Is dUTPase Enzymatic Activity Truly Essential for Viability?
Abstract
1. Introduction
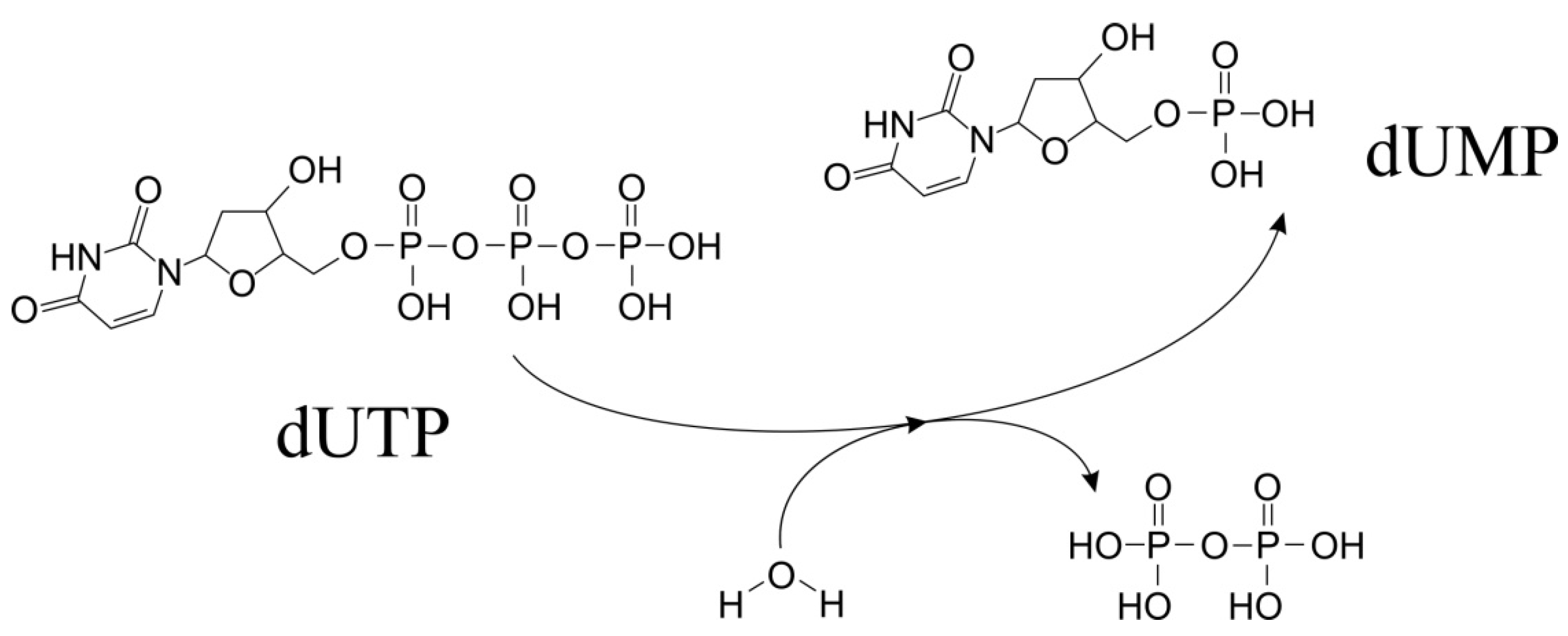
2. Structural Organization and Enzymatic Activity of dUTPase
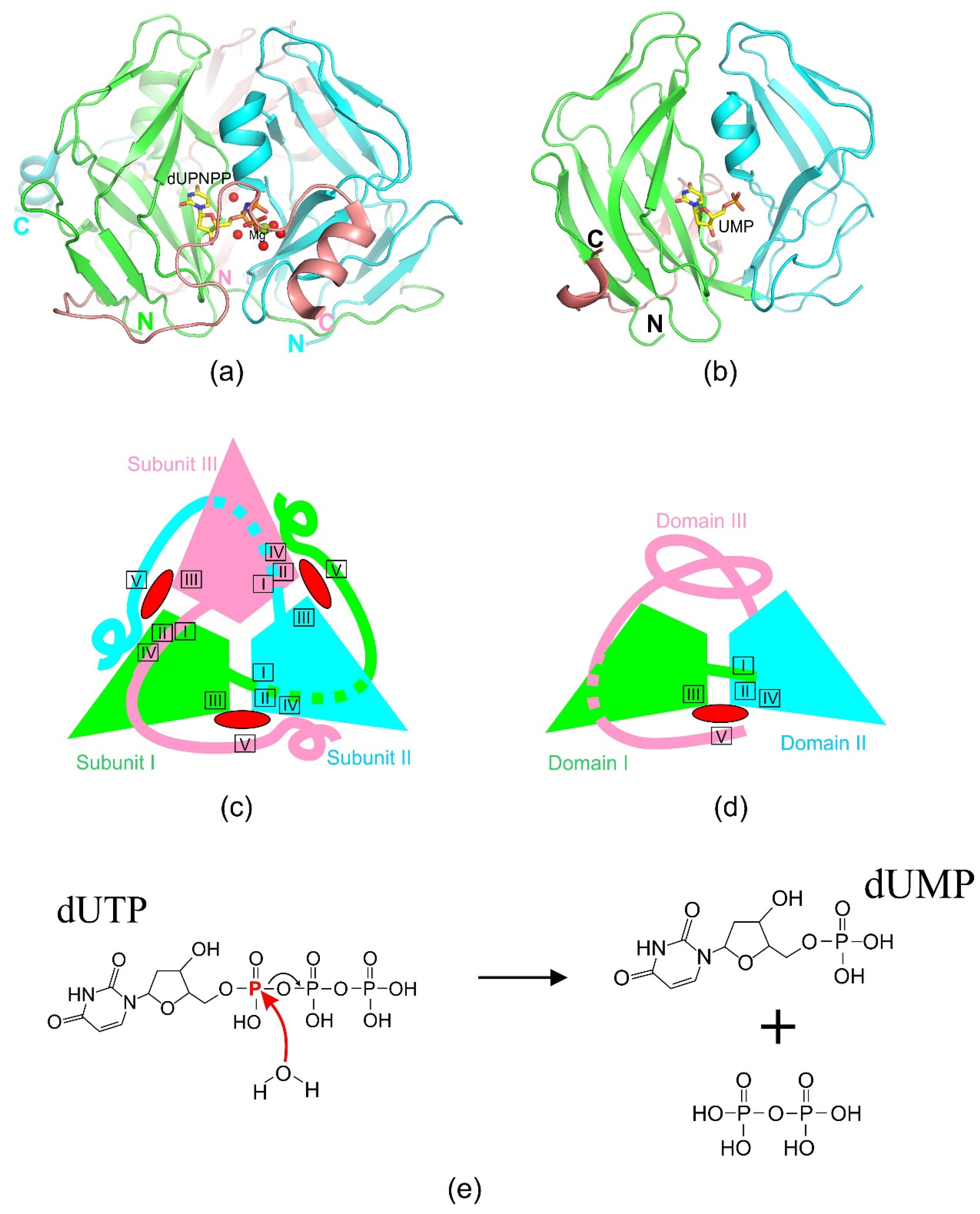
2.1. Homotrimeric dUTPases
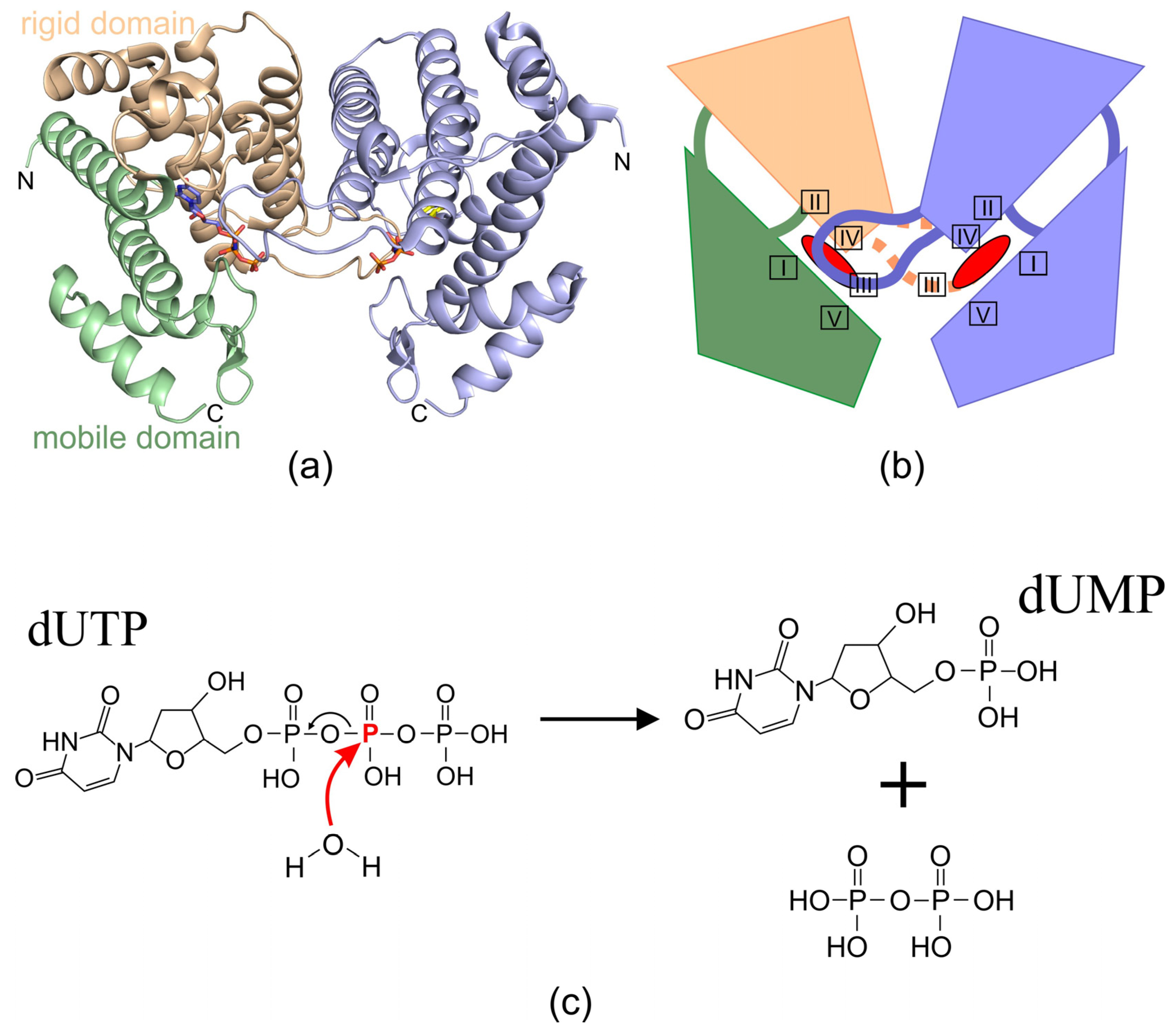
2.2. Homodimeric dUTPases
2.3. Monomeric dUTPases
3. dUTPase Functions in Living Organisms
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Camacho, A.; Arrebola, R.; Pena-Diaz, J.; Ruiz-Perez, L.M.; Gonzalez-Pacanowska, D. Description of a novel eukaryotic deoxyuridine 5′-triphosphate nucleotidohydrolase in Leishmania major. Biochem. J. 1997, 325, 441–447. [Google Scholar] [CrossRef]
- Tormo-Mas, M.A.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, I.; Barbe, J.; Novick, R.P.; Christie, G.E.; Penades, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef]
- Gadsden, M.H.; McIntosh, E.M.; Game, J.C.; Wilson, P.J.; Haynes, R.H. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993, 12, 4425–4431. [Google Scholar] [CrossRef]
- el-Hajj, H.H.; Zhang, H.; Weiss, B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 1988, 170, 1069–1075. [Google Scholar] [CrossRef]
- McGeoch, D.J.; Cook, S.; Dolan, A.; Jamieson, F.E.; Telford, E.A. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 1995, 247, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Kerepesi, C.; Szabo, J.E.; Papp-Kadar, V.; Dobay, O.; Szabo, D.; Grolmusz, V.; Vertessy, B.G. Life without dUTPase. Front Microbiol. 2016, 7, 1768. [Google Scholar] [CrossRef] [PubMed]
- Vertessy, B.G.; Toth, J. Keeping uracil out of DNA: Physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 2009, 42, 97–106. [Google Scholar] [CrossRef]
- Glukhov, A.; Marchenkov, V.; Dzhus, U.; Krutilina, A.; Selikhanov, G.; Gabdulkhakov, A. Bacteriophage T5 dUTPase: Combination of Common Enzymatic and Novel Functions. Int. J. Mol. Sci. 2024, 25, 892. [Google Scholar] [CrossRef]
- Larsson, G.; Svensson, L.A.; Nyman, P.O. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP). Nat. Struct. Biol. 1996, 3, 532–538. [Google Scholar] [CrossRef]
