Mangrove-Derived Endophytic Bacteria Enhance Growth, Yield, and Stress Resilience in Rice
Abstract
1. Introduction
2. Results
2.1. Mangrove Bacteria Enhance the Resilience of Arabidopsis thaliana to Salinity Under Waterlogging Conditions
2.2. Mangrove Bacteria Enhance the Resilience of O. sativa cv. Nipponbare Using Hydroponics
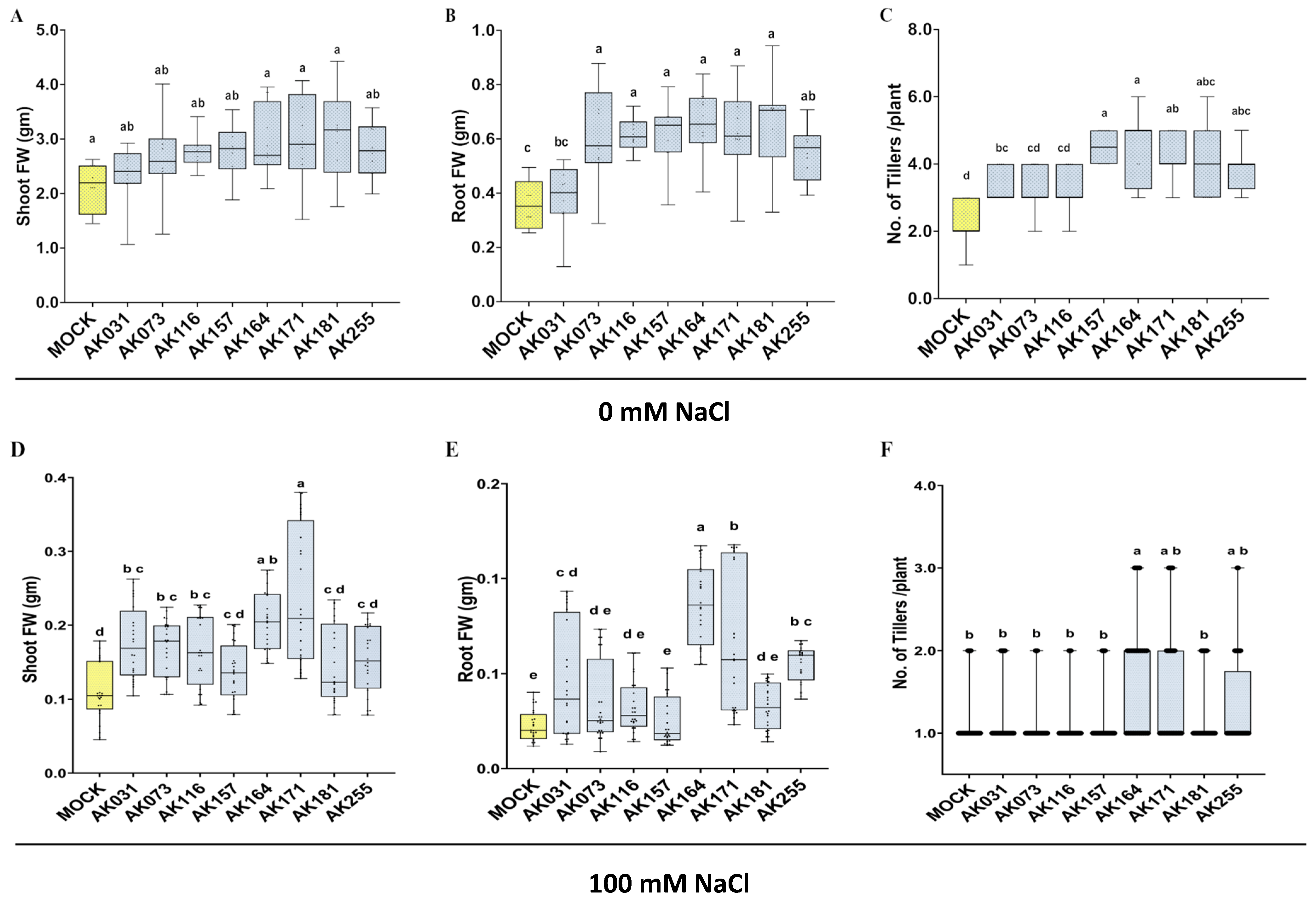
