Synergistic Biocontrol of Agrobacterium tumefaciens by Phage PAT1 and Ascaphin-8: Enhanced Antimicrobial Activity and Virulence Attenuation via HupB Loss
Abstract
1. Introduction
2. Results
2.1. Sublethal Concentration of Ascaphin 8 Against A. tumefaciens
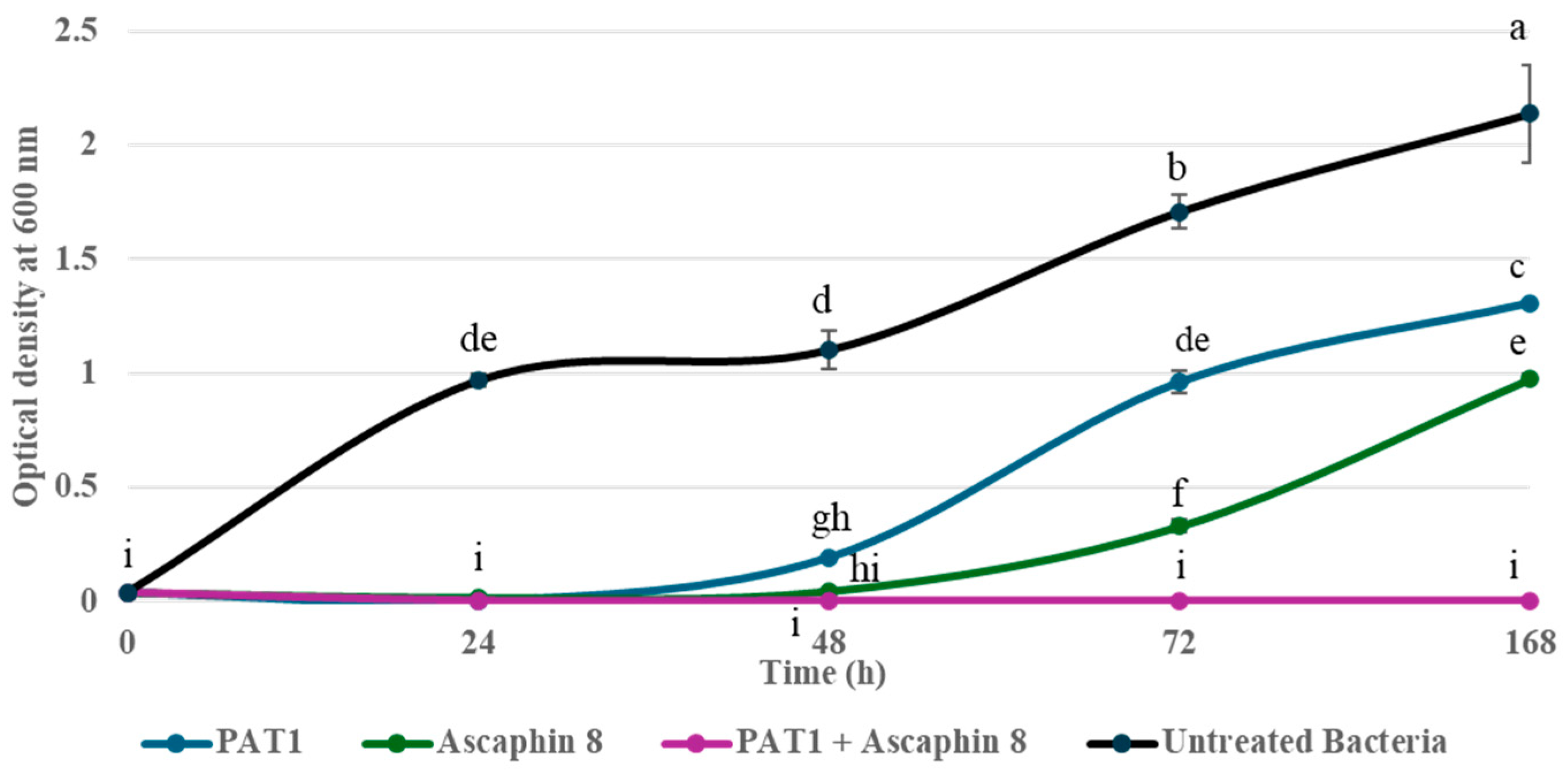
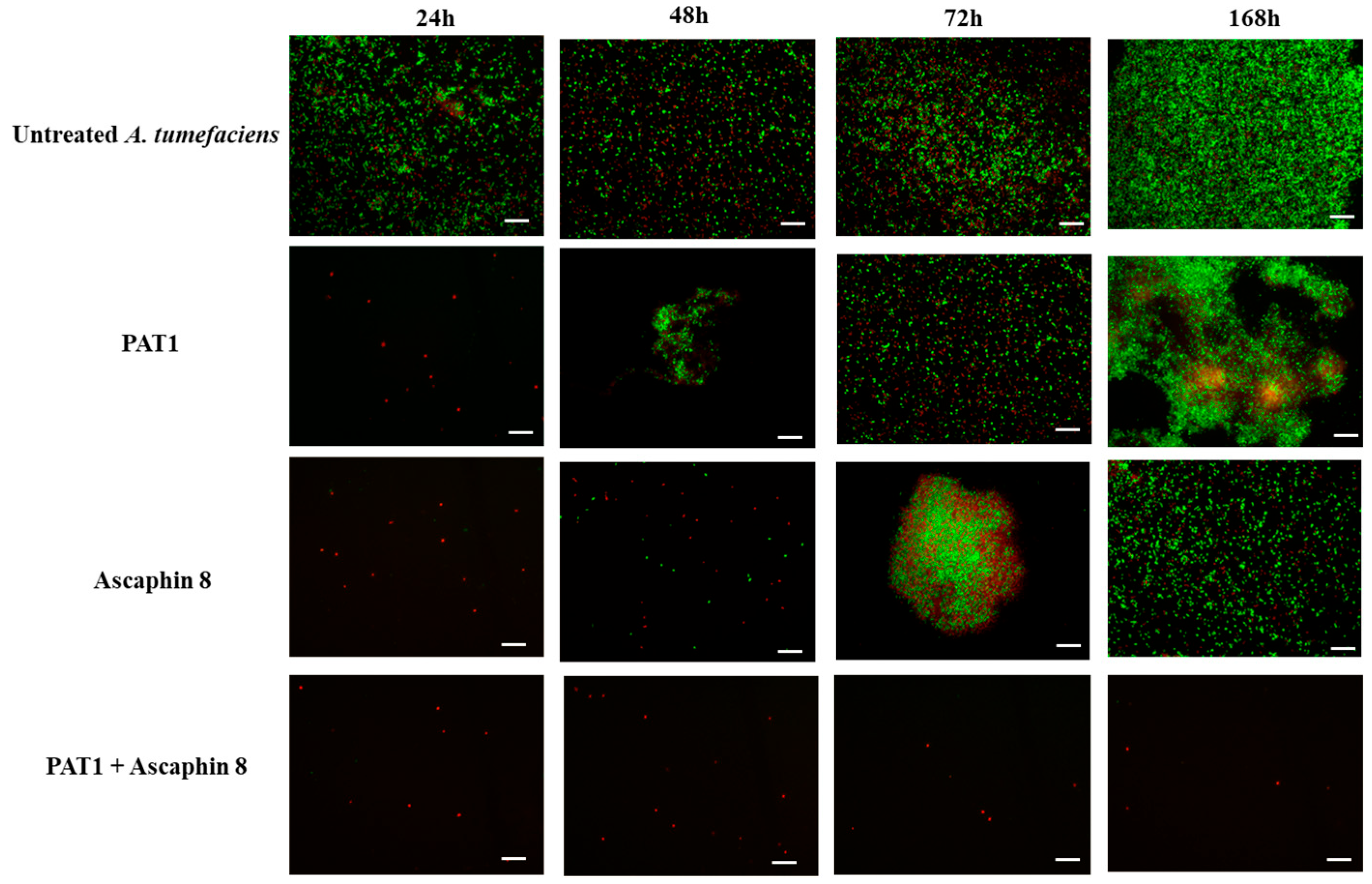
2.2. Synergistic Antibacterial Activity of PAT1-Ascaphin 8 Combination Against A. tumefaciens
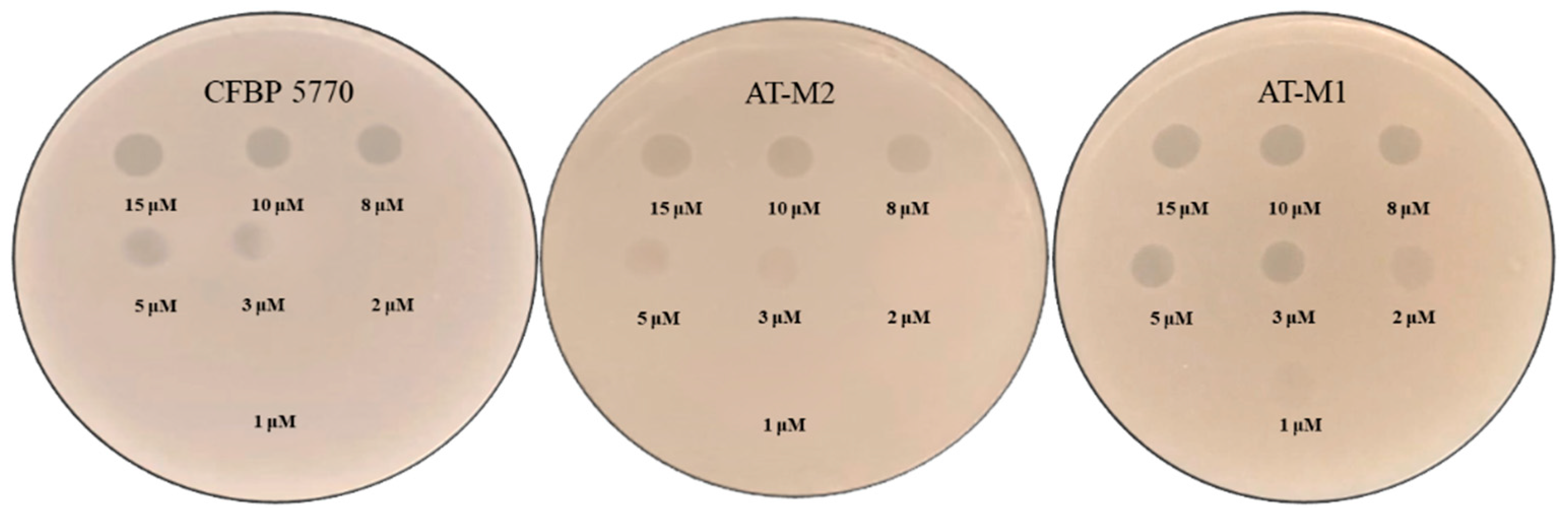
2.3. AT-M1 and AT-M2 Susceptibility to PAT1 and Ascaphin 8
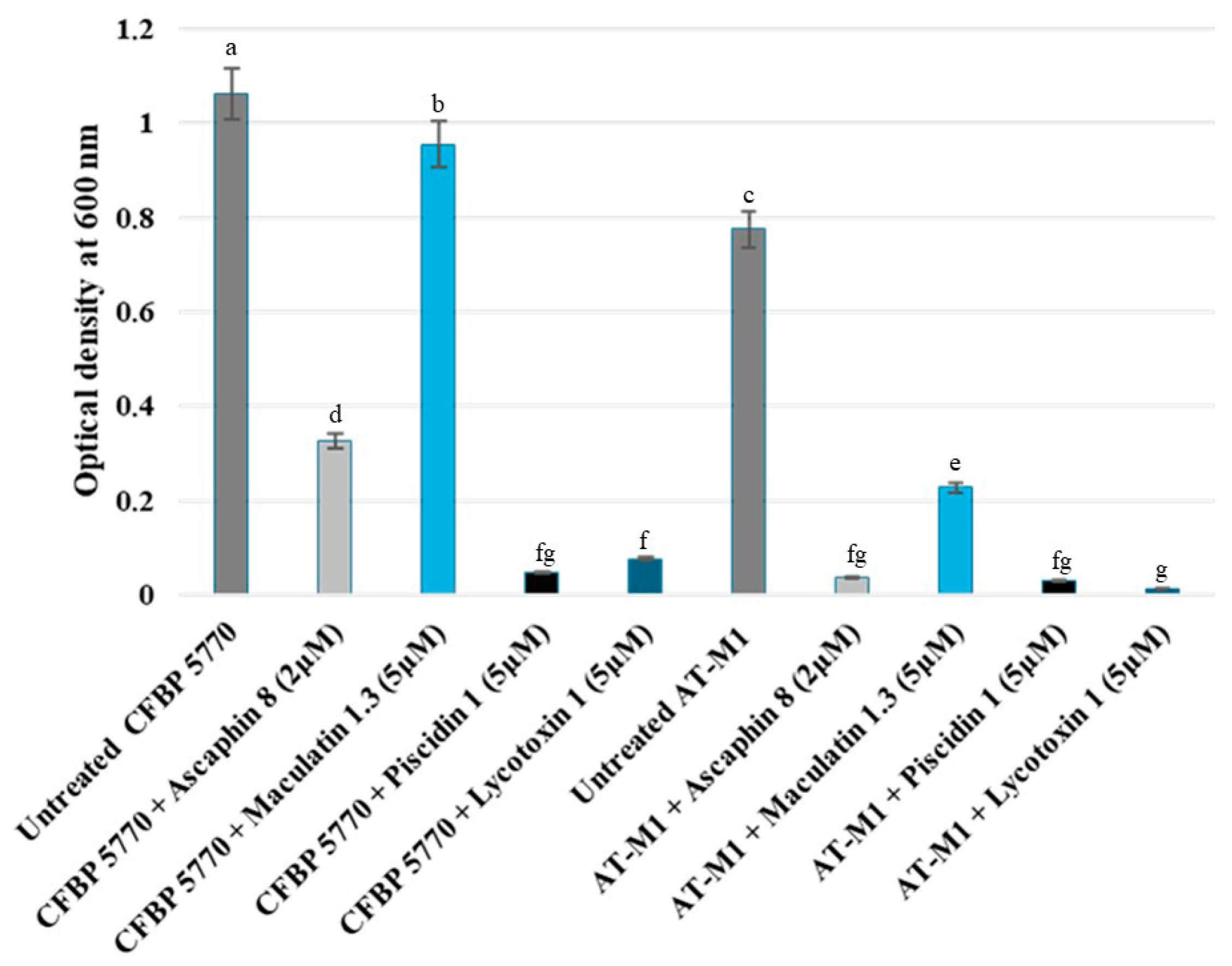
2.4. Comparative Sensitivity of Wild-Type Strain CFBP 5770 and AT-M1 Mutant to Different Antimicrobial Peptides
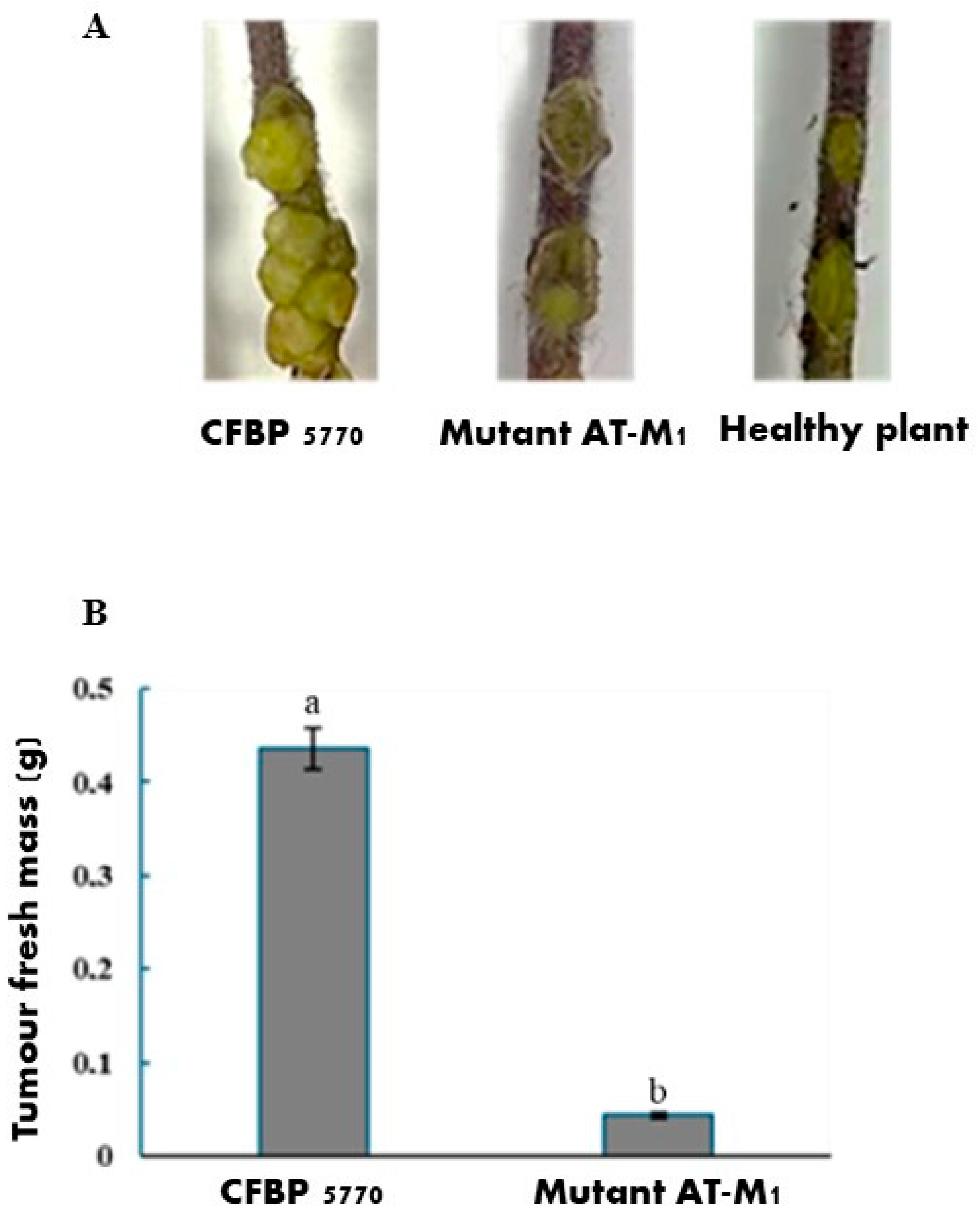
2.5. Impact of PAT1 Resistance on the Virulence of A. tumefaciens
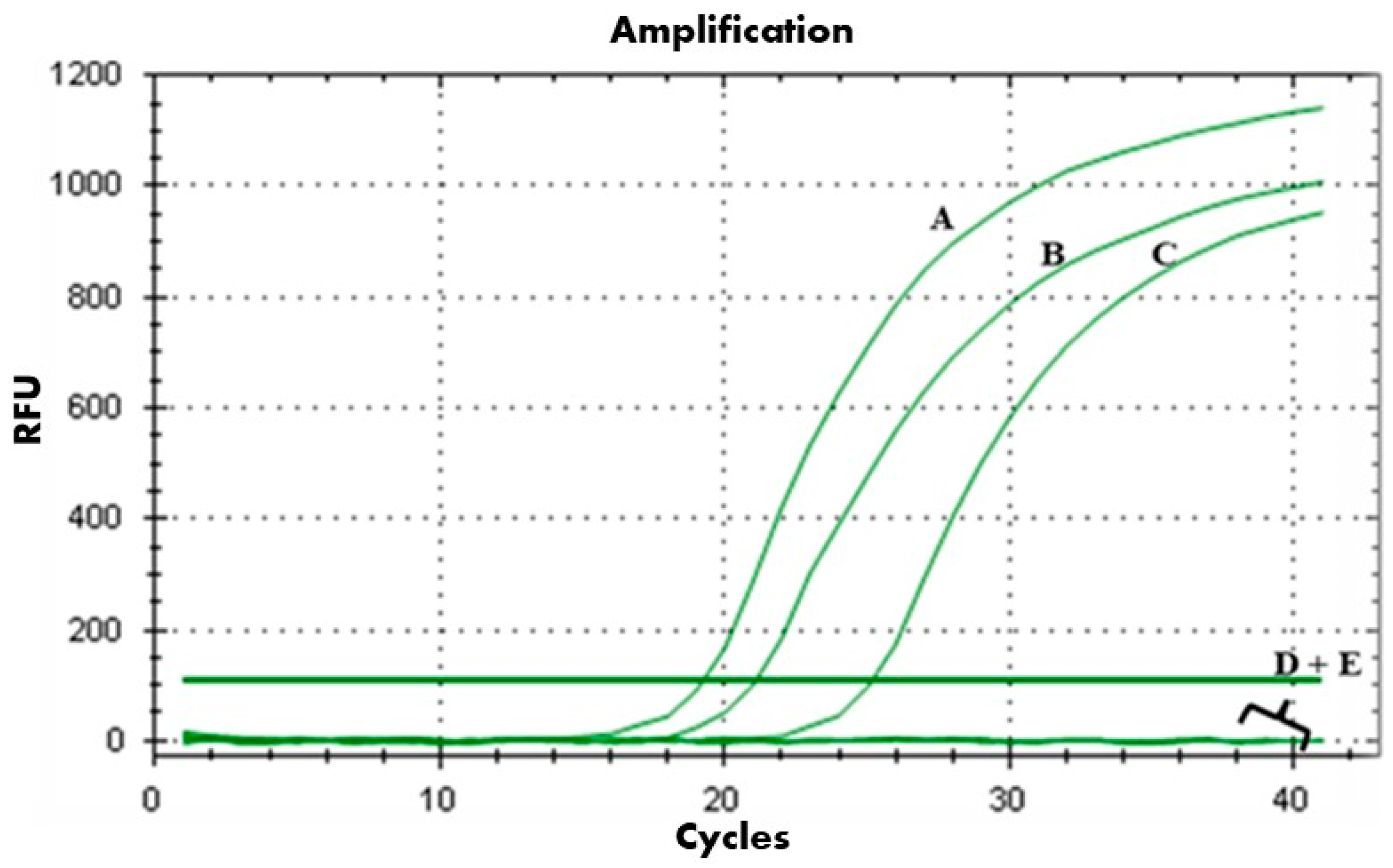
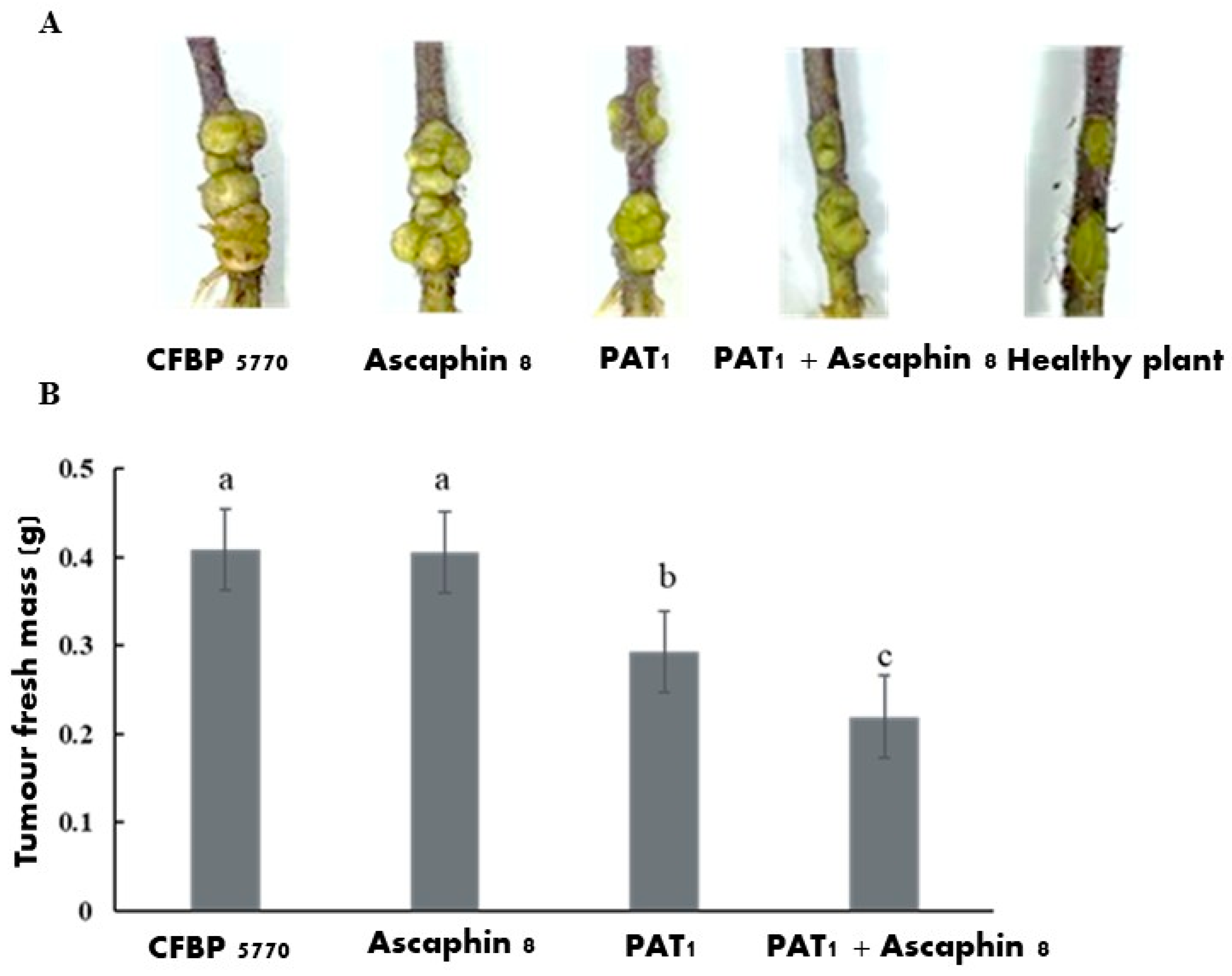
2.6. The In Planta Antibacterial Efficacy of PAT1, Ascaphin 8, and Their Combination Against A. tumefaciens
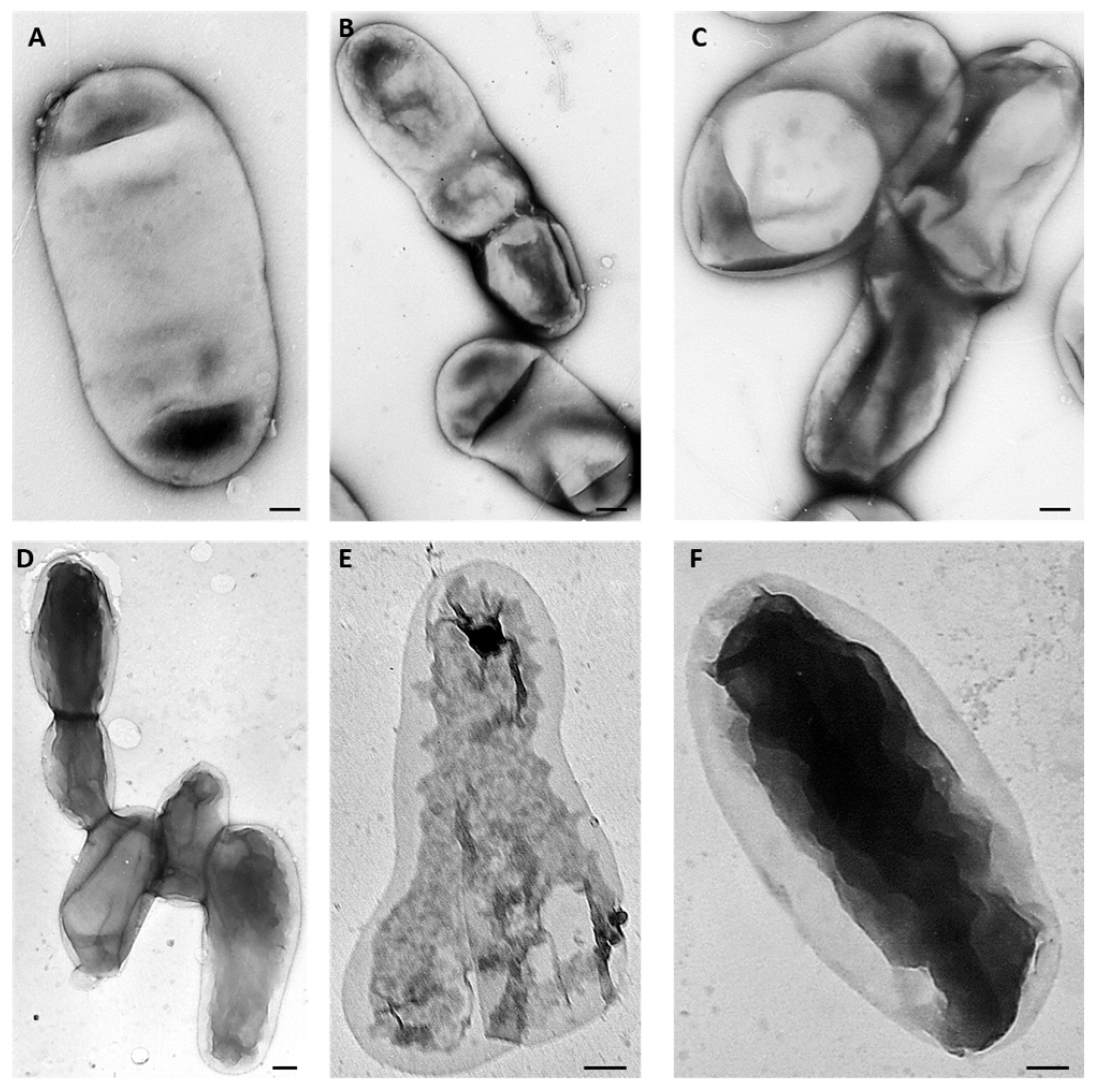
2.7. Protein Identification in Wild-Type Strain and AT-M1 Mutant by Mass Spectrometry
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis, Bacteriophage, and Bacterial Growth Conditions
4.2. Bactericidal Activity of Ascaphin 8 Against A. tumefaciens
4.3. In Vitro Antibacterial Potency of PAT1 and Ascaphin 8 Combination Against A. tumefaciens
4.4. Susceptibility Test of AT-M1 and AT-M2 to PAT1 and Ascaphin 8
4.5. Antimicrobial Peptide Sensitivity of the AT-M1 Mutant
4.6. In Planta Antibacterial Efficacy of PAT1 and Ascaphin 8 Against A. tumefaciens and Virulence Assessment of the AT-M1 Mutant in Tomato Plants
4.7. Statistical Data Analysis
4.8. Proteomic Analysis of AT-M1 and Wild-Type Bacteria
4.8.1. Protein Extraction and Purification
4.8.2. SDS-PAGE and In-Gel Digestion
4.8.3. MALDI-MS and RPLC-ESI-MS of Tryptic Digests
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escobar, M.A.; Leslie, C.A.; McGranahan, G.H.; Dandekar, A.M. Silencing Crown Gall Disease in Walnut (Juglans regia L.). Plant Sci. 2002, 163, 591–597. [Google Scholar] [CrossRef]
