Non-Canonical Compartmentalization of DROSHA Protein at the Golgi Apparatus: miRNA Biogenesis-Independent Functionality in Human Cancer Cells of Diverse Tissue Origin
Abstract
1. Introduction
2. Results
2.1. Non-Canonical DROSHA Distribution in the Cytoplasm of Human Immortalized Cells During Interphase
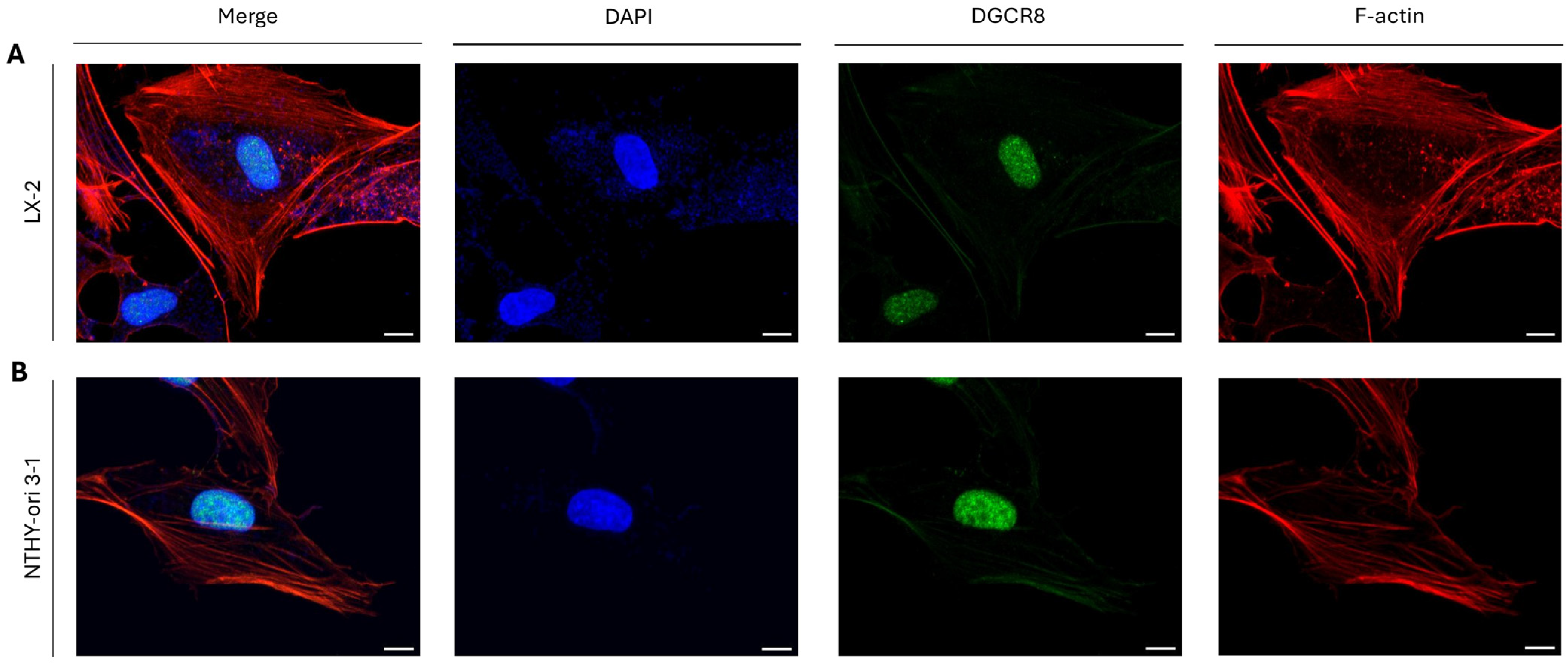
2.2. DGCR8-Independent Role(s) of DROSHA Protein in the Cytoplasm of Immortalized Cells at the Interphase Stage
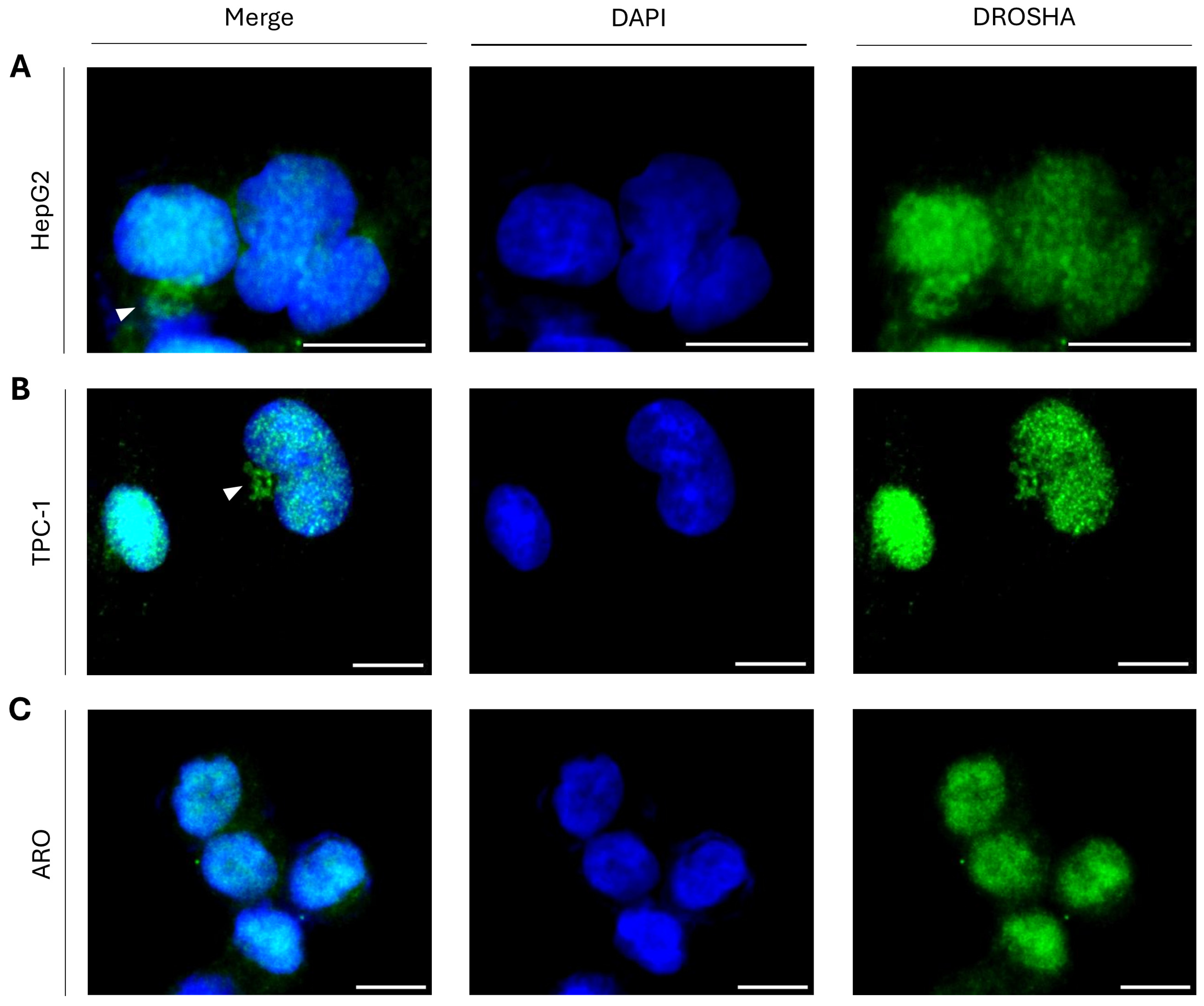
2.3. DROSHA Compartmentalization in the Cytoplasm of Liver and Thyroid Cancer Cells During Interphase
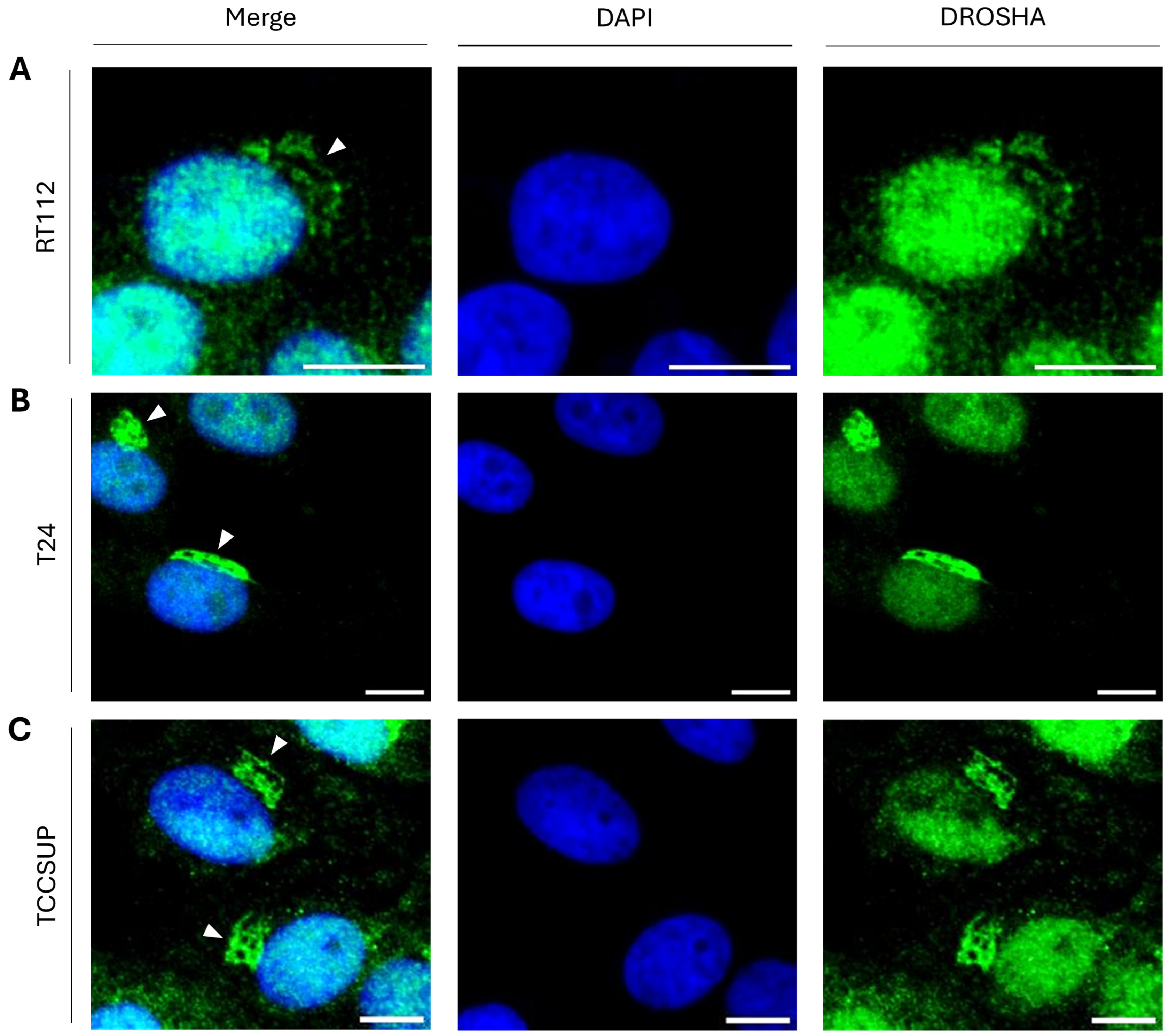
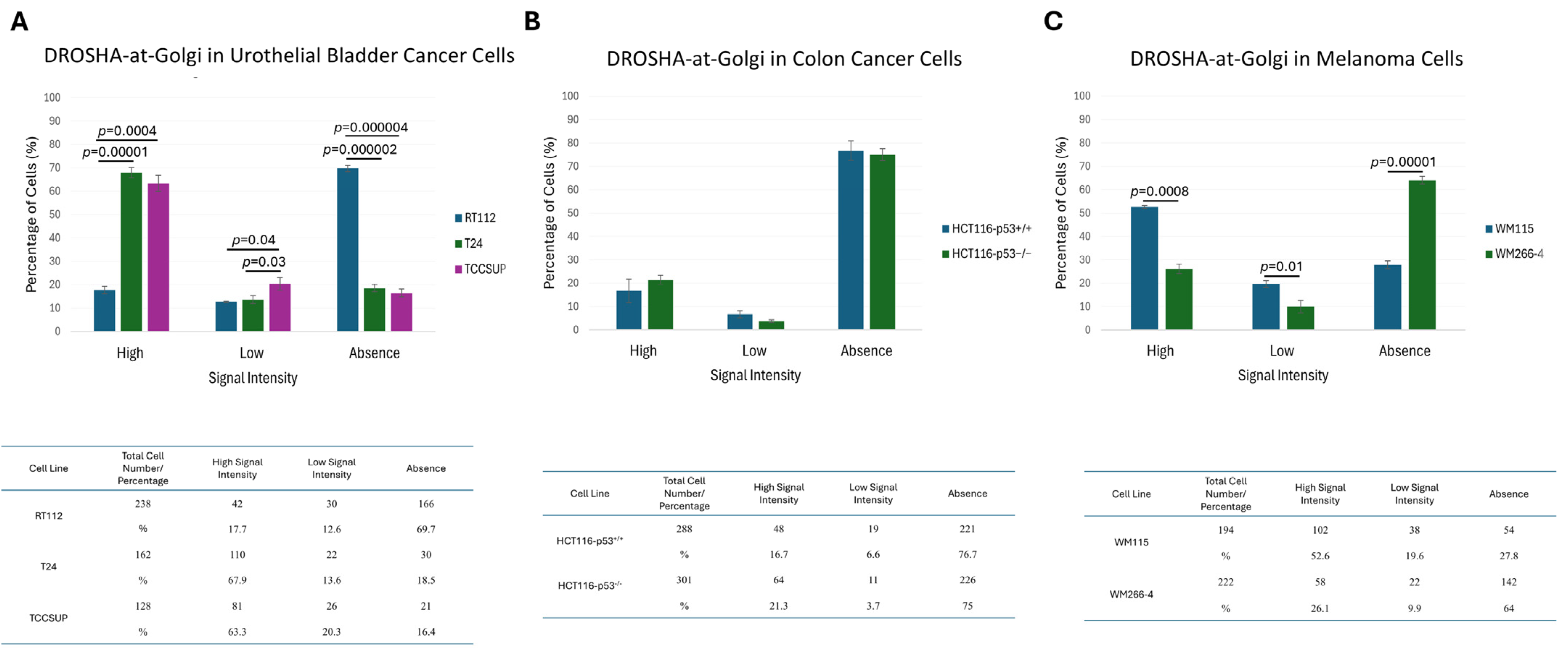
2.4. Formation of “DROSHA-Bodies” in the Cytoplasm of Human Urothelial Bladder Cancer Cells: A Phenotypic Universality Unmasking
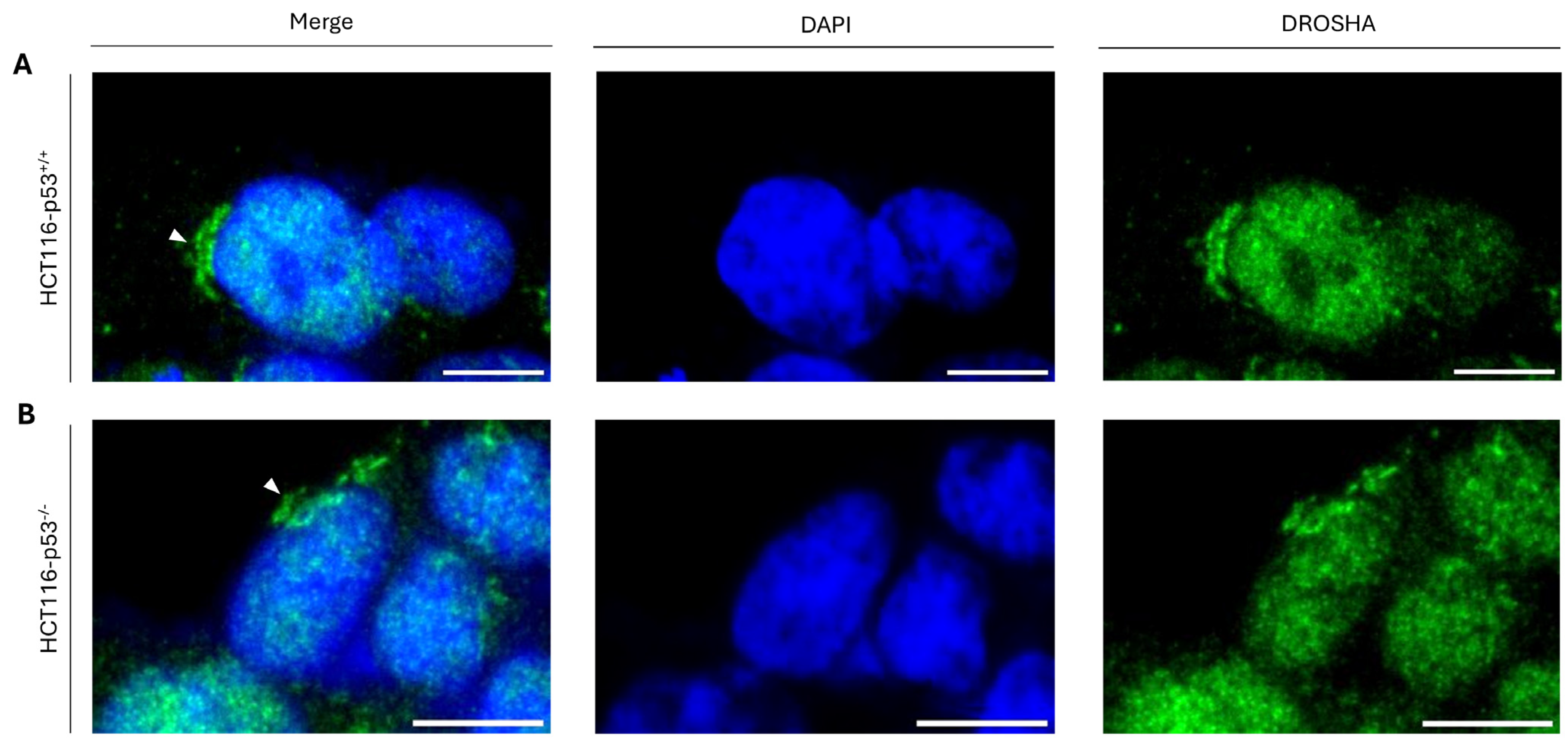
2.5. Cytoplasmic Accumulation of DROSHA Does Not Require the p53 Protein in Colon Cancer Cells
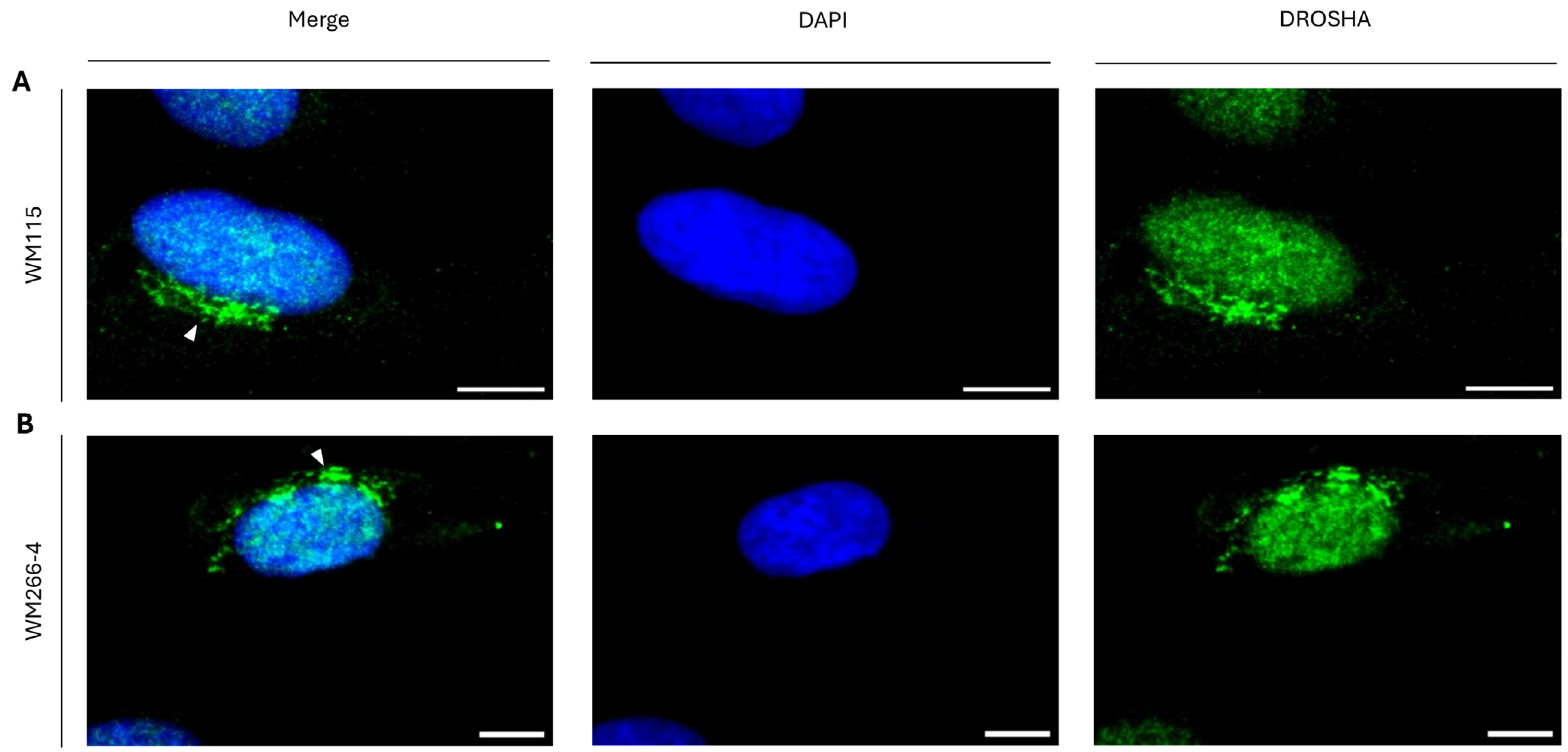
2.6. Metastasis-Independent Assembly of Cytoplasmic “DROSHA-Bodies” in Human Melanoma Cells
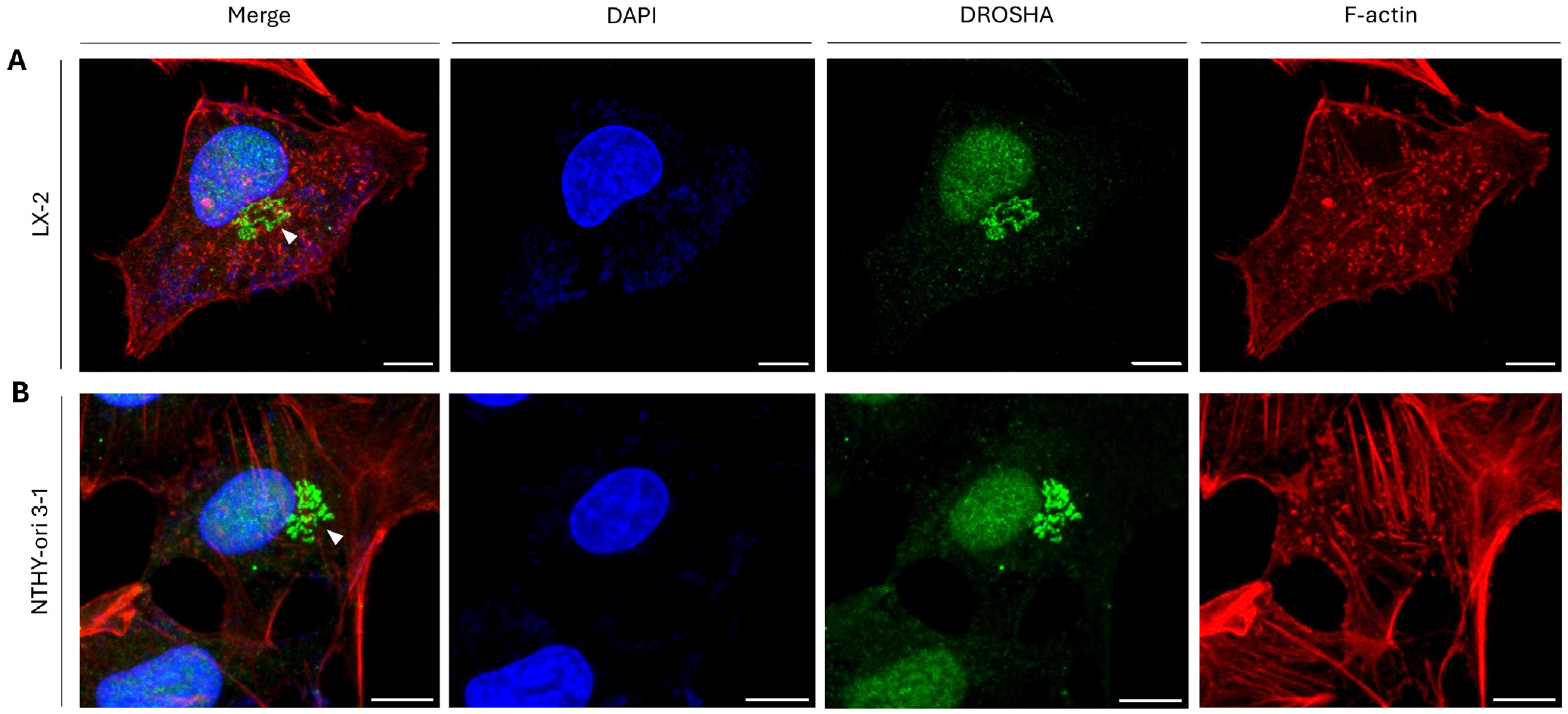
2.7. DROSHA Compartmentalization at the Golgi Apparatus in Human Immortalized and Cancer Cells During Interphase
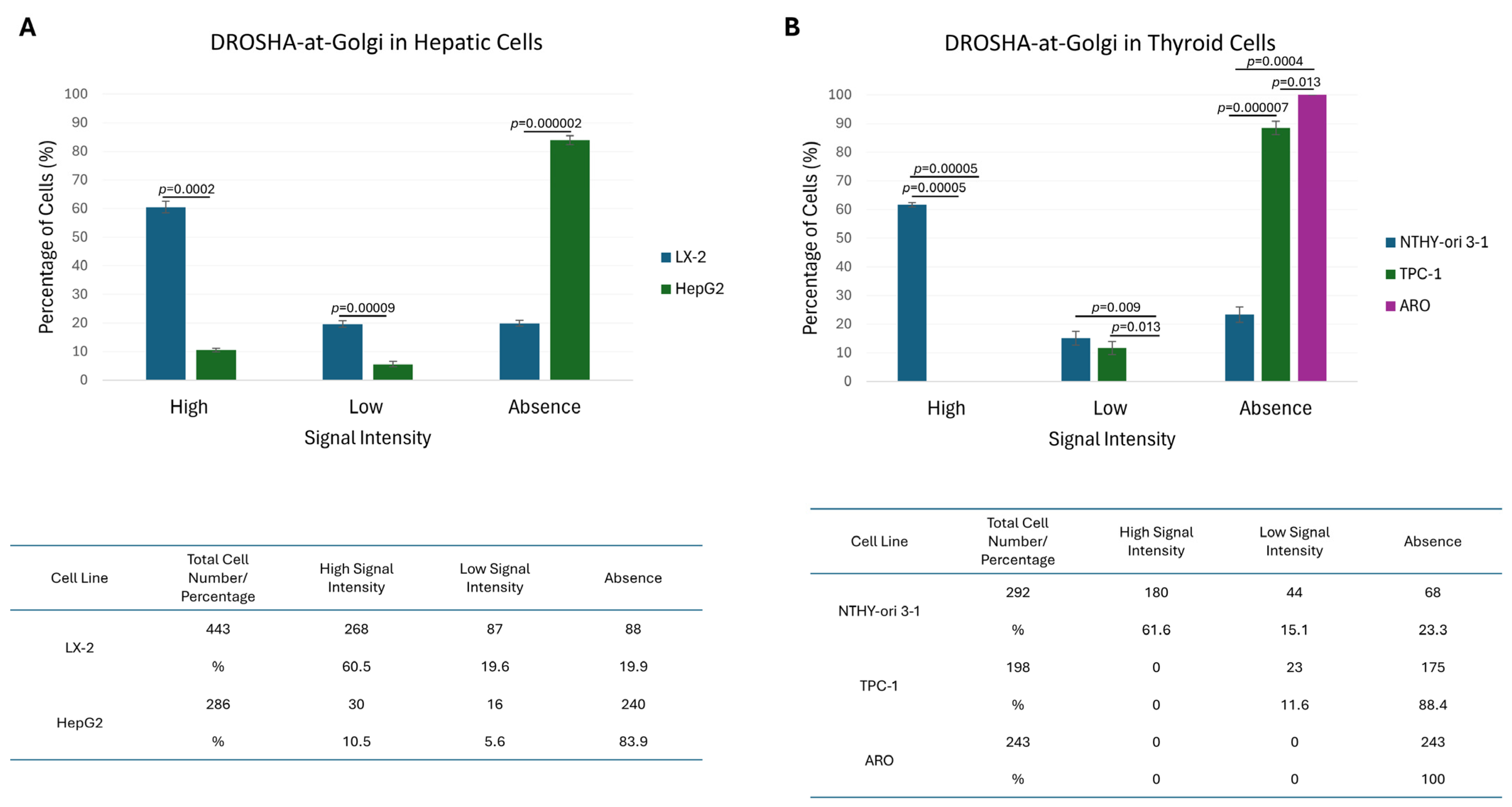
2.8. Immunophenotypic Variations of Golgi Apparatus-Containing DROSHA Protein in Human Immortalized Cells