2.3. AK164, AK171, and BiCom Enhance Rice Growth Under Hydroponic Normal and Salt Stress Conditions
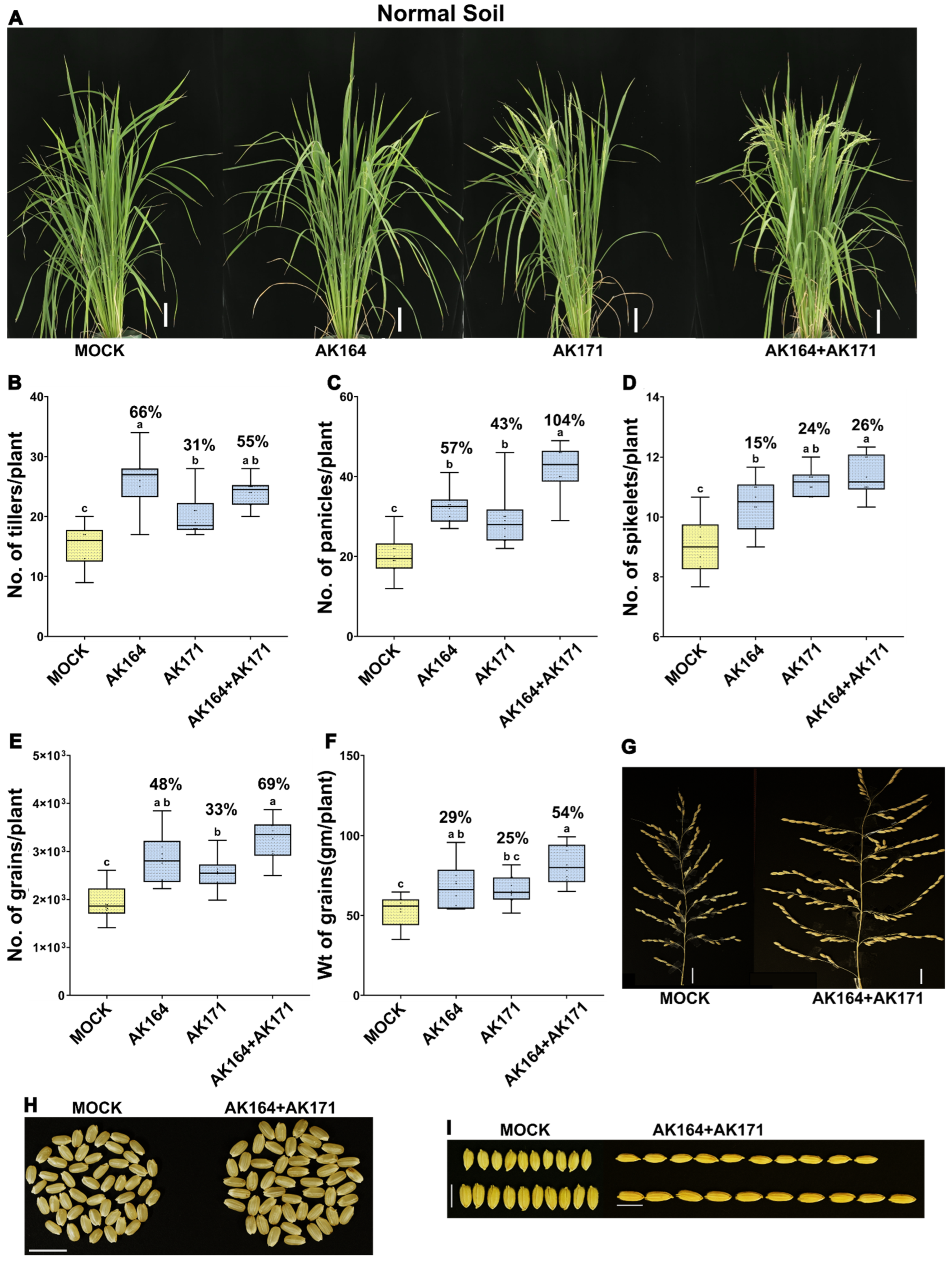
2.4. AK164 and AK171 Enhance Soil-Grown Rice Performance and Yield
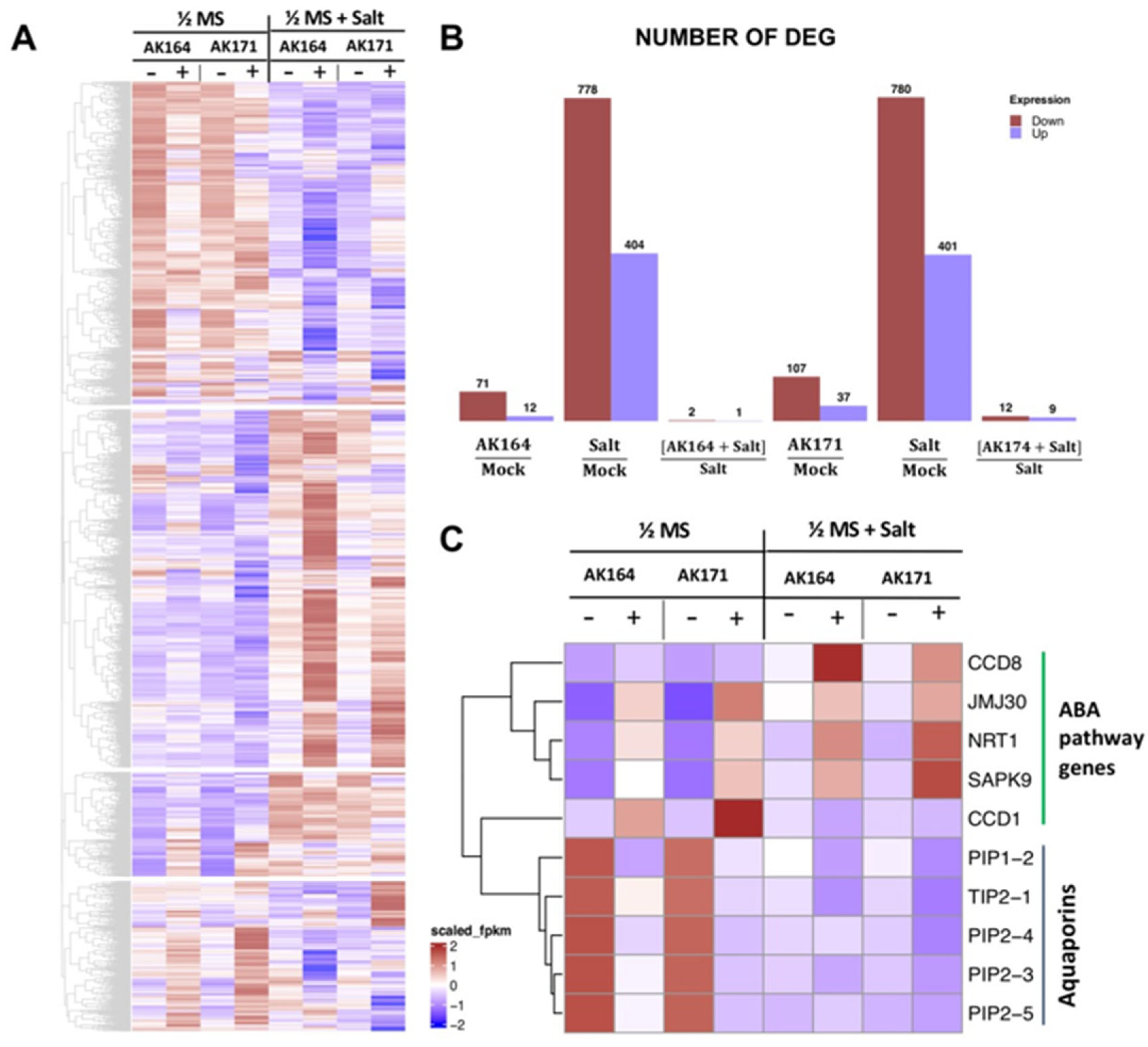
2.5. AK164 and AK171 Induce Upregulation of Rice ABA and Carotenoid Signaling Pathways
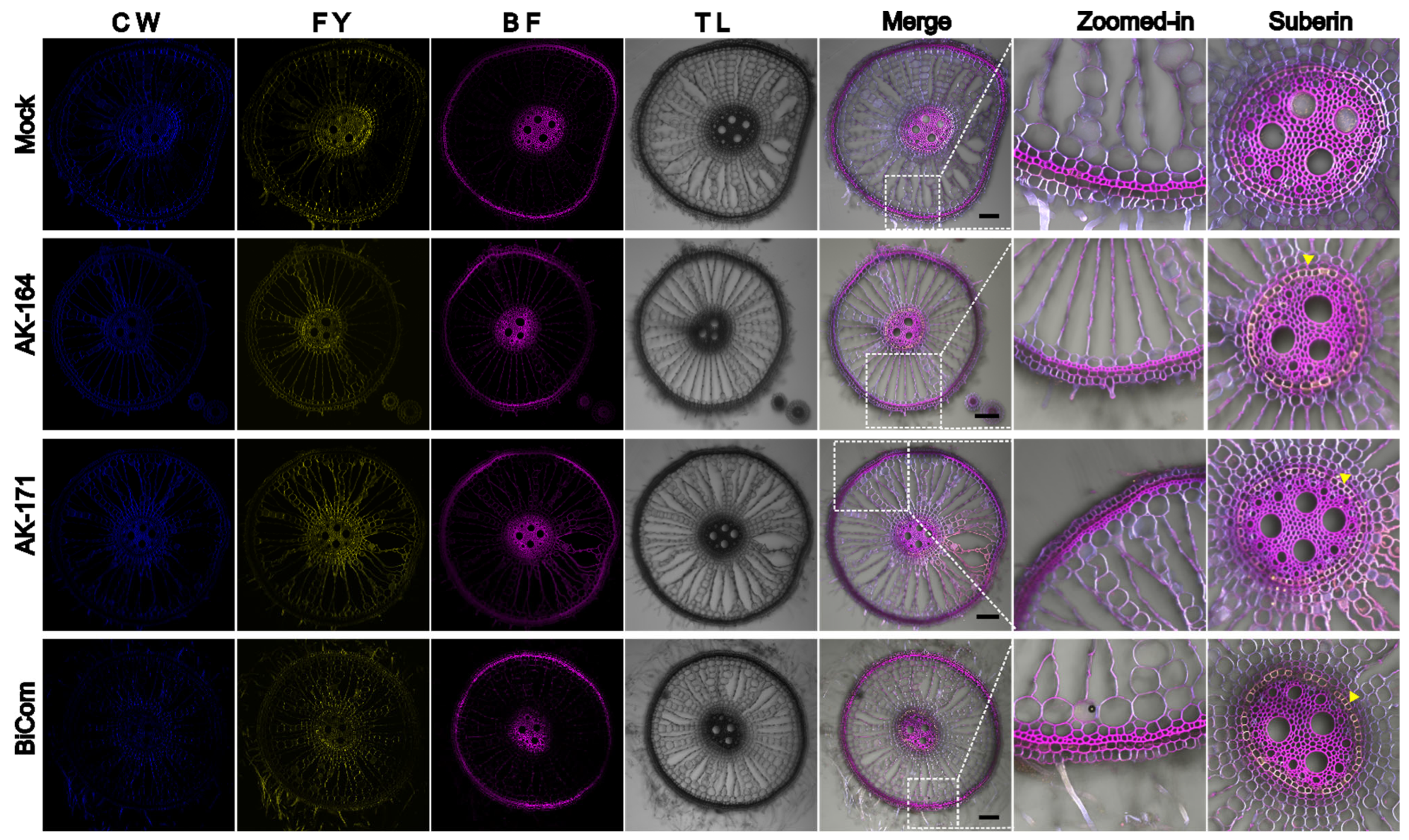