- Anand, A.; Uppalapati, S.R.; Ryu, C.-M.; Allen, S.N.; Kang, L.; Tang, Y.; Mysore, K.S. Salicylic Acid and Systemic Acquired Resistance Play a Role in Attenuating Crown Gall Disease Caused by Agrobacterium tumefaciens. Plant Physiol. 2008, 146, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Frikha-Gargouri, O.; Ben Abdallah, D.; Ghorbel, I.; Charfeddine, I.; Jlaiel, L.; Triki, M.A.; Tounsi, S. Lipopeptides from a Novel Bacillus Methylotrophicus 39b Strain Suppress Agrobacterium Crown Gall Tumours on Tomato Plants. Pest Manag. Sci. 2017, 73, 568–574. [Google Scholar] [CrossRef]
- Torres, M.; Jiquel, A.; Jeanne, E.; Naquin, D.; Dessaux, Y.; Faure, D. Agrobacterium tumefaciens Fitness Genes Involved in the Colonization of Plant Tumors and Roots. New Phytol. 2022, 233, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, Y.; Yu, K.; Gao, S.; Zhao, X.; Cao, A.; Han, Q. Technology for Distribution and Control of Agrobacterium tumefaciens in Cherry Tree Soil. Agriculture 2024, 14, 1857. [Google Scholar] [CrossRef]
- Attai, H.; Boon, M.; Phillips, K.; Noben, J.-P.; Lavigne, R.; Brown, P.J.B. Larger Than Life: Isolation and Genomic Characterization of a Jumbo Phage That Infects the Bacterial Plant Pathogen, Agrobacterium tumefaciens. Front. Microbiol. 2018, 9, 1861. [Google Scholar] [CrossRef]
- Fujiwara, A.; Fujisawa, M.; Hamasaki, R.; Kawasaki, T.; Fujie, M.; Yamada, T. Biocontrol of Ralstonia Solanacearum by Treatment with Lytic Bacteriophages. Appl. Environ. Microbiol. 2011, 77, 4155–4162. [Google Scholar] [CrossRef]
- Clavijo-Coppens, F.; Ginet, N.; Cesbron, S.; Briand, M.; Jacques, M.-A.; Ansaldi, M. Novel Virulent Bacteriophages Infecting Mediterranean Isolates of the Plant Pest Xylella fastidiosa and Xanthomonas albilineans. Viruses 2021, 13, 725. [Google Scholar] [CrossRef]
- Rendueles, C.; Duarte, A.C.; Escobedo, S.; Fernández, L.; Rodríguez, A.; García, P.; Martínez, B. Combined Use of Bacteriocins and Bacteriophages as Food Biopreservatives. A Review. Int. J. Food Microbiol. 2022, 368, 109611. [Google Scholar] [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and Cons of Phage Therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Feng, Y.; McNally, A.; Zong, Z. Characterization of Phage Resistance and Phages Capable of Intestinal Decolonization of Carbapenem-Resistant Klebsiella Pneumoniae in Mice. Commun. Biol. 2022, 5, 48. [Google Scholar] [CrossRef]
