable Access to Clinical Trials: How Do We Achieve It? Am. Soc. Clin. Oncol. Educ. Book 2023, 43, e389838. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, N.; Mai, Y.; Ren, L.; Chen, Q.; Cao, Z.; Chen, Q.; Liu, Y.; Hou, W.; Yang, J.; et al. Correcting batch effects in large-scale multiomics studies using a reference-material-based ratio method. Genome Biol. 2023, 24, 201. [Google Scholar] [CrossRef]
- Svinin, G.; Loo, R.T.J.; Soudy, M.; Nasta, F.; Le Bars, S.; Glaab, E. Computational Systems Biology Methods for Cross-Disease Comparison of Omics Data. WIREs Comput. Mol. Sci. 2025, 15, e70042. [Google Scholar] [CrossRef]
- Zhang, Z.; Hernandez, K.; Savage, J.; Li, S.; Miller, D.; Agrawal, S.; Ortuno, F.; Staudt, L.M.; Heath, A.; Grossman, R.L. Uniform genomic data analysis in the NCI Genomic Data Commons. Nat. Commun. 2021, 12, 1226. [Google Scholar] [CrossRef]
- Rončević, A.; Koruga, N.; Soldo Koruga, A.; Rončević, R. Artificial Intelligence in Glioblastoma—Transforming Diagnosis and Treatment. Chin. Neurosurg. J. 2025, 11, 10. [Google Scholar] [CrossRef]
- Kuo, P.-C.; Weiner, E.B.; Dankwa-Mullan, I.; Nelson, W.A.; Hassanpour, S. Ethical challenges and evolving strategies in the integration of artificial intelligence into clinical practice. PLOS Digit. Health 2025, 4, e0000810. [Google Scholar] [CrossRef]
- Boudi, A.L.; Boudi, M.; Chan, C.; Boudi, F.B. Ethical Challenges of Artificial Intelligence in Medicine. Cureus 2024, 16, e74495. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morello, G.; La Cognata, V.; Guarnaccia, M.; Gentile, G.; Cavallaro, S. Artificial Intelligence-Driven Multi-Omics Approaches in Glioblastoma. Int. J. Mol. Sci. 2025, 26, 9362. https://doi.org/10.3390/ijms26199362
Morello G, La Cognata V, Guarnaccia M, Gentile G, Cavallaro S. Artificial Intelligence-Driven Multi-Omics Approaches in Glioblastoma. International Journal of Molecular Sciences. 2025; 26(19):9362. https://doi.org/10.3390/ijms26199362
Chicago/Turabian StyleMorello, Giovanna, Valentina La Cognata, Maria Guarnaccia, Giulia Gentile, and Sebastiano Cavallaro. 2025. "Artificial Intelligence-Driven Multi-Omics Approaches in Glioblastoma" International Journal of Molecular Sciences 26, no. 19: 9362. https://doi.org/10.3390/ijms26199362
APA StyleMorello, G., La Cognata, V., Guarnaccia, M., Gentile, G., & Cavallaro, S. (2025). Artificial Intelligence-Driven Multi-Omics Approaches in Glioblastoma. International Journal of Molecular Sciences, 26(19), 9362. https://doi.org/10.3390/ijms26199362