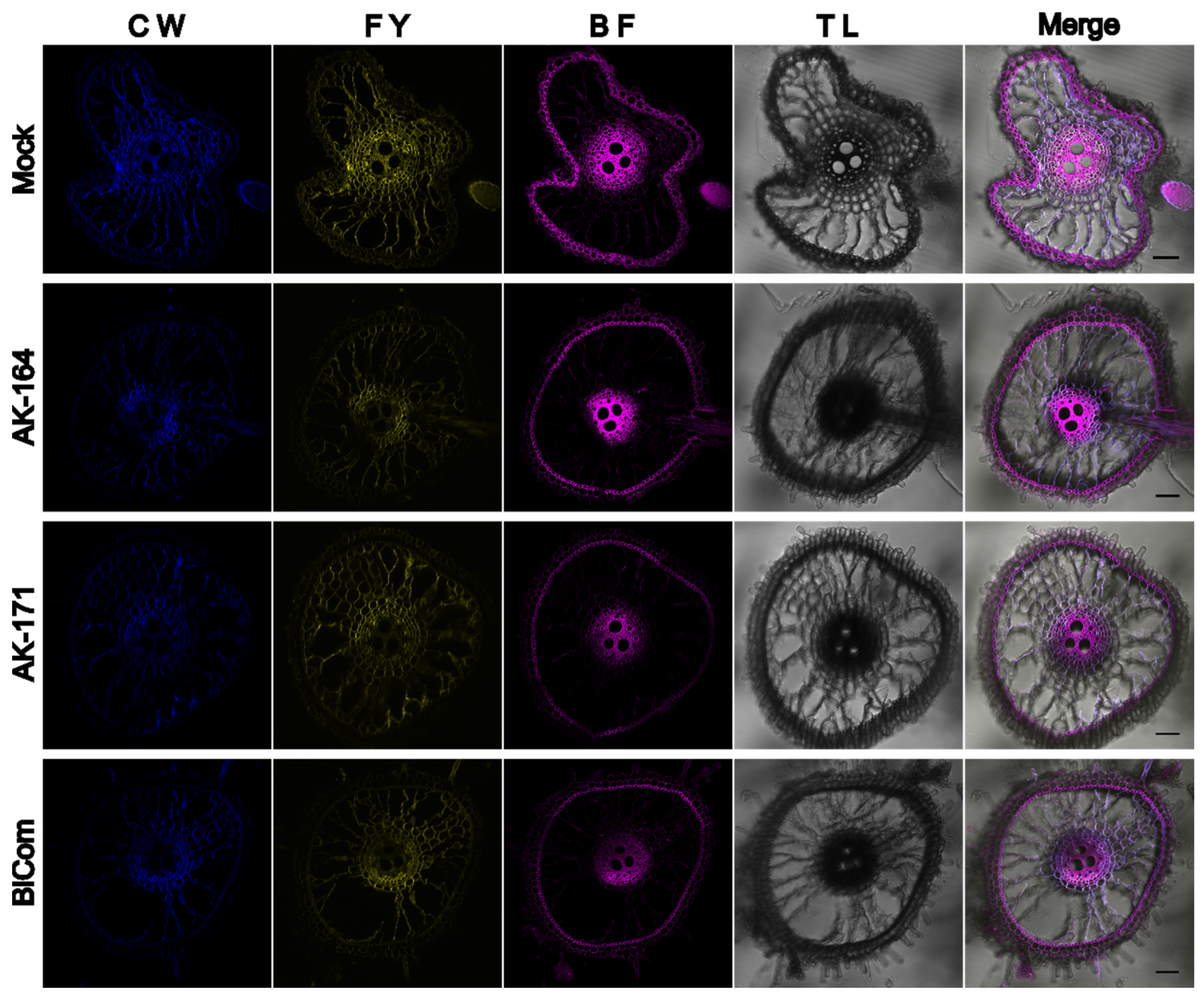
2.6. AK164 and AK171 Promote the Deposition of Secondary Cell Wall Components in Colonized Rice
3. Discussion
4. Methods
4.1. Bacterial Culture Conditions
4.2. Plant Material and Growth Conditions
4.3. Hydroponic Growth Conditions of Oryza sativa cv. Nipponbare
4.4. Greenhouse Experiments
4.5. RNA-Seq Library Preparation and Transcriptome Analysis
4.6. Histochemical Staining of Rice Roots
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. Leveraging Food Systems for Inclusive Rural Transformation; FAO: Rome, Italy, 2017; p. 181. [Google Scholar]
- FAO. FAOSTAT. Online Statistical Database; FAO: Rome, Italy, 2017; Available online: https://www.fao.org/statistics/en (accessed on 26 August 2025).