2.9. “DROSHA-At-Golgi” Is Being Unveiled as a Novel, Compelling Biomarker for Human Malignancies in an Oncogenic Signature-Specific Manner
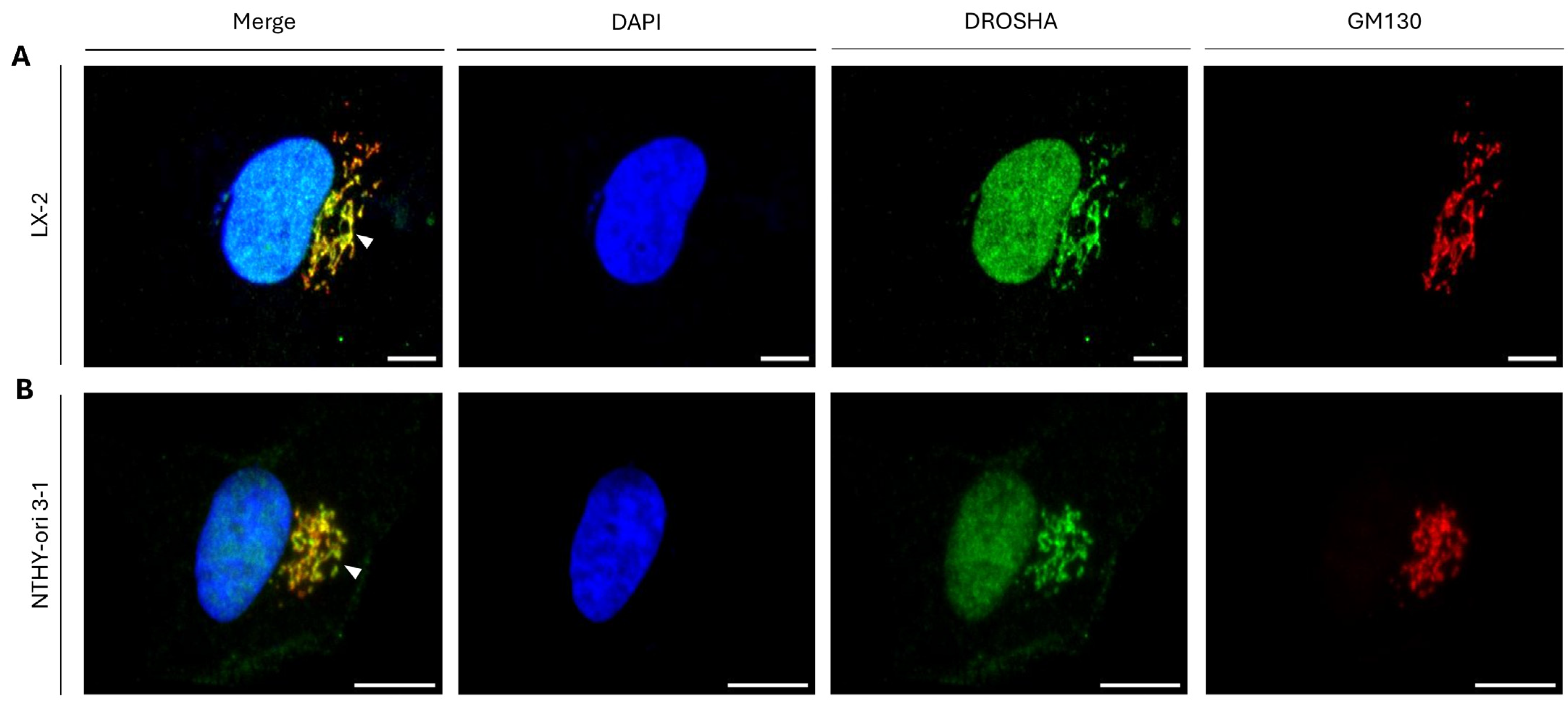
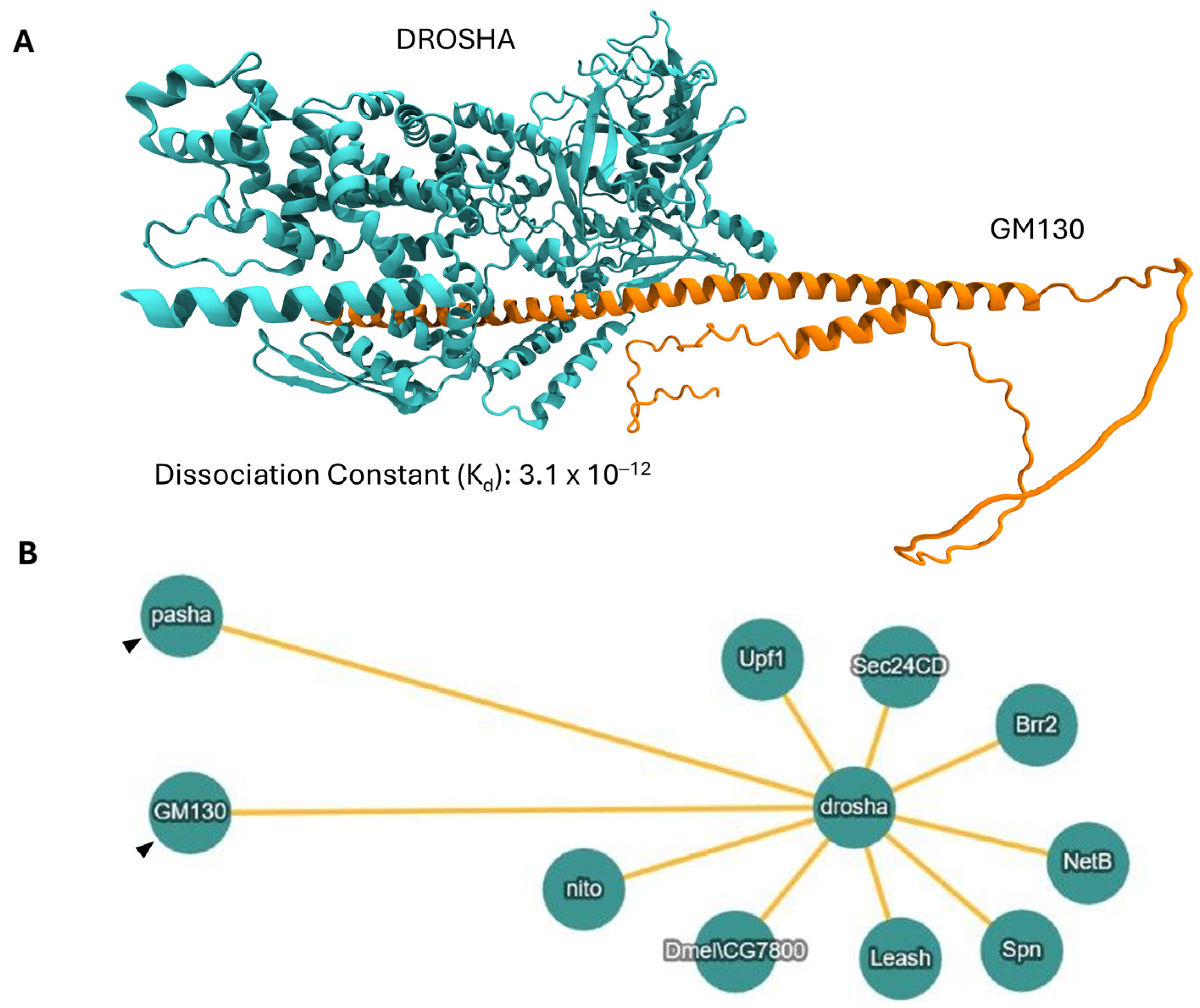
2.10. DROSHA/GM130 Protein/Protein Interaction (PPI): A Molecular Mechanism for DROSHA’s Compartmentalization at the Golgi Apparatus
2.11. Impact of DROSHA Gene Alterations in Diverse Human Malignancies
3. Discussion
4. Materials and Methods
4.1. Cell Lines—Cell Cultures
4.2. Transfection
4.3. Immunofluorescence
4.4. Confocal Laser Scanning Microscopy
4.5. Molecular Modeling
4.6. Morphometric Measurements—Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lett, K.E.; Logan, M.K.; McLaurin, D.M.; Hebert, M.D. Coilin enhances phosphorylation and stability of DGCR8 and promotes miRNA biogenesis. Mol. Biol. Cell 2021, 32, br4. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Partin, A.C.; Ngo, T.D.; Herrell, E.; Jeong, B.C.; Hon, G.; Nam, Y. Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs. Nat. Commun. 2017, 8, 1737. [Google Scholar] [CrossRef]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Theotoki, E.I.; Kakoulidis, P.; Velentzas, A.D.; Nikolakopoulos, K.S.; Angelis, N.V.; Tsitsilonis, O.E.; Anastasiadou, E.; Stravopodis, D.J. TRBP2, a Major Component of the RNAi Machinery, Is Subjected to Cell Cycle-Dependent Regulation in Human Cancer Cells of Diverse Tissue Origin. Cancers 2024, 16, 3701. