Global and Sex-Stratified Genome-Wide Association Study of Long COVID Based on Patient-Driven Symptom Recall
Abstract
1. Introduction
2. Results
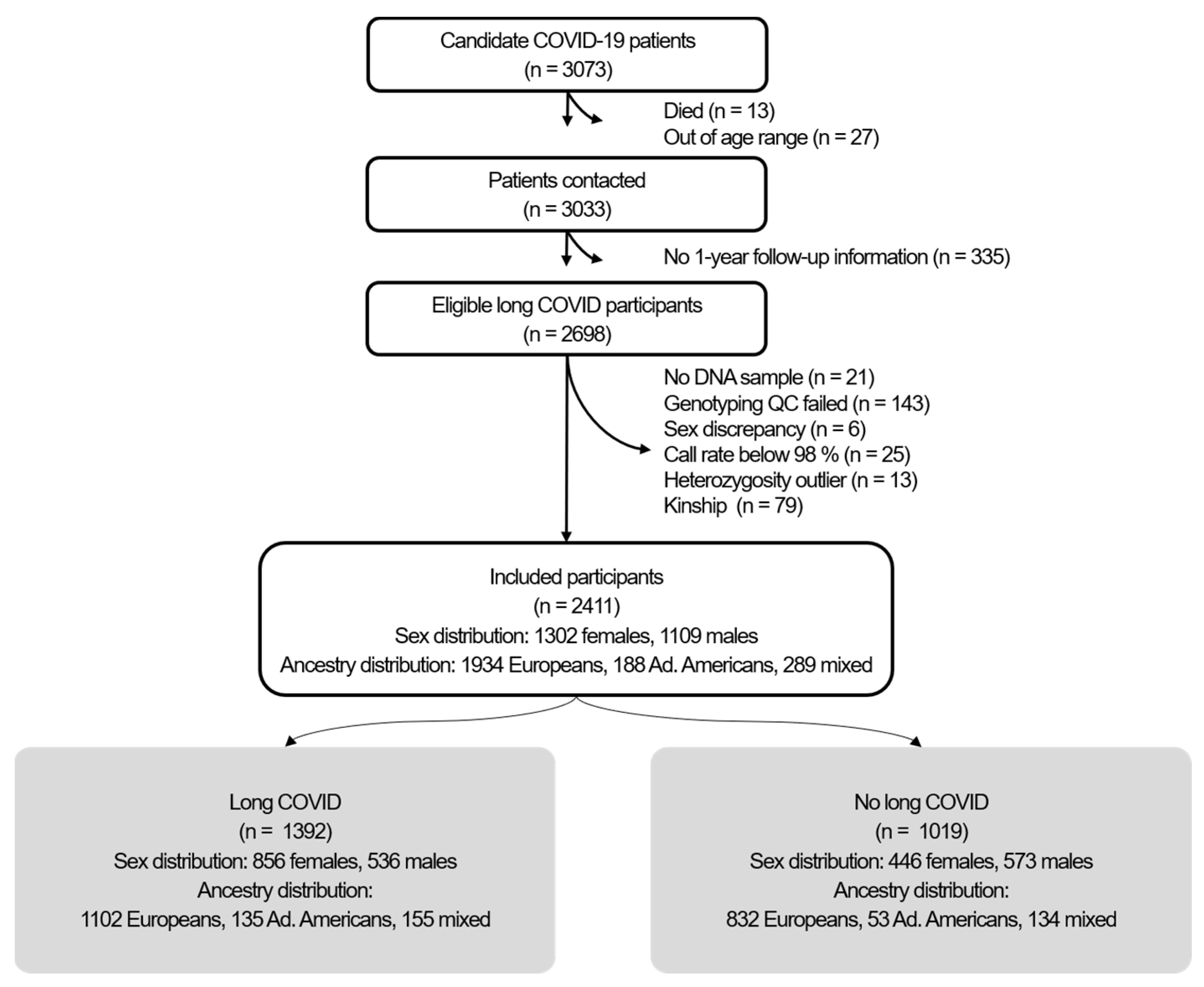
2.1. Participants
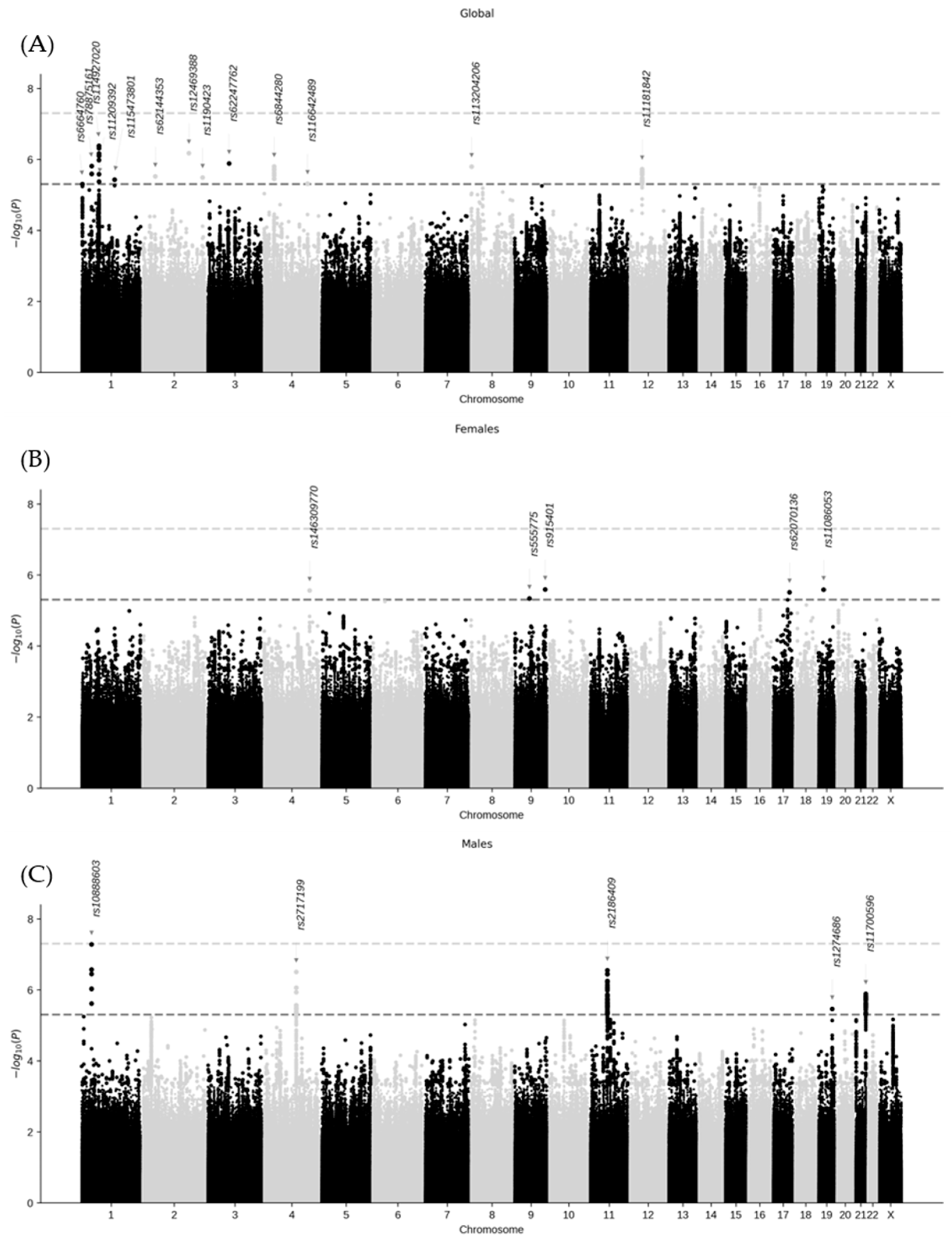
2.2. Genome-Wide Association Analysis and Variant–Sex Interaction
2.3. Functional Annotation
3. Discussion
4. Materials and Methods
4.1. Study Design and Participants
4.2. Definition of Long COVID
4.3. DNA Collection
4.4. Genotyping
4.5. Quality Control of Genotype Data
4.6. Variant Imputation
4.7. Clinical Data
4.8. Statistical Analyses
4.9. Functional Annotation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| COVID-19 | Coronavirus Disease 2019 |
| eQTL | Expression Quantitative Trait Locus |
| GWAS | Genome-Wide Association Study |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
References
- Tsilingiris, D.; Vallianou, N.G.; Karampela, I.; Christodoulatos, G.S.; Papavasileiou, G.; Petropoulou, D.; Magkos, F.; Dalamaga, M. Laboratory Findings and Biomarkers in Long COVID: What Do We Know So Far? Insights into Epidemiology, Pathogenesis, Therapeutic Perspectives and Challenges. Int. J. Mol. Sci. 2023, 24, 10458. [Google Scholar] [CrossRef]
- Greenhalgh, T.; Sivan, M.; Perlowski, A.; Nikolich, J.Ž. Long COVID: A Clinical Update. Lancet 2024, 404, 707–724. [Google Scholar] [CrossRef]
- Robertson, M.M.; Qasmieh, S.A.; Kulkarni, S.G.; Teasdale, C.A.; Jones, H.E.; McNairy, M.; Borrell, L.N.; Nash, D. The Epidemiology of Long Coronavirus Disease in US Adults. Clin. Infect. Dis. 2023, 76, 1636–1645. [Google Scholar] [CrossRef]
- Kim, Y.; Bae, S.; Chang, H.H.; Kim, S.W. Long COVID Prevalence and Impact on Quality of Life 2 Years after Acute COVID-19. Sci. Rep. 2023, 13, 11207, Erratum in Sci. Rep. 2023, 13, 11960. [Google Scholar]
- O’Mahoney, L.L.; Routen, A.; Gillies, C.; Jenkins, S.A.; Almaqhawi, A.; Ayoubkhani, D.; Banerjee, A.; Brightling, C.; Calvert, M.; Cassambai, S.; et al. The Risk of Long Covid Symptoms: A Systematic Review and Meta-Analysis of Controlled Studies. Nat. Commun. 2025, 16, 4249. [Google Scholar] [CrossRef]
- Constantinescu-Bercu, A.; Lobiuc, A.; Căliman-Sturdza, O.A.; Oiţă, R.C.; Iavorschi, M.; Pavăl, N.-E.; Șoldănescu, I.; Dimian, M.; Covasa, M. Long COVID: Molecular Mechanisms and Detection Techniques. Int. J. Mol. Sci. 2023, 25, 408. [Google Scholar] [CrossRef]