- Maiques, E.; Quiles-Puchalt, N.; Donderis, J.; Ciges-Tomas, J.R.; Alite, C.; Bowring, J.Z.; Humphrey, S.; Penades, J.R.; Marina, A. Another look at the mechanism involving trimeric dUTPases in Staphylococcus aureus pathogenicity island induction involves novel players in the party. Nucleic Acids Res. 2016, 44, 5457–5469. [Google Scholar] [CrossRef] [PubMed]
- Mol, C.D.; Harris, J.M.; McIntosh, E.M.; Tainer, J.A. Human dUTP pyrophosphatase: Uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure 1996, 4, 1077–1092. [Google Scholar] [CrossRef]
- Bernier-Villamor, V.; Camacho, A.; Hidalgo-Zarco, F.; Perez, J.; Ruiz-Perez, L.M.; Gonzalez-Pacanowska, D. Characterization of deoxyuridine 5′-triphosphate nucleotidohydrolase from Trypanosoma cruzi. FEBS Lett. 2002, 526, 147–150. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Gonzalez-Pacanowska, D.; Wilson, K.S. On the catalytic mechanism of dimeric dUTPases. Biochem. J. 2013, 456, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Wren, B.W.; Mungall, K.; Ketley, J.M.; Churcher, C.; Basham, D.; Chillingworth, T.; Davies, R.M.; Feltwell, T.; Holroyd, S.; et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 2000, 403, 665–668. [Google Scholar] [CrossRef]
- Camacho, A.; Hidalgo-Zarco, F.; Bernier-Villamor, V.; Ruiz-Perez, L.M.; Gonzalez-Pacanowska, D. Properties of Leishmania major dUTP nucleotidohydrolase, a distinct nucleotide-hydrolysing enzyme in kinetoplastids. Biochem. J. 2000, 346, 163–168. [Google Scholar] [CrossRef]
- Donderis, J.; Bowring, J.; Maiques, E.; Ciges-Tomas, J.R.; Alite, C.; Mehmedov, I.; Tormo-Mas, M.A.; Penades, J.R.; Marina, A. Convergent evolution involving dimeric and trimeric dUTPases in pathogenicity island mobilization. PLoS Pathog. 2017, 13, e1006581. [Google Scholar] [CrossRef]
- Caradonna, S.J.; Cheng, Y.C. Induction of uracil-DNA glycosylase and dUTP nucleotidohydrolase activity in herpes simplex virus-infected human cells. J. Biol. Chem. 1981, 256, 9834–9837. [Google Scholar] [CrossRef]