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Bose, M.; Chatterjee, S.; Chakrabarty, Y.; Barman, B.; Bhattacharyya, S.N. Retrograde trafficking of Argonaute 2 acts as a rate-limiting step for de novo miRNP formation on endoplasmic reticulum-attached polysomes in mammalian cells. Life Sci. Alliance 2020, 3, e201800161. [Google Scholar] [CrossRef]
- Filippov, V.; Solovyev, V.; Filippova, M.; Gill, S.S. A novel type of RNase III family proteins in eukaryotes. Gene 2000, 245, 213–221. [Google Scholar] [CrossRef]
- Wu, H.; Xu, H.; Miraglia, L.J.; Crooke, S.T. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000, 275, 36957–36965. [Google Scholar] [CrossRef]
- Fortin, K.R.; Nicholson, R.H.; Nicholson, A.W. Mouse ribonuclease III. cDNA structure, expression analysis, and chromosomal location. BMC Genom. 2002, 3, 26. [Google Scholar] [CrossRef]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Nam, J.W.; Heo, I.; Rhee, J.K.; Sohn, S.Y.; Cho, Y.; Zhang, B.T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Lee, Y.; Han, J.; Yeom, K.H.; Jin, H.; Kim, V.N. Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 51–57. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef] [PubMed]
- Shiohama, A.; Sasaki, T.; Noda, S.; Minoshima, S.; Shimizu, N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 2003, 304, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 1996, 65, 113–132. [Google Scholar] [CrossRef]