- Mateu, L.; Tebe, C.; Loste, C.; Santos, J.R.; Lladós, G.; López, C.; España-Cueto, S.; Toledo, R.; Font, M.; Chamorro, A.; et al. Determinants of the Onset and Prognosis of the Post-COVID-19 Condition: A 2-Year Prospective Observational Cohort Study. Lancet Reg. Health Eur. 2023, 33, 100724. [Google Scholar] [CrossRef] [PubMed]
- Dennis, A.; Wamil, M.; Alberts, J.; Oben, J.; Cuthbertson, D.J.; Wootton, D.; Crooks, M.; Gabbay, M.; Brady, M.; Hishmeh, L.; et al. Multiorgan Impairment in Low-Risk Individuals with Post-COVID-19 Syndrome: A Prospective, Community-Based Study. BMJ Open 2021, 11, e048391. [Google Scholar] [CrossRef] [PubMed]
- Hein, Z.M.; Thazin; Kumar, S.; Che Ramli, M.D.; Che Mohd Nassir, C.M.N. Immunomodulatory Mechanisms Underlying Neurological Manifestations in Long COVID: Implications for Immune-Mediated Neurodegeneration. Int. J. Mol. Sci. 2025, 26, 6214. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.; Sarapultsev, A. Exploring the Pathophysiology of Long COVID: The Central Role of Low-Grade Inflammation and Multisystem Involvement. Int. J. Mol. Sci. 2024, 25, 6389. [Google Scholar] [CrossRef]
- Varillas-Delgado, D.; Jimenez-Antona, C.; Lizcano-Alvarez, A.; Cano-de-la-Cuerda, R.; Molero-Sanchez, A.; Laguarta-Val, S. Predictive Factors and ACE-2 Gene Polymorphisms in Susceptibility to Long COVID-19 Syndrome. Int. J. Mol. Sci. 2023, 24, 16717. [Google Scholar] [CrossRef]
- Lammi, V.; Nakanishi, T.; Jones, S.E.; Andrews, S.J.; Karjalainen, J.; Cortés, B.; O’Brien, H.E.; Ochoa-Guzman, A.; Fulton-Howard, B.E.; Broberg, M.; et al. Genome-Wide Association Study of Long COVID. Nat. Genet. 2025, 57, 1402–1417. [Google Scholar] [CrossRef]
- Soldevila, G.; Raman, C.; Lozano, F. The Immunomodulatory Properties of the CD5 Lymphocyte Receptor in Health and Disease. Curr. Opin. Immunol. 2011, 23, 310–318. [Google Scholar] [CrossRef]
- Ali, E.; Badawi, M.; Abdelmahmuod, E.; Kohla, S.; Yassin, M.A. Chronic Lymphocytic Leukemia Concomitant with COVID 19: A Case Report. Am. J. Case Rep. 2020, 21, e926062. [Google Scholar] [CrossRef] [PubMed]
- Saluja, P.; Gautam, N.; Amisha, F.; Safar, M.; Bartter, T. Emergence of Chronic Lymphocytic Leukemia During Admission for COVID-19: Cause or Coincidence? Cureus 2022, 14, e23470. [Google Scholar] [CrossRef] [PubMed]