- Bartnik, P.; Lewtak, K.; Fiołka, M.; Czaplewska, P.; Narajczyk, M.; Czajkowski, R. Resistance of Dickeya solani Strain IPO 2222 to Lytic Bacteriophage ΦD5 Results in Fitness Tradeoffs for the Bacterium during Infection. Sci. Rep. 2022, 12, 10725. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a Bacteriophage Cocktail to Constrain the Emergence of Phage-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [PubMed]
- Koderi Valappil, S.; Shetty, P.; Deim, Z.; Terhes, G.; Urbán, E.; Váczi, S.; Patai, R.; Polgár, T.; Pertics, B.Z.; Schneider, G.; et al. Survival Comes at a Cost: A Coevolution of Phage and Its Host Leads to Phage Resistance and Antibiotic Sensitivity of Pseudomonas aeruginosa Multidrug Resistant Strains. Front. Microbiol. 2021, 12, 783722. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.-L.; Wang, C.-H.; Liao, Y.-T.; Chan, F.-Y.; Kanaoka, Y.; Uchihashi, T.; Kato, K.; Lai, L.; Chang, Y.-W.; Ho, M.-C.; et al. Visualizing the Membrane Disruption Action of Antimicrobial Peptides by Cryo-Electron Tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial Peptides (AMPs): Ancient Compounds That Represent Novel Weapons in the Fight against Bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Szymczak, P.; Możejko, M.; Grzegorzek, T.; Jurczak, R.; Bauer, M.; Neubauer, D.; Sikora, K.; Michalski, M.; Sroka, J.; Setny, P.; et al. Discovering Highly Potent Antimicrobial Peptides with Deep Generative Model HydrAMP. Nat. Commun. 2023, 14, 1453, Erratum in Nat. Commun. 2023, 14, 5129. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, X. Biosynthesis, Bioactivity, Biotoxicity and Applications of Antimicrobial Peptides for Human Health. Biosaf. Health 2022, 4, 118–134. [Google Scholar] [CrossRef]