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef]
- Hsu, F.C.; Chou, M.Y.; Peng, H.P.; Chou, S.J.; Shih, M.C. Insights into hypoxic systemic responses based on analyses of transcriptional regulation in Arabidopsis. PLoS ONE 2011, 6, e28888. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.B.; Ram, P.C. Physiological and molecular basis of susceptibility and tolerance of rice plants to complete submergence. Ann. Bot. 2003, 91, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Yeung, E.; van Veen, H.; Vashisht, D.; Sobral Paiva, A.L.; Hummel, M.; Rankenberg, T.; Steffens, B.; Steffen-Heins, A.; Sauter, M.; de Vries, M.; et al. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2018, 115, E6085–E6094. [Google Scholar] [CrossRef]
- Tsai, K.J.; Lin, C.Y.; Ting, C.Y.; Shih, M.C. Ethylene-Regulated Glutamate Dehydrogenase Fine-Tunes Metabolism during Anoxia-Reoxygenation. Plant Physiol. 2016, 172, 1548–1562. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef] [PubMed]
- Mustroph, A.; Lee, S.C.; Oosumi, T.; Zanetti, M.E.; Yang, H.; Ma, K.; Yaghoubi-Masihi, A.; Fukao, T.; Bailey-Serres, J. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010, 152, 1484–1500. [Google Scholar] [CrossRef]
- Juntawong, P.; Hummel, M.; Bazin, J.; Bailey-Serres, J. Ribosome profiling: A tool for quantitative evaluation of dynamics in mRNA translation. Methods 2015, 1284, 139–173. [Google Scholar] [CrossRef]
- Tsuji, H.; Saika, H.; Tsutsumi, N.; Hirai, A.; Nakazono, M. Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 2006, 47, 995–1003. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Romano, S.; Vanni, F.; Viaggi, D. Economics of culture and food in evolving agri-food systems and rural areas. Bio-Based Appl. Econ. 2020, 9, 127–136. [Google Scholar] [CrossRef]
- Saad, M.M.; Eida, A.A.; Hirt, H. Tailoring plant-associated microbial inoculants in agriculture: A roadmap for successful application. J. Exp. Bot. 2020, 71, 3878–3901. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yuan, R.; Yang, S.; Dai, Z.; Di, N.; Yang, H.; He, Z.; Wei, M. A salt-tolerant growth-promoting phyllosphere microbial combination from mangrove plants and its mechanism for promoting salt tolerance in rice. Microbiome 2024, 12, 270. [Google Scholar] [CrossRef]
- Dai, Z.; Yuan, R.; Yang, X.; Xi, H.; Zhuo, M.; Wei, M. Salinity-responsive key endophytic bacteria in the propagules of Kandelia obovata enhance salt tolerance in rice. J. Integr. Agric. 2025, 24, 1738–1753. [Google Scholar] [CrossRef]
- Upadhyay, S.; Singh, J.; Singh, D. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Thakur, R.; Yadav, S. Biofilm forming, exopolysaccharide producing and halotolerant, bacterial consortium mitigates salinity stress in Triticum aestivum. Int. J. Biol. Macromol. 2024, 262, 130049. [Google Scholar] [CrossRef]
- Chen, E.; Yang, C.; Tao, W.; Li, S. Polysaccharides Produced by Plant Growth-Promoting Rhizobacteria Strain Burkholderia sp. BK01 Enhance Salt Stress Tolerance to Arabidopsis thaliana. Polymers 2024, 16, 145. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Shahid, M.; Singh, U.B.; Khan, M.S.; Singh, P.; Kumar, R.; Singh, R.N.; Kumar, A.; Singh, H.V. Bacterial ACC deaminase: Insights into enzymology, biochemistry, genetics, and potential role in amelioration of environmental stress in crop plants. Front. Microbiol. 2023, 14, 1132770. [Google Scholar] [CrossRef]
- Li, C.-H.; Wang, G.; Zhao, J.-L.; Zhang, L.-Q.; Ai, L.-F.; Han, Y.-F.; Sun, D.-Y.; Zhang, S.-W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef]
- Alghamdi, A.K.; Parween, S.; Hirt, H.; Saad, M.M. Complete genome sequence analysis of plant growth-promoting bacterium, Isoptericola sp. AK164 isolated from the rhizosphere of Avicennia marina growing at the Red Sea coast. Arch. Microbiol. 2023, 205, 307, Erratum in Arch. Microbiol. 2023, 205, 341. [Google Scholar] [CrossRef]