- Lanza, L.; Koroveshi, B.; Barducchi, F.; Lorenzo, A.; Venturino, E.; Cappelli, E.; Lillo, F.; Bain, B.J. A New Diagnosis of Monoclonal B-cell Lymphocytosis with Cytoplasmic Inclusions in a Patient with COVID-19. Am. J. Hematol. 2022, 97, 1372–1373. [Google Scholar] [CrossRef]
- Jackson, H.R.; Miglietta, L.; Habgood-Coote, D.; D’Souza, G.; Shah, P.; Nichols, S.; Vito, O.; Powell, O.; Davidson, M.S.; Shimizu, C.; et al. Diagnosis of Multisystem Inflammatory Syndrome in Children by a Whole-Blood Transcriptional Signature. J. Pediatr. Infect. Dis. Soc. 2023, 12, 322–331. [Google Scholar] [CrossRef]
- Delgado-Vega, A.M.; Martínez-Bueno, M.; Oparina, N.Y.; López Herráez, D.; Kristjansdottir, H.; Steinsson, K.; Kozyrev, S.V.; Alarcón-Riquelme, M.E. Whole Exome Sequencing of Patients from Multicase Families with Systemic Lupus Erythematosus Identifies Multiple Rare Variants. Sci. Rep. 2018, 8, 8775. [Google Scholar] [CrossRef]
- Shen, S.; Li, H.; Liu, J.; Sun, L.; Yuan, Y. The Panoramic Picture of Pepsinogen Gene Family with Pan-Cancer. Cancer Med. 2020, 9, 9064–9080. [Google Scholar] [CrossRef] [PubMed]
- Huo, Q.; Li, Z.; Chen, S.; Wang, J.; Li, J.; Xie, N. VWCE as a Potential Biomarker Associated with Immune Infiltrates in Breast Cancer. Cancer Cell Int. 2021, 21, 272. [Google Scholar] [CrossRef]
- Xie, S.A.; Zhang, W.; Du, F.; Liu, S.; Ning, T.T.; Zhang, N.; Zhang, S.-T.; Zhu, S.-T. PTOV1 Facilitates Colorectal Cancer Cell Proliferation through Activating AKT1 Signaling Pathway. Heliyon 2024, 10, e36017. [Google Scholar] [CrossRef]
- Giannitrapani, L.; Mirarchi, L.; Amodeo, S.; Licata, A.; Soresi, M.; Cavaleri, F.; Casalicchio, S.; Ciulla, G.; Ciuppa, M.E.; Cervello, M.; et al. Can Baseline IL-6 Levels Predict Long COVID in Subjects Hospitalized for SARS-CoV-2 Disease? Int. J. Mol. Sci. 2023, 24, 1731. [Google Scholar] [CrossRef]
- Kaiyrzhanov, R.; Rocca, C.; Suri, M.; Gulieva, S.; Zaki, M.S.; Henig, N.Z.; Siquier, K.; Guliyeva, U.; Mounir, S.M.; Marom, D.; et al. Biallelic Loss of EMC10 Leads to Mild to Severe Intellectual Disability. Ann. Clin. Transl. Neurol. 2022, 9, 1080–1089. [Google Scholar] [CrossRef]