- Mirski, T.; Lidia, M.; Nakonieczna, A.; Gryko, R. Bacteriophages, Phage Endolysins and Antimicrobial Peptides—The Possibilities for Their Common Use to Combat Infections and in the Design of New Drugs. Ann. Agric. Environ. Med. 2019, 26, 203–209. [Google Scholar] [CrossRef]
- Alisigwe, C.V.; Ikpa, C.S.; Otuonye, U.J. Examining Alternative Approaches to Antibiotic Utilisation: A Critical Evaluation of Phage Therapy and Antimicrobial Peptides Combination as Potential Alternatives. Microbe 2025, 6, 100254. [Google Scholar] [CrossRef]
- Sabri, M.; Handi, K.E.; Cara, O.; Stradis, A.D.; Elbeaino, T. Isolation, Characterization and Genomic Analysis of a Novel Lytic Bacteriophage Infecting Agrobacterium tumefaciens. Phytopathol. Mediterr. 2024, 63, 323–334. [Google Scholar] [CrossRef]
- Ninawe, A.S.; Sivasankari, S.; Ramasamy, P.; Kiran, G.S.; Selvin, J. Bacteriophages for Aquaculture Disease Control. Aquac. Int. 2020, 28, 1925–1938. [Google Scholar] [CrossRef]
- Tang, M.; Huang, Z.; Zhang, X.; Kong, J.; Zhou, B.; Han, Y.; Zhang, Y.; Chen, L.; Zhou, T. Phage Resistance Formation and Fitness Costs of Hypervirulent Klebsiella pneumoniae Mediated by K2 Capsule-Specific Phage and the Corresponding Mechanisms. Front. Microbiol. 2023, 14, 1156292. [Google Scholar] [CrossRef] [PubMed]
- Eley, A.; Ibrahim, M.; Kurdi, S.E.; Conlon, J.M. Activities of the Frog Skin Peptide, Ascaphin-8 and Its Lysine-Substituted Analogs against Clinical Isolates of Extended-Spectrum Beta-Lactamase (ESBL) Producing Bacteria. Peptides 2008, 29, 25–30. [Google Scholar] [CrossRef]
- Van Nieuwenhuyse, B.; Van der Linden, D.; Chatzis, O.; Lood, C.; Wagemans, J.; Lavigne, R.; Schroven, K.; Paeshuyse, J.; de Magnée, C.; Sokal, E.; et al. Bacteriophage-Antibiotic Combination Therapy against Extensively Drug-Resistant Pseudomonas aeruginosa Infection to Allow Liver Transplantation in a Toddler. Nat. Commun. 2022, 13, 5725. [Google Scholar] [CrossRef]
- Barache, N.; Belguesmia, Y.; Martinez, B.; Seal, B.S.; Drider, D. Bacteriocins and Bacteriophages as Dual Biological Players for Food Safety Applications. Encyclopedia 2024, 4, 79–90. [Google Scholar] [CrossRef]