- Alghamdi, A.K.; Parween, S.; Hirt, H.; Saad, M.M. Unraveling the genomic secrets of Tritonibacter mobilis AK171: A plant growth-promoting bacterium isolated from Avicennia marina. BMC Genom. 2024, 25, 672. [Google Scholar] [CrossRef]
- Jhan, L.H.; Yang, C.Y.; Huang, C.M.; Lai, M.C.; Huang, Y.H.; Baiya, S.; Kao, C.F. Integrative pathway and network analysis provide insights on flooding-tolerance genes in soybean. Sci. Rep. 2023, 13, 1980. [Google Scholar] [CrossRef]
- Singh, T.; Bisht, N.; Ansari, M.M.; Chauhan, P.S. The hidden harmony: Exploring ROS-phytohormone nexus for shaping plant root architecture in response to environmental cues. Plant Physiol. Biochem. 2024, 206, 108273. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Sasidharan, R.; Hartman, S.; Liu, Z.; Martopawiro, S.; Sajeev, N.; Van Veen, H.; Yeung, E.; Voesenek, L.A.C.J. Signal dynamics and interactions during flooding stress. Plant Physiol. 2018, 176, 1106–1117. [Google Scholar] [CrossRef]
- Qin, S.; Zhang, Y.-J.; Yuan, B.; Xu, P.-Y.; Xing, K.; Wang, J.; Jiang, J.-H. Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil. 2014, 374, 753–766, Erratum in Plant and Soil. 2014, 378, 413–414. [Google Scholar] [CrossRef]
- Shayanthan, A.; Ordoñez, P.A.C.; Oresnik, I.J. The Role of Synthetic Microbial Communities (SynCom) in Sustainable Agriculture. Front. Agron. 2022, 4, 896307. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Zahir, Z.A.; Javaid, A.; Ashraf, M. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol. Adv. 2014, 32, 429–448. [Google Scholar] [CrossRef]
- Duarte, B.; Carreiras, J.A.; Cruz-Silva, A.; Mateos-Naranjo, E.; Rodríguez-Llorente, I.D.; Pajuelo, E.; Redondo-Gómez, S.; Mesa-Marín, J.; Figueiredo, A. Marine Plant Growth Promoting Bacteria (PGPB) inoculation technology: Testing the effectiveness of different application methods to improve tomato plants tolerance against acute heat wave stress. Plant Stress. 2024, 11, 100434. [Google Scholar] [CrossRef]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134, Erratum in Proc. Natl. Acad. Sci. USA 2015, 122, e2508245122. [Google Scholar] [CrossRef]
- Lu, S.X.; Knowles, S.M.; Webb, C.J.; Celaya, R.B.; Cha, C.; Siu, J.P.; Tobin, E.M. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011, 155, 906–915. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Singh, M.; Tripathi, R.D.; Trivedi, P.K. Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 2014, 4, 3964. [Google Scholar] [CrossRef]
- McClung, C.R. McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Seo, P.J. JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 2018, 95, 961–975. [Google Scholar] [CrossRef]
- Li, K.; Zhang, S.; Tang, S.; Zhang, J.; Dong, H.; Yang, S.; Qu, H.; Xuan, W.; Gu, M.; Xu, G. The rice transcription factor Nhd1 regulates root growth and nitrogen uptake by activating nitrogen transporters. Plant Physiol. 2022, 189, 1608–1624. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Samanta, M.K.; Gayen, S.; Maiti, M.K. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016, 16, 158. [Google Scholar] [CrossRef]
- Liu, X.; Li, Z.; Hou, Y.; Wang, Y.; Wang, H.; Tong, X.; Ao, H.; Zhang, J. Protein Interactomic Analysis of SAPKs and ABA-Inducible bZIPs Revealed Key Roles of SAPK10 in Rice Flowering. Int. J. Mol. Sci. 2019, 20, 1427. [Google Scholar] [CrossRef]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef]
- Vishal, B.; Krishnamurthy, P.; Ramamoorthy, R.; Kumar, P.P. OsTPS8 controls yield-related traits and confers salt stress tolerance in rice by enhancing suberin deposition. New Phytol. 2019, 221, 1369–1386. [Google Scholar] [CrossRef]
- Liu, C.; Yu, H.; Rao, X.; Li, L.; Dixon, R.A. Abscisic acid regulates secondary cell-wall formation and lignin deposition in Arabidopsis thaliana through phosphorylation of NST1. Proc. Natl. Acad. Sci. 2021, 118, e2010911118, Erratum in Proc. Natl. Acad. Sci. USA 2021, 118, e2106367118. https://doi.org/10.1073/pnas.2106367118. [Google Scholar] [CrossRef]