- Saarela, A.; Timonen, O.; Kirjavainen, J.; Liu, Y.; Silvennoinen, K.; Mervaala, E.; Kälviäinen, R. Novel LAMC3 Pathogenic Variant Enriched in Finnish Population Causes Malformations of Cortical Development and Severe Epilepsy. Epileptic Disord. 2024, 26, 498–509. [Google Scholar] [CrossRef]
- Premraj, L.; Kannapadi, N.V.; Briggs, J.; Seal, S.M.; Battaglini, D.; Fanning, J.; Suen, J.; Robba, C.; Fraser, J.; Cho, S.-M. Mid and Long-Term Neurological and Neuropsychiatric Manifestations of Post-COVID-19 Syndrome: A Meta-Analysis. J. Neurol. Sci. 2022, 434, 120162. [Google Scholar] [CrossRef] [PubMed]
- Pulliam, L.; Sun, B.; McCafferty, E.; Soper, S.A.; Witek, M.A.; Hu, M.; Ford, J.M.; Song, S.; Kapogiannis, D.; Glesby, M.J.; et al. Microfluidic Isolation of Neuronal-Enriched Extracellular Vesicles Shows Distinct and Common Neurological Proteins in Long COVID, HIV Infection and Alzheimer’s Disease. Int. J. Mol. Sci. 2024, 25, 3830. [Google Scholar] [CrossRef]
- Chagas, L.D.S.; Serfaty, C.A. The Influence of Microglia on Neuroplasticity and Long-Term Cognitive Sequelae in Long COVID: Impacts on Brain Development and Beyond. Int. J. Mol. Sci. 2024, 25, 3819. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Zeng, C.; Liu, J.; Zhao, C.; Ge, F.; Li, Y.; Qian, M.; Du, J.; Wang, W.; Li, Y.; et al. Josephin Domain Containing 2 (JOSD2) Promotes Lung Cancer by Inhibiting LKB1 (Liver Kinase B1) Activity. Signal Transduct. Target. Ther. 2024, 9, 11. [Google Scholar] [CrossRef]
- Wang, W.P.; Shi, D.; Yun, D.; Hu, J.; Wang, J.F.; Liu, J.; Yang, Y.-P.; Li, M.-R.; Wang, J.-F.; Kong, D.-L. Role of Deubiquitinase JOSD2 in the Pathogenesis of Esophageal Squamous Cell Carcinoma. World J. Gastroenterol. 2024, 30, 565–578. [Google Scholar] [CrossRef]
- Huang, Y.; Zeng, J.; Liu, T.; Xu, Q.; Song, X.; Zeng, J. Deubiquitinating Enzyme JOSD2 Promotes Hepatocellular Carcinoma Progression through Interacting with and Inhibiting CTNNB1 Degradation. Cell Biol. Int. 2022, 46, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Yang, L.; Wang, Y.; Zou, Z.; Liu, M.; Xu, H.; Wu, Y. JOSD2 Regulates PKM2 Nuclear Translocation and Reduces Acute Myeloid Leukemia Progression. Exp. Hematol. Oncol. 2022, 11, 42. [Google Scholar] [CrossRef]