- Manohar, P.; Loh, B.; Nachimuthu, R.; Leptihn, S. Phage-Antibiotic Combinations to Control Pseudomonas aeruginosa–Candida Two-Species Biofilms. Sci. Rep. 2024, 14, 9354. [Google Scholar] [CrossRef]
- Gu Liu, C.; Green, S.I.; Min, L.; Clark, J.R.; Salazar, K.C.; Terwilliger, A.L.; Kaplan, H.B.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Phage-Antibiotic Synergy Is Driven by a Unique Combination of Antibacterial Mechanism of Action and Stoichiometry. mBio 2020, 11, e01462-20. [Google Scholar] [CrossRef]
- Osman, A.-H.; Kotey, F.C.N.; Odoom, A.; Darkwah, S.; Yeboah, R.K.; Dayie, N.T.K.D.; Donkor, E.S. The Potential of Bacteriophage-Antibiotic Combination Therapy in Treating Infections with Multidrug-Resistant Bacteria. Antibiotics 2023, 12, 1329. [Google Scholar] [CrossRef]
- Saenkham, P.; Eiamphungporn, W.; Farrand, S.K.; Vattanaviboon, P.; Mongkolsuk, S. Multiple Superoxide Dismutases in Agrobacterium tumefaciens: Functional Analysis, Gene Regulation, and Influence on Tumorigenesis. J. Bacteriol. 2007, 189, 8807–8817. [Google Scholar] [CrossRef] [PubMed]
- Conforte, V.P.; Malamud, F.; Yaryura, P.M.; Toum Terrones, L.; Torres, P.S.; De Pino, V.; Chazarreta, C.N.; Gudesblat, G.E.; Castagnaro, A.P.; Marano, M.R.; et al. The Histone-like Protein HupB Influences Biofilm Formation and Virulence in Xanthomonas citri Ssp. citri through the Regulation of Flagellar Biosynthesis. Mol. Plant Pathol. 2019, 20, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Sharma, N.; Singh, P.; Pandey, M.; Ilyas, M.; Sisodiya, L.; Choudhury, T.; Gosain, T.P.; Singh, R.; Atmakuri, K. HupB, a Nucleoid-Associated Protein, Is Critical for Survival of Mycobacterium tuberculosis under Host-Mediated Stresses and for Enhanced Tolerance to Key First-Line Antibiotics. Front. Microbiol. 2022, 13, 937970. [Google Scholar] [CrossRef]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and Short-Chain Dehydrogenase/Reductase Gene and Protein Families: The SDR Superfamily: Functional and Structural Diversity within a Family of Metabolic and Regulatory Enzymes. Cell Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef]