- Fuchs, W.; Ziemann, K.; Teifke, J.P.; Werner, O.; Mettenleiter, T.C. The non-essential UL50 gene of avian infectious laryngotracheitis virus encodes a functional dUTPase which is not a virulence factor. J. Gen. Virol. 2000, 81, 627–638. [Google Scholar] [CrossRef]
- Kremmer, E.; Sommer, P.; Holzer, D.; Galetsky, S.A.; Molochkov, V.A.; Gurtsevitch, V.; Winkelmann, C.; Lisner, R.; Niedobitek, G.; Gr Sser, F.A. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) ORF54 encodes a functional dUTPase expressed in the lytic replication cycle. J. Gen. Virol. 1999, 80 Pt 5, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Preston, V.G.; Fisher, F.B. Identification of the herpes simplex virus type 1 gene encoding the dUTPase. Virology 1984, 138, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Stow, N.D. New genes from old: Redeployment of dUTPase by herpesviruses. J. Virol. 2005, 79, 12880–12892. [Google Scholar] [CrossRef]
- McGeoch, D.J. Protein sequence comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990, 18, 4105–4110. [Google Scholar] [CrossRef]
- McGeehan, J.E.; Depledge, N.W.; McGeoch, D.J. Evolution of the dUTPase gene of mammalian and avian herpesviruses. Curr. Protein Pept. Sci. 2001, 2, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Kremmer, E.; Sommer, P.; Grasser, F.A. Epstein-Barr virus-encoded deoxyuridine triphosphate nucleotidohydrolase (dUTPase): A potential target for drug therapy. Transplant. Proc. 1997, 29, 812–814. [Google Scholar] [CrossRef] [PubMed]
- Whittingham, J.L.; Leal, I.; Nguyen, C.; Kasinathan, G.; Bell, E.; Jones, A.F.; Berry, C.; Benito, A.; Turkenburg, J.P.; Dodson, E.J.; et al. dUTPase as a platform for antimalarial drug design: Structural basis for the selectivity of a class of nucleoside inhibitors. Structure 2005, 13, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Studebaker, A.W.; Balendiran, G.K.; Williams, M.V. The herpesvirus encoded dUTPase as a potential chemotherapeutic target. Curr. Protein Pept. Sci. 2001, 2, 371–379. [Google Scholar] [CrossRef]