- Oh, S.; Nam, S.K.; Lee, K.W.; Lee, H.S.; Park, Y.; Kwak, Y.; Lee, K.S.; Kim, J.-W.; Kim, J.W.; Kang, M.; et al. Genomic and Transcriptomic Characterization of Gastric Cancer with Bone Metastasis. Cancer Res. Treat. 2024, 56, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Stabenau, K.A.; Samuels, T.L.; Lam, T.K.; Mathison, A.J.; Wells, C.; Altman, K.W.; Battle, M.A.; Johnston, N. Pepsinogen/Proton Pump Co-Expression in Barrett’s Esophageal Cells Induces Cancer-Associated Changes. Laryngoscope 2023, 133, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Almorox, L.; Antequera, L.; Rojas, I.; Herrera, L.J.; Ortuño, F.M. Gene Expression Analysis for Uterine Cervix and Corpus Cancer Characterization. Genes 2024, 15, 312. [Google Scholar] [CrossRef]
- Kunitomi, H.; Kobayashi, Y.; Wu, R.C.; Takeda, T.; Tominaga, E.; Banno, K.; Aoki, D. LAMC1 Is a Prognostic Factor and a Potential Therapeutic Target in Endometrial Cancer. J. Gynecol. Oncol. 2020, 31, e11. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, X.; Xie, W.; Li, Z.; Liu, W.; Liu, A. Identification of a Basement Membrane-Related Gene Signature for Predicting Prognosis and Estimating the Tumor Immune Microenvironment in Breast Cancer. Front. Endocrinol. 2022, 13, 1065530. [Google Scholar] [CrossRef]
- Takenami, T.; Maeda, S.; Karasawa, H.; Suzuki, T.; Furukawa, T.; Morikawa, T.; Takadate, T.; Hayashi, H.; Nakagawa, K.; Motoi, F.; et al. Novel Biomarkers Distinguishing Pancreatic Head Cancer from Distal Cholangiocarcinoma Based on Proteomic Analysis. BMC Cancer 2019, 19, 318. [Google Scholar] [CrossRef]
- Camps-Vilaró, A.; Pinsach-Abuin, M.L.; Degano, I.R.; Ramos, R.; Martí-Lluch, R.; Elosua, R.; Subirana, I.; Solà-Richarte, C.; Puigmulé, M.; Pérez, A.; et al. Genetic Characteristics Involved in COVID-19 Severity. The CARGENCORS Case-Control Study and Meta-Analysis. J. Med. Virol. 2024, 96, e29404. [Google Scholar] [CrossRef]
- World Health Organization Post COVID-19 Condition (Long COVID). Available online: https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition (accessed on 18 December 2024).
- Ceban, F.; Ling, S.; Lui, L.M.W.; Lee, Y.; Gill, H.; Teopiz, K.M.; Rodrigues, N.B.; Subramaniapillai, M.; Di Vincenzo, J.D.; Cao, B.; et al. Fatigue and Cognitive Impairment in Post-COVID-19 Syndrome: A Systematic Review and Meta-Analysis. Brain Behav. Immun. 2022, 101, 93–135. [Google Scholar] [CrossRef]