- El Handi, K.; Sabri, M.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Hafidi, M.; El Moujabber, M.; Elbeaino, T. Exploring Active Peptides with Antimicrobial Activity in Planta against Xylella fastidiosa. Biology 2022, 11, 1685. [Google Scholar] [CrossRef]
- Sabri, M.; El Handi, K.; Valentini, F.; De Stradis, A.; Achbani, E.H.; Benkirane, R.; Elbeaino, T. Exploring Antimicrobial Peptides Efficacy against Fire Blight (Erwinia amylovora). Plants 2023, 12, 113. [Google Scholar] [CrossRef]
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database Screening and in Vivo Efficacy of Antimicrobial Peptides against Meticillin-Resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef]
- Ishibashi, N.; Himeno, K.; Fujita, K.; Masuda, Y.; Perez, R.H.; Zendo, T.; Wilaipun, P.; Leelawatcharamas, V.; Nakayama, J.; Sonomoto, K. Purification and Characterization of Multiple Bacteriocins and an Inducing Peptide Produced by Enterococcus faecium NKR-5-3 from Thai Fermented Fish. Biosci. Biotechnol. Biochem. 2012, 76, 947–953. [Google Scholar] [CrossRef]
- Baró, A.; Badosa, E.; Montesinos, L.; Feliu, L.; Planas, M.; Montesinos, E.; Bonaterra, A. Screening and Identification of BP100 Peptide Conjugates Active against Xylella fastidiosa Using a Viability-qPCR Method. BMC Microbiol. 2020, 20, 229. [Google Scholar] [CrossRef] [PubMed]
- Ganjeh, A.; Rahimian, H.; Babaeizad, V.; Basavand, E. Stem and Crown Gall Disease Caused by Agrobacterium tumefaciens on Golden Euonymus in Iran. J. Plant Pathol. 2020, 102, 1307. [Google Scholar] [CrossRef]
- Stocks, S.M. Mechanism and Use of the Commercially Available Viability Stain, BacLight. Cytom. A 2004, 61, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem. 2016, 88, 5438–5443. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, S.R. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE). Curr. Protoc. 2012, 6, 7.3.1–7.3.28. [Google Scholar] [CrossRef]
- Calvano, C.D.; Rigante, E.; Picca, R.A.; Cataldi, T.R.I.; Sabbatini, L. An Easily Transferable Protocol for In-Situ Quasi-Non-Invasive Analysis of Protein Binders in Works of Art. Talanta 2020, 215, 120882. [Google Scholar] [CrossRef]