- Stout, C.D. Induced fit, drug design, and dUTPase. Structure 2004, 12, 2–3. [Google Scholar] [CrossRef]
- Canman, C.E.; Radany, E.H.; Parsels, L.A.; Davis, M.A.; Lawrence, T.S.; Maybaum, J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994, 54, 2296–2298. [Google Scholar]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef]
- Tormo-Mas, M.A.; Donderis, J.; Garcia-Caballer, M.; Alt, A.; Mir-Sanchis, I.; Marina, A.; Penades, J.R. Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. Mol. Cell 2013, 49, 947–958. [Google Scholar] [CrossRef]
- Hill, R.L.L.; Vlach, J.; Parker, L.K.; Christie, G.E.; Saad, J.S.; Dokland, T. Derepression of SaPIbov1 Is Independent of phiNM1 Type 2 dUTPase Activity and Is Inhibited by dUTP and dUMP. J. Mol. Biol. 2017, 429, 1570–1580. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Ma, S.; Yang, R.; Xu, J.; Wang, Y.; Ye, S. Structural definition of pseudorabies virus dUTPase reveals a novel folding dimer in the herpesvirus family. Int. J. Biol. Macromol. 2024, 280, 135696. [Google Scholar] [CrossRef]
- Dauter, Z.; Persson, R.; Rosengren, A.M.; Nyman, P.O.; Wilson, K.S.; Cedergren-Zeppezauer, E.S. Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex. J. Mol. Biol. 1999, 285, 655–673. [Google Scholar] [CrossRef]
- Barabas, O.; Pongracz, V.; Kovari, J.; Wilmanns, M.; Vertessy, B.G. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004, 279, 42907–42915. [Google Scholar] [CrossRef]
- Larsson, G.; Nyman, P.O.; Kvassman, J.O. Kinetic characterization of dUTPase from Escherichia coli. J. Biol. Chem. 1996, 271, 24010–24016. [Google Scholar] [CrossRef]
- Penades, J.R.; Donderis, J.; Garcia-Caballer, M.; Tormo-Mas, M.A.; Marina, A. dUTPases, the unexplored family of signalling molecules. Curr. Opin. Microbiol. 2013, 16, 163–170. [Google Scholar] [CrossRef] [PubMed]
- el-Hajj, H.H.; Wang, L.; Weiss, B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 1992, 174, 4450–4456. [Google Scholar] [CrossRef] [PubMed]