- R Core Team. R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Price, A.L.; Weale, M.E.; Patterson, N.; Myers, S.R.; Need, A.C.; Shianna, K.V.; Ge, D.; Rotter, J.I.; Torres, E.; Taylor, K.D.; et al. Long-Range LD Can Confound Genome Scans in Admixed Populations. Am. J. Hum. Genet. 2008, 83, 132–138. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- 1000 Genomes Project Consortium. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Taliun, D.; Harris, D.N.; Kessler, M.D.; Carlson, J.; Szpiech, Z.A.; Torres, R.; Taliun, S.A.G.; Corvelo, A.; Gogarten, S.M.; Kang, H.M.; et al. Sequencing of 53,831 Diverse Genomes from the NHLBI TOPMed Program. Nature 2021, 590, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Taskesen, E.; van Bochoven, A.; Posthuma, D. Functional Mapping and Annotation of Genetic Associations with FUMA. Nat. Commun. 2017, 8, 1826. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of Functional Variation in Personal Genomes Using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Castanza, A.S.; Recla, J.M.; Eby, D.; Thorvaldsdóttir, H.; Bult, C.J.; Mesirov, J.P. Extending Support for Mouse Data in the Molecular Signatures Database (MSigDB). Nat. Methods 2023, 20, 1619–1620. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Agrawal, A.; Balci, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next Generation Pathway Database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef] [PubMed]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Güneş, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and Deposition Resource. Nucleic Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
| Females (n = 1302) | Males (n = 1109) | |||||
|---|---|---|---|---|---|---|
| With Long COVID (n = 856) | Without Long COVID (n = 446) | p-Value | With Long COVID (n = 536) | Without Long COVID (n = 573) | p-Value | |
| Age (mean ± SD) | 56.4 ± 12.0 | 58.3 ± 12.6 | 0.009 | 57.8 ± 12.2 | 60.1 ± 12.7 | 0.002 |
| Severe (hospitalized) acute phase (n, %) | 196 (23.4%) | 91 (20.5%) | 0.261 | 242 (47.3%) | 164 (29.6%) | <0.001 |
| Current smokers (n, %) | 57 (6.70%) | 43 (9.71%) | 0.070 | 49 (9.23%) | 72 (12.7%) | 0.086 |
| Diabetes mellitus (n, %) | 86 (10.0%) | 44 (9.87%) | 0.995 | 80 (14.9%) | 99 (17.3%) | 0.320 |
| Hypertension (n, %) | 202 (23.6%) | 144 (32.3%) | 0.001 | 209 (39.0%) | 225 (39.3%) | 0.956 |
| Dyslipidemia (n, %) | 201 (23.5%) | 121 (27.1%) | 0.175 | 179 (33.4%) | 194 (34.0%) | 0.888 |