- Dickinson, A.J.; Lehner, K.; Mi, J.; Jia, K.P.; Mijar, M.; Dinneny, J.; Al-Babili, S.; Benfey, P.N. beta-Cyclocitral is a conserved root growth regulator. Proc. Natl. Acad. Sci. USA 2019, 116, 10563–10567. [Google Scholar] [CrossRef]
- Ko, M.R.; Song, M.H.; Kim, J.K.; Baek, S.A.; You, M.K.; Lim, S.H.; Ha, S.H. RNAi-mediated suppression of three carotenoid-cleavage dioxygenase genes, OsCCD1, 4a, and 4b, increases carotenoid content in rice. J. Exp. Bot. 2018, 69, 5105–5116. [Google Scholar] [CrossRef]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Toth, P.; Haider, I.; Pozo, M.J.; de Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Boursiac, Y.; Chen, S.; Luu, D.T.; Sorieul, M.; van den Dries, N.; Maurel, C. Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol. 2005, 139, 790–805. [Google Scholar] [CrossRef]
- Martinez-Ballesta, M.C.; Diaz, R.; Martinez, V.; Carvajal, M. Different blocking effects of HgCl2 and NaCl on aquaporins of pepper plants. J. Plant Physiol. 2003, 160, 1487–1492. [Google Scholar] [CrossRef]
- Merlaen, B.; De Keyser, E.; Ding, L.; Leroux, O.; Chaumont, F.; Van Labeke, M.C. Physiological responses and aquaporin expression upon drought and osmotic stress in a conservative vs prodigal Fragaria x ananassa cultivar. Plant Physiol. Biochem. 2019, 145, 95–106. [Google Scholar] [CrossRef]
- Qiao, Y.; Ren, J.; Yin, L.; Liu, Y.; Deng, X.; Liu, P.; Wang, S. Exogenous melatonin alleviates PEG-induced short-term water deficiency in maize by increasing hydraulic conductance. BMC Plant Biol. 2020, 20, 218. [Google Scholar] [CrossRef]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef]
- Zwiazek, J.J.; Xu, H.; Tan, X.; Navarro-Rodenas, A.; Morte, A. Significance of oxygen transport through aquaporins. Sci. Rep. 2017, 7, 40411. [Google Scholar] [CrossRef]
- Martre, P.; Morillon, R.; Barrieu, F.; North, G.B.; Nobel, P.S.; Chrispeels, M.J. Plasma membrane aquaporins play a significant role during recovery from water deficit. Plant Physiol. 2002, 130, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fukumoto, T.; Matsumoto, T.; Gena, P.; Frascaria, D.; Kaneko, T.; Katsuhara, M.; Zhong, S.; Sun, X.; Zhu, Y.; et al. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 2013, 63, 151–158. [Google Scholar] [CrossRef]
- Ding, L.; Uehlein, N.; Kaldenhoff, R.; Guo, S.; Zhu, Y.; Kai, L. Aquaporin PIP2;1 affects water transport and root growth in rice (Oryza sativa L.). Plant Physiol. Biochem. 2019, 139, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Bienert, M.D.; Diehn, T.A.; Richet, N.; Chaumont, F.; Bienert, G.P. Heterotetramerization of Plant PIP1 and PIP2 Aquaporins Is an Evolutionary Ancient Feature to Guide PIP1 Plasma Membrane Localization and Function. Front. Plant Sci. 2018, 9, 382. [Google Scholar] [CrossRef]
- Kudoyarova, G.; Veselov, D.; Yemelyanov, V.; Shishova, M. The Role of Aquaporins in Plant Growth under Conditions of Oxygen Deficiency. Int. J. Mol. Sci. 2022, 23, 10159. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Z.; Liu, F.; Sun, L.; Hao, F. Versatile Roles of Aquaporins in Plant Growth and Development. Int. J. Mol. Sci. 2020, 21, 9485. [Google Scholar] [CrossRef] [PubMed]
- Li, G.W.; Peng, Y.H.; Yu, X.; Zhang, M.H.; Cai, W.M.; Sun, W.N.; Su, W.A. Transport functions and expression analysis of vacuolar membrane aquaporins in response to various stresses in rice. J. Plant Physiol. 2008, 165, 1879–1888. [Google Scholar] [CrossRef]
- Alexandersson, E.; Fraysse, L.; Sjovall-Larsen, S.; Gustavsson, S.; Fellert, M.; Karlsson, M.; Johanson, U.; Kjellbom, P. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 2005, 59, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Wang, Z.Y.; Lin, H.; Cui, W.E.; Chen, J.; Liu, M.; Chen, Z.L.; Qu, L.J.; Gu, H. Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 2006, 16, 277–286. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Voesenek, L.A. Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 2008, 59, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef]