- Yeom, K.H.; Lee, Y.; Han, J.; Suh, M.R.; Kim, V.N. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006, 34, 4622–4629. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, Y.; Tucker, L.; Ramratnam, B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010, 38, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Chang, H.M.; Lapierre, R.J.; Gregory, R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef]
- Kim, B.; Jeong, K.; Kim, V.N. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol. Cell 2017, 66, 258–269.e5. [Google Scholar] [CrossRef]
- Knuckles, P.; Vogt, M.A.; Lugert, S.; Milo, M.; Chong, M.M.; Hautbergue, G.M.; Wilson, S.A.; Littman, D.R.; Taylor, V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012, 15, 962–969. [Google Scholar] [CrossRef]
- Johanson, T.M.; Keown, A.A.; Cmero, M.; Yeo, J.H.; Kumar, A.; Lew, A.M.; Zhan, Y.; Chong, M.M. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat. Immunol. 2015, 16, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Heras, S.R.; Macias, S.; Plass, M.; Fernandez, N.; Cano, D.; Eyras, E.; Garcia-Perez, J.L.; Caceres, J.F. The Microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 2013, 20, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Gromak, N.; Dienstbier, M.; Macias, S.; Plass, M.; Eyras, E.; Caceres, J.F.; Proudfoot, N.J. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013, 5, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Reich, A.A.; Hastings, M.L. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014, 10, e1004312. [Google Scholar] [CrossRef]