| BMI (categorized): | 0.534 | 0.234 | ||||
| <18.5 kg/m2 (n, %) | 6 (0.72%) | 6 (1.40%) | 2 (0.38%) | 1 (0.18%) | ||
| 18.5–24.9 kg/m2 (n, %) | 286 (34.3%) | 155 (36.1%) | 109 (20.7%) | 137 (25.0%) | ||
| 25.0–29.9 kg/m2 (n, %) | 268 (32.1%) | 127 (29.6%) | 266 (50.5%) | 277 (50.5%) | ||
| ≥30.0 kg/m2 (n, %) | 275 (32.9%) | 141 (32.9%) | 150 (28.5%) | 133 (24.3%) | ||
| Ancestry: | 0.112 | 0.002 | ||||
| Admixed Americans (n, %) | 89 (10.4%) | 31 (6.95%) | 46 (8.58%) | 22 (3.84%) | ||
| Europeans (n, %) | 677 (79.1%) | 370 (83.0%) | 425 (79.3%) | 462 (80.6%) | ||
| Mixed (n, %) | 90 (10.5%) | 45 (10.1%) | 65 (12.1%) | 89 (15.5%) | ||
| Gene Variant | Chrom. | Position (GRCh38) | Non-Effect Allele | Effect Allele | Effect Allele Frequency | β | Standard Error | GWAS p-Value | Gene Variant– Sex Interaction p-Value 1 |
|---|---|---|---|---|---|---|---|---|---|
| Females and males combined | |||||||||
| rs6664760 | 1 | 3,731,704 | T | C | 0.035 | 0.857 | 0.188 | 5.0 × 10−6 | 1 |
| rs78875161 | 1 | 39,627,261 | C | T | 0.034 | −0.800 | 0.167 | 1.5 × 10−6 | 1 |
| rs114927020 | 1 | 68,967,153 | G | A | 0.099 | −0.518 | 0.102 | 4.2 × 10−6 | 1 |
| rs11209392 | 1 | 69,122,167 | T | G | 0.016 | −1.194 | 0.260 | 4.3 × 10−6 | 1 |
| rs115473801 | 1 | 157,613,190 | G | A | 0.027 | −0.891 | 0.193 | 3.7 × 10−6 | 1 |
| rs62144353 | 2 | 42,755,360 | C | A | 0.111 | 0.467 | 0.100 | 3.0 × 10−6 | 1 |
| rs12469388 | 2 | 177,824,304 | G | T | 0.030 | −0.908 | 0.183 | 6.7 × 10−6 | 1 |
| rs1190423 | 2 | 232,449,952 | A | C | 0.341 | −0.298 | 0.064 | 3.3 × 10−6 | 1 |
| rs62247762 | 3 | 73,537,975 | C | T | 0.024 | −0.989 | 0.204 | 1.3 × 10−6 | 1 |
| rs6844280 | 4 | 31,385,522 | C | T | 0.487 | −0.293 | 0.061 | 1.6 × 10−6 | 1 |
| rs116642489 | 4 | 152,949,693 | C | G | 0.065 | −0.558 | 0.122 | 4.8 × 10−6 | 1 |
| rs113204206 | 8 | 1,585,368 | C | G | 0.105 | −0.474 | 0.099 | 1.6 × 10−6 | 1 |
| rs11181842 | 12 | 43,007,535 | C | T | 0.161 | −0.387 | 0.081 | 1.9 × 10−6 | 1 |
| Only females | |||||||||
| rs146309770 | 4 | 160,217,229 | T | TA | 0.035 | 0.613 | 0.131 | 2.7 × 10−6 | 1.6 × 10−2 |
| rs555775 | 9 | 75,445,184 | C | T | 0.034 | −0.535 | 0.117 | 4.6 × 10−6 | 1.0 × 10−1 |
| rs915401 | 9 | 131,013,384 | A | G | 0.099 | 0.419 | 0.089 | 2.6 × 10−6 | 9.1 × 10−5 |