- Benedek, A.; Horvath, A.; Hirmondo, R.; Ozohanics, O.; Bekesi, A.; Modos, K.; Revesz, A.; Vekey, K.; Nagy, G.N.; Vertessy, B.G. Potential steps in the evolution of a fused trimeric all-beta dUTPase involve a catalytically competent fused dimeric intermediate. FEBS J. 2016, 283, 3268–3286. [Google Scholar] [CrossRef]
- Rona, G.; Palinkas, H.L.; Borsos, M.; Horvath, A.; Scheer, I.; Benedek, A.; Nagy, G.N.; Zagyva, I.; Vertessy, B.G. NLS copy-number variation governs efficiency of nuclear import—Case study on dUTPases. FEBS J. 2014, 281, 5463–5478. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chai, Y.; Song, H.; Weng, C.; Qi, J.; Sun, Y.; Gao, G.F. Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target. mBio 2019, 10, e02483-19. [Google Scholar] [CrossRef] [PubMed]
- Baragana, B.; McCarthy, O.; Sanchez, P.; Bosch-Navarrete, C.; Kaiser, M.; Brun, R.; Whittingham, J.L.; Roberts, S.M.; Zhou, X.X.; Wilson, K.S.; et al. beta-Branched acyclic nucleoside analogues as inhibitors of Plasmodium falciparum dUTPase. Bioorg. Med. Chem. 2011, 19, 2378–2391. [Google Scholar] [CrossRef] [PubMed]
- Zang, K.; Li, F.; Ma, Q. The dUTPase of white spot syndrome virus assembles its active sites in a noncanonical manner. J. Biol. Chem. 2018, 293, 1088–1099. [Google Scholar] [CrossRef]
- Tarbouriech, N.; Buisson, M.; Seigneurin, J.M.; Cusack, S.; Burmeister, W.P. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure 2005, 13, 1299–1310. [Google Scholar] [CrossRef]
- Mustafi, D.; Bekesi, A.; Vertessy, B.G.; Makinen, M.W. Catalytic and structural role of the metal ion in dUTP pyrophosphatase. Proc. Natl. Acad. Sci. USA 2003, 100, 5670–5675. [Google Scholar] [CrossRef]
- Vertessy, B.G.; Persson, R.; Rosengren, A.M.; Zeppezauer, M.; Nyman, P.O. Specific derivatization of the active site tyrosine in dUTPase perturbs ligand binding to the active site. Biochem. Biophys. Res. Commun. 1996, 219, 294–300. [Google Scholar] [CrossRef]
- Kumari, K.; Aggarwal, S.; Khan, F.M.; Munde, M.; Gourinath, S. Structural analysis of dUTPase from Helicobacter pylori reveals unusual activity for dATP. Int. J. Biol. Macromol. 2024, 282, 136937. [Google Scholar] [CrossRef]