- Lee, D.; Nam, J.W.; Shin, C. DROSHA targets its own transcript to modulate alternative splicing. RNA 2017, 23, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Aguado, L.C.; Schmid, S.; May, J.; Sabin, L.R.; Panis, M.; Blanco-Melo, D.; Shim, J.V.; Sachs, D.; Cherry, S.; Simon, A.E.; et al. RNase III nucleases from diverse kingdoms serve as antiviral effectors. Nature 2017, 547, 114–117. [Google Scholar] [CrossRef]
- Shapiro, J.S.; Schmid, S.; Aguado, L.C.; Sabin, L.R.; Yasunaga, A.; Shim, J.V.; Sachs, D.; Cherry, S.; tenOever, B.R. Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. USA 2014, 111, 7108–7113. [Google Scholar] [CrossRef]
- Thomson, J.M.; Newman, M.; Parker, J.S.; Morin-Kensicki, E.M.; Wright, T.; Hammond, S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes. Dev. 2006, 20, 2202–2207. [Google Scholar] [CrossRef]
- Stravopodis, D.J.; Karkoulis, P.K.; Konstantakou, E.G.; Melachroinou, S.; Thanasopoulou, A.; Aravantinos, G.; Margaritis, L.H.; Anastasiadou, E.; Voutsinas, G.E. Thymidylate synthase inhibition induces p53-dependent and p53-independent apoptotic responses in human urinary bladder cancer cells. J. Cancer Res. Clin. Oncol. 2011, 137, 359–374. [Google Scholar] [CrossRef]
- Zhang, Y.; Seemann, J. RNA scaffolds the Golgi ribbon by forming condensates with GM130. Nat. Cell Biol. 2024, 26, 1139–1153. [Google Scholar] [CrossRef]