| rs62070136 | 17 | 72,195,002 | T | G | 0.016 | −0.884 | 0.189 | 3.1 × 10−6 | 2.1 × 10−1 |
| rs11086053 | 19 | 17,119,562 | A | C | 0.027 | −0.693 | 0.148 | 2.6 × 10−6 | 1 |
| Only males | |||||||||
| rs10888603 | 1 | 38,972,945 | A | C | 0.035 | −0.592 | 0.109 | 5.2 × 10−8 | 5.5 × 10−2 |
| rs2717199 | 4 | 111,595,181 | A | G | 0.034 | −0.478 | 0.094 | 3.1 × 10−7 | 1.8 × 10−3 |
| rs2186409 | 11 | 61,127,020 | T | G | 0.099 | −0.496 | 0.097 | 2.8 × 10−7 | 7.8 × 10−4 |
| rs1274686 | 19 | 50,495,701 | C | T | 0.016 | 0.832 | 0.179 | 3.5 × 10−6 | 1.0 × 10−5 |
| rs11700596 | 21 | 46,454,141 | G | C | 0.027 | 0.522 | 0.108 | 1.3 × 10−6 | 1.2 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polo-Alonso, S.; Hernáez, Á.; Dégano, I.R.; Martí-Lluch, R.; Pinsach-Abuin, M.; Elosua, R.; Subirana, I.; Puigmulé, M.; Pérez, A.; Cruz, R.; et al. Global and Sex-Stratified Genome-Wide Association Study of Long COVID Based on Patient-Driven Symptom Recall. Int. J. Mol. Sci. 2025, 26, 9252. https://doi.org/10.3390/ijms26189252
Polo-Alonso S, Hernáez Á, Dégano IR, Martí-Lluch R, Pinsach-Abuin M, Elosua R, Subirana I, Puigmulé M, Pérez A, Cruz R, et al. Global and Sex-Stratified Genome-Wide Association Study of Long COVID Based on Patient-Driven Symptom Recall. International Journal of Molecular Sciences. 2025; 26(18):9252. https://doi.org/10.3390/ijms26189252
Chicago/Turabian StylePolo-Alonso, Sara, Álvaro Hernáez, Irene R. Dégano, Ruth Martí-Lluch, Mel·lina Pinsach-Abuin, Roberto Elosua, Isaac Subirana, Marta Puigmulé, Alexandra Pérez, Raquel Cruz, and et al. 2025. "Global and Sex-Stratified Genome-Wide Association Study of Long COVID Based on Patient-Driven Symptom Recall" International Journal of Molecular Sciences 26, no. 18: 9252. https://doi.org/10.3390/ijms26189252
APA StylePolo-Alonso, S., Hernáez, Á., Dégano, I. R., Martí-Lluch, R., Pinsach-Abuin, M., Elosua, R., Subirana, I., Puigmulé, M., Pérez, A., Cruz, R., Diz-de Almeida, S., Puigdecant, E., Selga, E., Nogues, X., Masclans, J. R., Güerri-Fernández, R., Cubero-Gallego, H., Tizon-Marcos, H., Vaquerizo, B., ... Marrugat, J. (2025). Global and Sex-Stratified Genome-Wide Association Study of Long COVID Based on Patient-Driven Symptom Recall. International Journal of Molecular Sciences, 26(18), 9252. https://doi.org/10.3390/ijms26189252