- Moroz, O.V.; Murzin, A.G.; Makarova, K.S.; Koonin, E.V.; Wilson, K.S.; Galperin, M.Y. Dimeric dUTPases, HisE, and MazG belong to a new superfamily of all-alpha NTP pyrophosphohydrolases with potential “house-cleaning” functions. J. Mol. Biol. 2005, 347, 243–255. [Google Scholar] [CrossRef]
- Frigols, B.; Quiles-Puchalt, N.; Mir-Sanchis, I.; Donderis, J.; Elena, S.F.; Buckling, A.; Novick, R.P.; Marina, A.; Penades, J.R. Virus Satellites Drive Viral Evolution and Ecology. PLoS Genet. 2015, 11, e1005609. [Google Scholar] [CrossRef]
- Hemsworth, G.R.; Moroz, O.V.; Fogg, M.J.; Scott, B.; Bosch-Navarrete, C.; Gonzalez-Pacanowska, D.; Wilson, K.S. The crystal structure of the Leishmania major deoxyuridine triphosphate nucleotidohydrolase in complex with nucleotide analogues, dUMP, and deoxyuridine. J. Biol. Chem. 2011, 286, 16470–16481. [Google Scholar] [CrossRef] [PubMed]
- Moroz, O.V.; Harkiolaki, M.; Galperin, M.Y.; Vagin, A.A.; Gonzalez-Pacanowska, D.; Wilson, K.S. The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the “basic module” for dimeric d(C/U)TPases. J. Mol. Biol. 2004, 342, 1583–1597. [Google Scholar] [CrossRef]
- Harkiolaki, M.; Dodson, E.J.; Bernier-Villamor, V.; Turkenburg, J.P.; Gonzalez-Pacanowska, D.; Wilson, K.S. The crystal structure of Trypanosoma cruzi dUTPase reveals a novel dUTP/dUDP binding fold. Structure 2004, 12, 41–53. [Google Scholar] [CrossRef]
- Bjornberg, O.; Neuhard, J.; Nyman, P.O. A bifunctional dCTP deaminase-dUTP nucleotidohydrolase from the hyperthermophilic archaeon Methanocaldococcus jannaschii. J. Biol. Chem. 2003, 278, 20667–20672. [Google Scholar] [CrossRef]
- Huffman, J.L.; Li, H.; White, R.H.; Tainer, J.A. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J. Mol. Biol. 2003, 331, 885–896. [Google Scholar] [CrossRef]
- Chan, S.; Segelke, B.; Lekin, T.; Krupka, H.; Cho, U.S.; Kim, M.Y.; So, M.; Kim, C.Y.; Naranjo, C.M.; Rogers, Y.C.; et al. Crystal structure of the Mycobacterium tuberculosis dUTPase: Insights into the catalytic mechanism. J. Mol. Biol. 2004, 341, 503–517. [Google Scholar] [CrossRef]