- Nakamura, N. Emerging new roles of GM130, a cis-Golgi matrix protein, in higher order cell functions. J. Pharmacol. Sci. 2010, 112, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]
- Czubak, K.; Lewandowska, M.A.; Klonowska, K.; Roszkowski, K.; Kowalewski, J.; Figlerowicz, M.; Kozlowski, P. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget 2015, 6, 23399–23416. [Google Scholar] [CrossRef] [PubMed]
- Shirai, R.; Yamauchi, J. Emerging Evidence of Golgi Stress Signaling for Neuropathies. Neurol. Int. 2024, 16, 334–348. [Google Scholar] [CrossRef]
- Li, Y.; Mu, L.; Li, Y.; Mi, Y.; Hu, Y.; Li, X.; Tao, D.; Qin, J. Golgi dispersal in cancer stem cells promotes chemoresistance of colorectal cancer via the Golgi stress response. Cell Death Dis. 2024, 15, 417. [Google Scholar] [CrossRef]
- Wang, J.; Niu, S.; Hu, X.; Li, T.; Liu, S.; Tu, Y.; Shang, Z.; Zhao, L.; Xu, P.; Lin, J.; et al. Trans-Golgi network tethering factors regulate TBK1 trafficking and promote the STING-IFN-I pathway. Cell Discov. 2025, 11, 23. [Google Scholar] [CrossRef]
- Maruntelu, I.; Constantinescu, A.E.; Covache-Busuioc, R.A.; Constantinescu, I. The Golgi Apparatus: A Key Player in Innate Immunity. Int. J. Mol. Sci. 2024, 25, 4120. [Google Scholar] [CrossRef]