- Bianco, M.; Calvano, C.D.; Ventura, G.; Losito, I.; Cataldi, T.R.I. Determination of Hidden Milk Allergens in Meat-Based Foodstuffs by Liquid Chromatography Coupled to Electrospray Ionization and High-Resolution Tandem Mass Spectrometry. Food Control 2022, 131, 108443. [Google Scholar] [CrossRef]

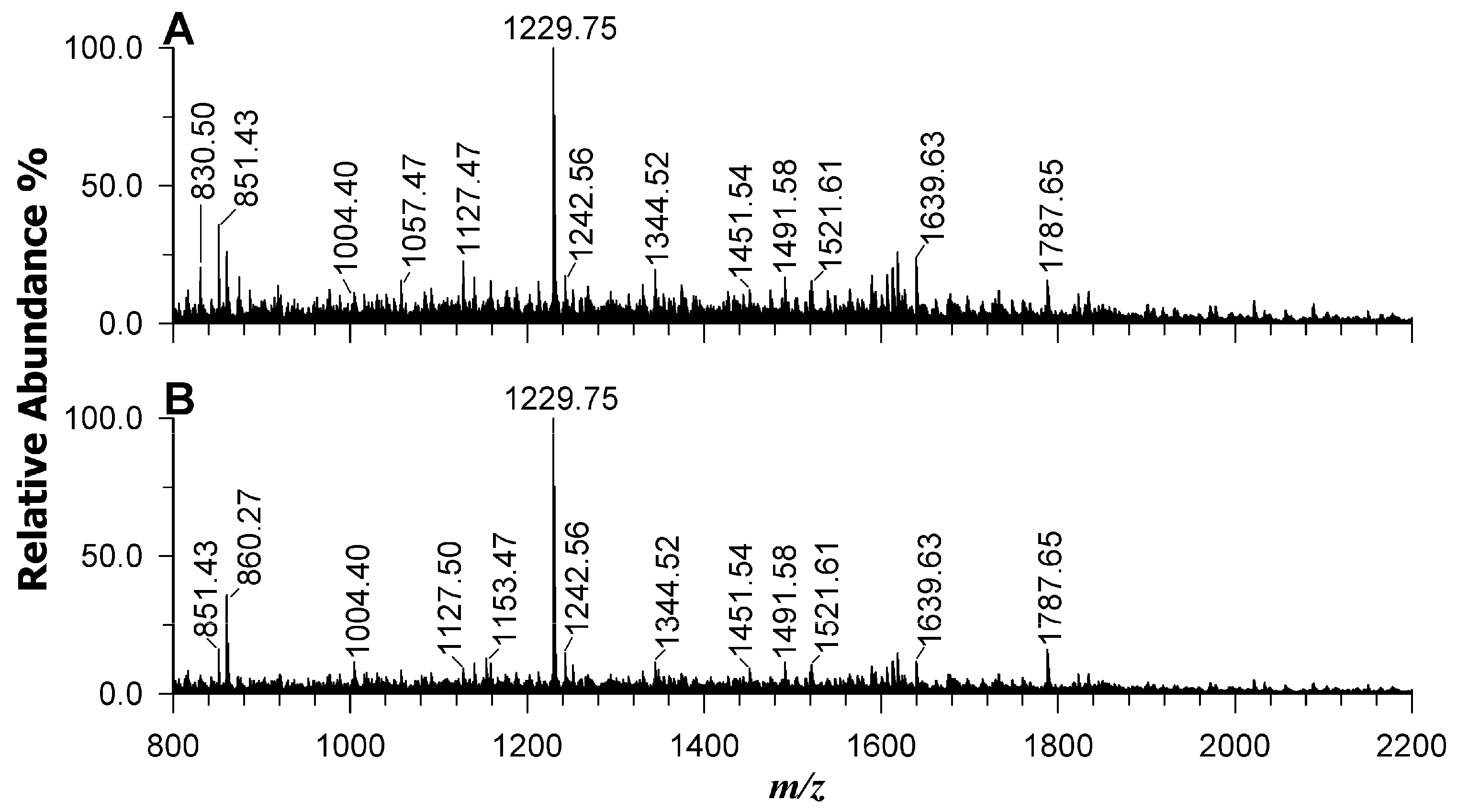
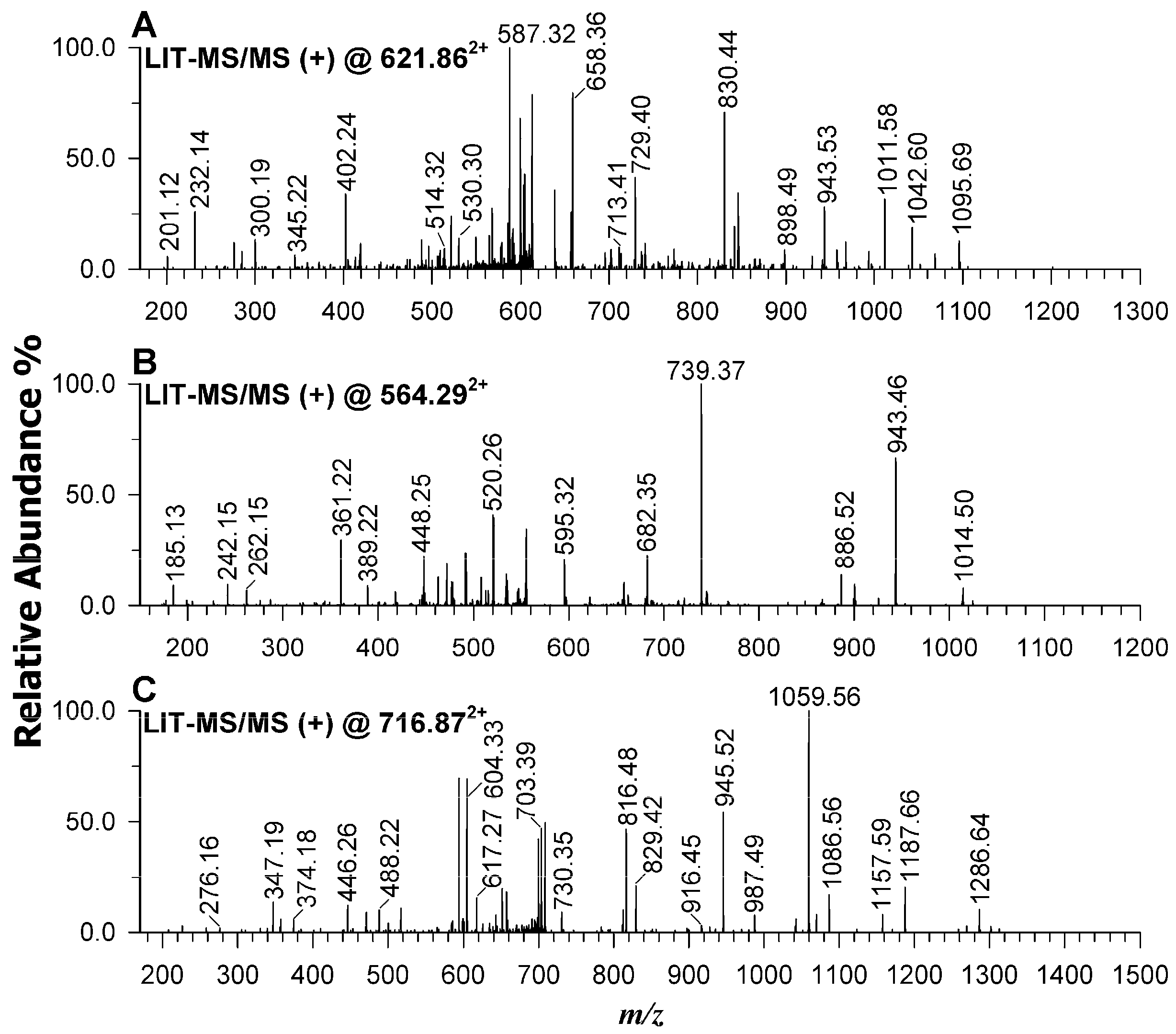
| Protein Families | Peptides | m/z | Band | Log2 Fold Change WT/ATM1 |
|---|---|---|---|---|
| Aspartate ammonia-lyase | LIESAALLHEINLGATAIGTGLNAPR | 872.4843+ | A-A′ | 0.76 |
| AVENFQITGVTIGHNPYLVR | 743.4033+ | 0.56 | ||
| GYLTEEQLQQALSPR | 866.9512+ | 0.49 | ||
| TFVVMLGEDQAR | 683.3492+ | 0.59 | ||
| TQLQDAVPMTLGQEFR | 917.4632+ | 0.18 | ||
| ATP synthase subunit beta | FTQAGSEVSALLGR | 718.3852+ | A-A′ | −0.36 |
| MLDPMIVGEEHYEVAR | 630.3063+ | −0.53 | ||
| TIAMDSTEGLVR | 646.8322+ | −0.52 | ||
| IMNVIGEPVDEAGPIVTAK | 977.0252+ | −0.24 | ||
| Chaperonin GroEL | AAVQEGIVPGGGVALLR | 803.9692+ | A-A′ | −0.09 |
| QIGLDIAEAMQR | 672.8542+ | −0.95 | ||
| TNDIAGDGTTTATVLAQAIVR | 1044.5552+ | −0.38 | ||
| Elongation factor | LLDQGQAGDNIGALVR | 820.4392+ | A-A′ B-B′ C-C′ | 0.20 |
| VDQVDDAELLELVELEVR | 1042.5472+ | 1.28 | ||
| VVIATEDNSYVER | 747.8792+ | 0.13 | ||
| ABC transporter substrate-binding protein | IDDLIFAITPDAAVR | 815.4522+ | A-A′ | 0.57 |
| DFNADDVIFSYNR | 788.3622+ | 0.73 | ||
| Flagellin | SIGNNMETTQGR | 654.3082+ | B-B′ | 0.74 |
| VGSASDNAAYWSIATTMR | 950.9492+ | 0.94 | ||
| QSVSNLDISDLSIYK | 841.4342+ | 0.72 | ||
| ASILTNASSMAALQTLR | 874.4732+ | 0.20 | ||
| SDASALSTVSDALGIGAAK | 867.4472+ | 1.04 | ||
| ALQTQQQLAIQALSIANSDSQNILSLFR | 1024.5603+ | 0.37 | ||
| Glyceraldehyde−3-phosphate dehydrogenase | GILGYTEEPLVSR | 717.3892+ | B-B′ | 1.12 |
| VLSWYDNEWGFSNR | 886.9042+ | 0.05 | ||
| Ribosomal subunit protein | VATVVAAPASQLAR | 677.4002+ | A-A′ C-C′ | −0.74 |
| IENALGEAVLSR | 636.3512+ | −1.22 | ||
| LLGLLNAPATR | 569.8542+ | −1.54 | ||
| SAGETGQLYGSVAAR | 733.8652+ | −1.46 | ||
| TLPEFSPGDTLR | 666.8432+ | 0.82 | ||
| SLATLPSLDELR | 657.8662+ | 1.75 | ||