- Pecsi, I.; Hirmondo, R.; Brown, A.C.; Lopata, A.; Parish, T.; Vertessy, B.G.; Toth, J. The dUTPase enzyme is essential in Mycobacterium smegmatis. PLoS ONE 2012, 7, e37461. [Google Scholar] [CrossRef]
- Monot, M.; Honore, N.; Garnier, T.; Zidane, N.; Sherafi, D.; Paniz-Mondolfi, A.; Matsuoka, M.; Taylor, G.M.; Donoghue, H.D.; Bouwman, A.; et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 2009, 41, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Muha, V.; Zagyva, I.; Venkei, Z.; Szabad, J.; Vertessy, B.G. Nuclear localization signal-dependent and -independent movements of Drosophila melanogaster dUTPase isoforms during nuclear cleavage. Biochem. Biophys. Res. Commun. 2009, 381, 271–275. [Google Scholar] [CrossRef]
- Bekesi, A.; Zagyva, I.; Hunyadi-Gulyas, E.; Pongracz, V.; Kovari, J.; Nagy, A.O.; Erdei, A.; Medzihradszky, K.F.; Vertessy, B.G. Developmental regulation of dUTPase in Drosophila melanogaster. J. Biol. Chem. 2004, 279, 22362–22370. [Google Scholar] [CrossRef] [PubMed]
- Chu, R.; Lin, Y.; Rao, M.S.; Reddy, J.K. Cloning and identification of rat deoxyuridine triphosphatase as an inhibitor of peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 1996, 271, 27670–27676. [Google Scholar] [CrossRef] [PubMed]
- Penades, J.R.; Christie, G.E. The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu. Rev. Virol. 2015, 2, 181–201. [Google Scholar] [CrossRef]
- Novick, R.P.; Christie, G.E.; Penades, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef]
- Penades, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Quiles-Puchalt, N.; Martinez-Rubio, R.; Ram, G.; Lasa, I.; Penades, J.R. Unravelling bacteriophage ϕ11 requirements for packaging and transfer of mobile genetic elements in Staphylococcus aureus. Mol. Microbiol. 2014, 91, 423–437. [Google Scholar] [CrossRef]