- Son, S.; Kim, B.; Yang, J.; Kim, V.N. Role of the proline-rich disordered domain of DROSHA in intronic microRNA processing. Genes. Dev. 2023, 37, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jimenez-Garcia, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Wojdyr, M. GEMMI: A library for structural biology. J. Open Source Softw. 2022, 7, 4200. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Thornton, J.M. PDBsum extras: SARS-CoV-2 and AlphaFold models. Protein Sci. 2022, 31, 283–289. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Jiang, W.; Wu, C.; Chen, T.; Chen, J.; Ding, X.; Zheng, S.; Piatkevich, K.D.; Zhu, Y.; Guo, T. Spatial proteomics of single cells and organelles on tissue slides using filter-aided expansion proteomics. Nat. Commun. 2024, 15, 9378. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Peng, D.; Todorova, V.; McCarthy, F.; Kim, K.; Liu, C.; Savy, L.; Januel, C.; Baltazar-Nunez, R.; Sekhar, M.; et al. Global organelle profiling reveals subcellular localization and remodeling at proteome scale. Cell 2025, 188, 1137–1155.e20. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theotoki, E.I.; Kakoulidis, P.; Papavassiliou, K.A.; Nikolakopoulos, K.-S.; Vlachou, E.N.; Basdra, E.K.; Papavassiliou, A.G.; Tsitsilonis, O.E.; Voutsinas, G.E.; Velentzas, A.D.; et al. Non-Canonical Compartmentalization of DROSHA Protein at the Golgi Apparatus: miRNA Biogenesis-Independent Functionality in Human Cancer Cells of Diverse Tissue Origin. Int. J. Mol. Sci. 2025, 26, 9319. https://doi.org/10.3390/ijms26199319