| Oxidoreductase | ATGNYEQALADFIAR | 820.4072+ | C-C′ | 2.26 |
| TVVITAAGQGIGR | 621.8652+ | 1.96 | ||
| Superoxide dismutase | AFELPELPYDYDALAPYMSR | 1181.0662+ | C-C’ | 2.01 |
| MAFELPELPYDYDALAPYMSR | 1246.5792+ | 1.18 | ||
| DNA-binding protein | NPSTGAEVDIPAR | 663.8362+ | D | >10 |
| LAGFGSFSVSR | 564.2952+ | >10 | ||
| GRNPSTGAEVDIPAR | 770.3972+ | >10 | ||
| MNKNELVSAVAEK | 716.8772+ | >10 |
| Peptide | Sequence | Origin | References |
|---|---|---|---|
| Ascaphin-8 | GFKDLLKGAAKALVKTVLF-NH2 | Frog | [37] |
| Lycotoxin I | IWLTALKFLGKHAAKHLAKQQLSKL | Spider | |
| Maculatin 1.3 | GLLGLLGSVVSHVVPAIVGHF-NH2 | Frog | |
| Piscidin 1 | FFHHIFRGIVHVGKTIHRLVTG | Fish |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabri, M.; El Handi, K.; Calvano, C.D.; Bianco, M.; De Stradis, A.; Elbeaino, T. Synergistic Biocontrol of Agrobacterium tumefaciens by Phage PAT1 and Ascaphin-8: Enhanced Antimicrobial Activity and Virulence Attenuation via HupB Loss. Int. J. Mol. Sci. 2025, 26, 9355. https://doi.org/10.3390/ijms26199355
Sabri M, El Handi K, Calvano CD, Bianco M, De Stradis A, Elbeaino T. Synergistic Biocontrol of Agrobacterium tumefaciens by Phage PAT1 and Ascaphin-8: Enhanced Antimicrobial Activity and Virulence Attenuation via HupB Loss. International Journal of Molecular Sciences. 2025; 26(19):9355. https://doi.org/10.3390/ijms26199355
Chicago/Turabian StyleSabri, Miloud, Kaoutar El Handi, Cosima Damiana Calvano, Mariachiara Bianco, Angelo De Stradis, and Toufic Elbeaino. 2025. "Synergistic Biocontrol of Agrobacterium tumefaciens by Phage PAT1 and Ascaphin-8: Enhanced Antimicrobial Activity and Virulence Attenuation via HupB Loss" International Journal of Molecular Sciences 26, no. 19: 9355. https://doi.org/10.3390/ijms26199355
APA StyleSabri, M., El Handi, K., Calvano, C. D., Bianco, M., De Stradis, A., & Elbeaino, T. (2025). Synergistic Biocontrol of Agrobacterium tumefaciens by Phage PAT1 and Ascaphin-8: Enhanced Antimicrobial Activity and Virulence Attenuation via HupB Loss. International Journal of Molecular Sciences, 26(19), 9355. https://doi.org/10.3390/ijms26199355