- Tormo, M.A.; Ferrer, M.D.; Maiques, E.; Ubeda, C.; Selva, L.; Lasa, I.; Calvete, J.J.; Novick, R.P.; Penades, J.R. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008, 190, 2434–2440. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Carpena, N.; Quiles-Puchalt, N.; Ram, G.; Novick, R.P.; Penades, J.R. Intra- and inter-generic transfer of pathogenicity island-encoded virulence genes by cos phages. ISME J. 2015, 9, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Novick, R.P. Phage-mediated intergeneric transfer of toxin genes. Science 2009, 323, 139–141. [Google Scholar] [CrossRef]
- Hill, R.L.L.; Dokland, T. The Type 2 dUTPase of Bacteriophage ϕNM1 Initiates Mobilization of Staphylococcus aureus Bovine Pathogenicity Island 1. J. Mol. Biol. 2016, 428, 142–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; et al. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef]
- Leang, R.S.; Wu, T.T.; Hwang, S.; Liang, L.T.; Tong, L.; Truong, J.T.; Sun, R. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS Pathog. 2011, 7, e1002292. [Google Scholar] [CrossRef]
- Madrid, A.S.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J. Virol. 2012, 86, 8693–8704. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glukhov, A.; Dzhus, U.; Kolyadenko, I.; Selikhanov, G.; Gabdulkhakov, A. Is dUTPase Enzymatic Activity Truly Essential for Viability? Int. J. Mol. Sci. 2025, 26, 9260. https://doi.org/10.3390/ijms26199260
Glukhov A, Dzhus U, Kolyadenko I, Selikhanov G, Gabdulkhakov A. Is dUTPase Enzymatic Activity Truly Essential for Viability? International Journal of Molecular Sciences. 2025; 26(19):9260. https://doi.org/10.3390/ijms26199260
Chicago/Turabian StyleGlukhov, Anatoly, Ulyana Dzhus, Ilya Kolyadenko, Georgii Selikhanov, and Azat Gabdulkhakov. 2025. "Is dUTPase Enzymatic Activity Truly Essential for Viability?" International Journal of Molecular Sciences 26, no. 19: 9260. https://doi.org/10.3390/ijms26199260
APA StyleGlukhov, A., Dzhus, U., Kolyadenko, I., Selikhanov, G., & Gabdulkhakov, A. (2025). Is dUTPase Enzymatic Activity Truly Essential for Viability? International Journal of Molecular Sciences, 26(19), 9260. https://doi.org/10.3390/ijms26199260