- Jang, J.Y.; Kim, D.G.; Kim, Y.O.; Kim, J.S.; Kang, H. An Expression Analysis of a Gene Family Encoding Plasma Membrane Aquaporins in Response to Abiotic Stresses in Arabidopsis Thaliana. Plant Mol. Biol. 2004, 54, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.L.; Yu, X.; Lane, D.; Sun, W.N.; Tang, Z.C.; Su, W.A. Upland rice and lowland rice exhibited different PIP expression under water deficit and ABA treatment. Cell Res. 2006, 16, 651–660. [Google Scholar] [CrossRef]
- Liu, Q.; Umeda, M.; Uchimiya, H. Isolation and expression analysis of two rice genes encoding the major intrinsic protein. Plant Mol. Biol. 1994, 26, 2003–2007. [Google Scholar] [CrossRef]
- Almahasheer, H.; Serrano, O.; Duarte, C.M.; Arias-Ortiz, A.; Masque, P.; Irigoien, X. Low Carbon sink capacity of Red Sea mangroves. Sci. Rep. 2017, 7, 9700. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. mprovement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Sexauer, M.; Shen, D.; Schon, M.; Andersen, T.G.; Markmann, K. Visualizing polymeric components that define distinct root barriers across plant lineages. Development 2021, 148, dev199820. [Google Scholar] [CrossRef] [PubMed]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
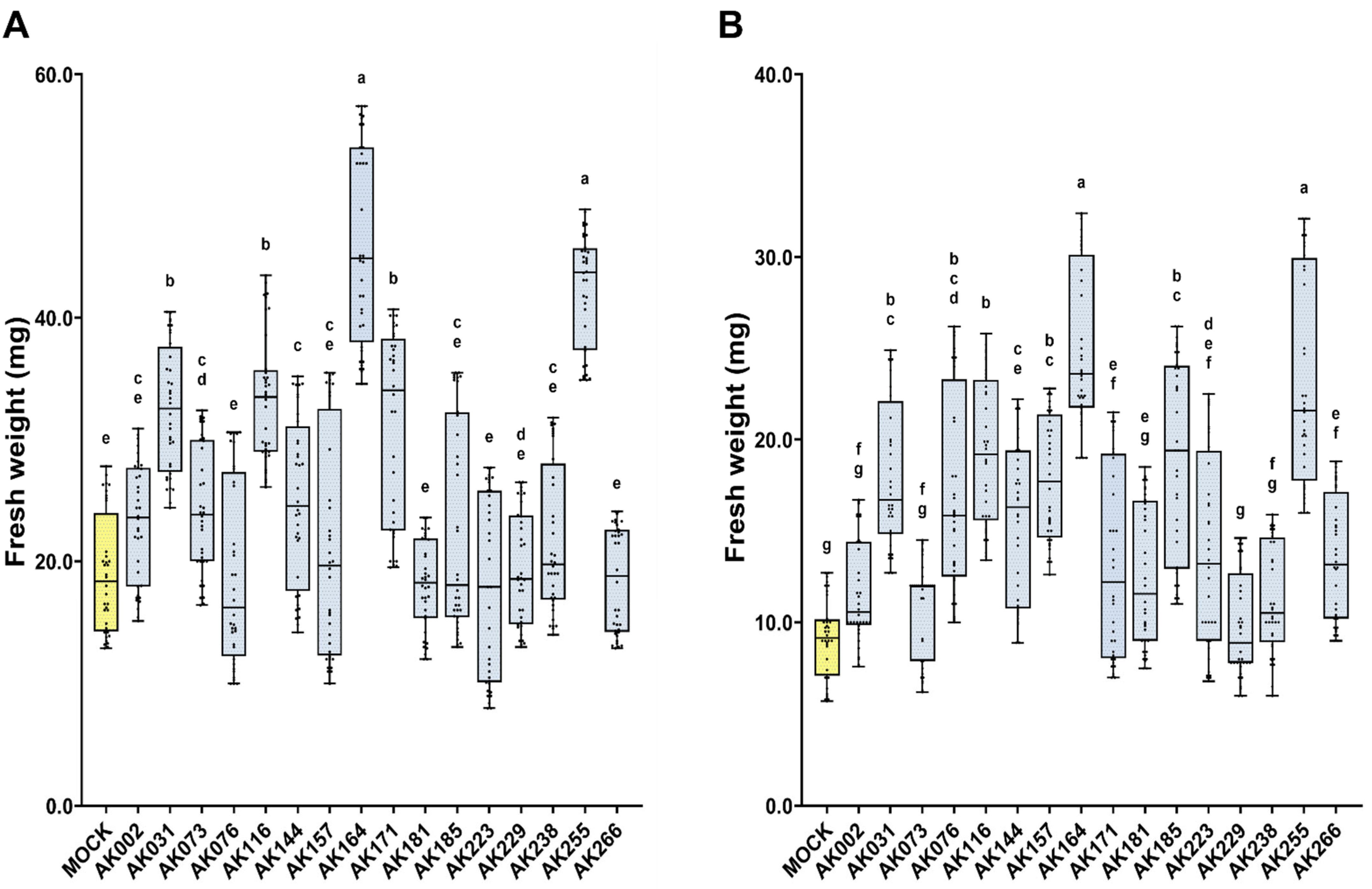


| No. | Strain | Name | ½ MS | ½ MS +100 mM NaCl |
|---|---|---|---|---|
| 1 | AK002 | Paenibacillus lautus (OR447938) | 9.5 | 21.8 |
| 2 | AK031 | Bacillus seohaeanensis (OR447774) | 45.8 **** | 96.1 *** |
| 3 | AK073 | Thalassospira tepidiphila (OR447979) | 12.8 | 24.4 |
| 4 | AK076 | Tritonibacter mobilis (OR447785) | −8.2 | 87.3 **** |
| 5 | AK116 | Halobacillus locisalis (OR447813) | 43.0 *** | 125.2 *** |
| 6 | AK144 | Microbulbifer elongatus (OR447909) | 17.5 * | 59.7 ** |
| 7 | AK157 | Demequina activiva (OR447779) | −1.9 | 78.5 **** |
| 8 | AK164 | Isoptericola chiayiensis (OR447867) | 107.0 **** | 165.3 **** |
| 9 | AK171 | T. mobilis (OR447787) | 35.2 * | 47.3 * |
| 10 | AK181 | Pseudomonas azotoformans (OR447934) | −20.7 | 57.4 * |
| 11 | AK185 | T. mobilis (OR447793) | 1.6 | 102.5 **** |
| 12 | AK223 | P. azotoformans (OR447928) | −10.1 | 58.0 * |
| 13 | AK229 | Martelella mangrovi (OR447916) | −3.1 | 11.2 |
| 14 | AK238 | Celerinatantimonas diazotrophica (OR447776) | 6.9 | 17.9 |
| 15 | AK255 | I. chiayiensis (OR447854) | 101.4 **** | 146.1 **** |
| 16 | AK266 | P. lautus (OR448017) | 9.5 | 48.7 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alghamdi, A.K.; Rawat, A.; Alzayed, W.; Parween, S.; Nagarajan, A.P.; Saad, M.M.; Hirt, H. Mangrove-Derived Endophytic Bacteria Enhance Growth, Yield, and Stress Resilience in Rice. Int. J. Mol. Sci. 2025, 26, 9317. https://doi.org/10.3390/ijms26199317
Alghamdi AK, Rawat A, Alzayed W, Parween S, Nagarajan AP, Saad MM, Hirt H. Mangrove-Derived Endophytic Bacteria Enhance Growth, Yield, and Stress Resilience in Rice. International Journal of Molecular Sciences. 2025; 26(19):9317. https://doi.org/10.3390/ijms26199317
Chicago/Turabian StyleAlghamdi, Amal Khalaf, Anamika Rawat, Waad Alzayed, Sabiha Parween, Arun Prasanna Nagarajan, Maged M. Saad, and Heribert Hirt. 2025. "Mangrove-Derived Endophytic Bacteria Enhance Growth, Yield, and Stress Resilience in Rice" International Journal of Molecular Sciences 26, no. 19: 9317. https://doi.org/10.3390/ijms26199317
APA StyleAlghamdi, A. K., Rawat, A., Alzayed, W., Parween, S., Nagarajan, A. P., Saad, M. M., & Hirt, H. (2025). Mangrove-Derived Endophytic Bacteria Enhance Growth, Yield, and Stress Resilience in Rice. International Journal of Molecular Sciences, 26(19), 9317. https://doi.org/10.3390/ijms26199317