Theotoki EI, Kakoulidis P, Papavassiliou KA, Nikolakopoulos K-S, Vlachou EN, Basdra EK, Papavassiliou AG, Tsitsilonis OE, Voutsinas GE, Velentzas AD, et al. Non-Canonical Compartmentalization of DROSHA Protein at the Golgi Apparatus: miRNA Biogenesis-Independent Functionality in Human Cancer Cells of Diverse Tissue Origin. International Journal of Molecular Sciences. 2025; 26(19):9319. https://doi.org/10.3390/ijms26199319
Chicago/Turabian StyleTheotoki, Eleni I., Panos Kakoulidis, Kostas A. Papavassiliou, Konstantinos-Stylianos Nikolakopoulos, Eleni N. Vlachou, Efthimia K. Basdra, Athanasios G. Papavassiliou, Ourania E. Tsitsilonis, Gerassimos E. Voutsinas, Athanassios D. Velentzas, and et al. 2025. "Non-Canonical Compartmentalization of DROSHA Protein at the Golgi Apparatus: miRNA Biogenesis-Independent Functionality in Human Cancer Cells of Diverse Tissue Origin" International Journal of Molecular Sciences 26, no. 19: 9319. https://doi.org/10.3390/ijms26199319
APA StyleTheotoki, E. I., Kakoulidis, P., Papavassiliou, K. A., Nikolakopoulos, K.-S., Vlachou, E. N., Basdra, E. K., Papavassiliou, A. G., Tsitsilonis, O. E., Voutsinas, G. E., Velentzas, A. D., Anastasiadou, E., & Stravopodis, D. J. (2025). Non-Canonical Compartmentalization of DROSHA Protein at the Golgi Apparatus: miRNA Biogenesis-Independent Functionality in Human Cancer Cells of Diverse Tissue Origin. International Journal of Molecular Sciences, 26(19), 9319. https://doi.org/10.3390/ijms26199319









