Plastid Phylogenomics of Camphora officinarum Nees: Unraveling Genetic Diversity and Geographic Differentiation in East Asian Subtropical Forests
Abstract
1. Introduction
2. Results
2.1. Fundamental Attributes of the Chloroplast Genomes in Camphor Trees
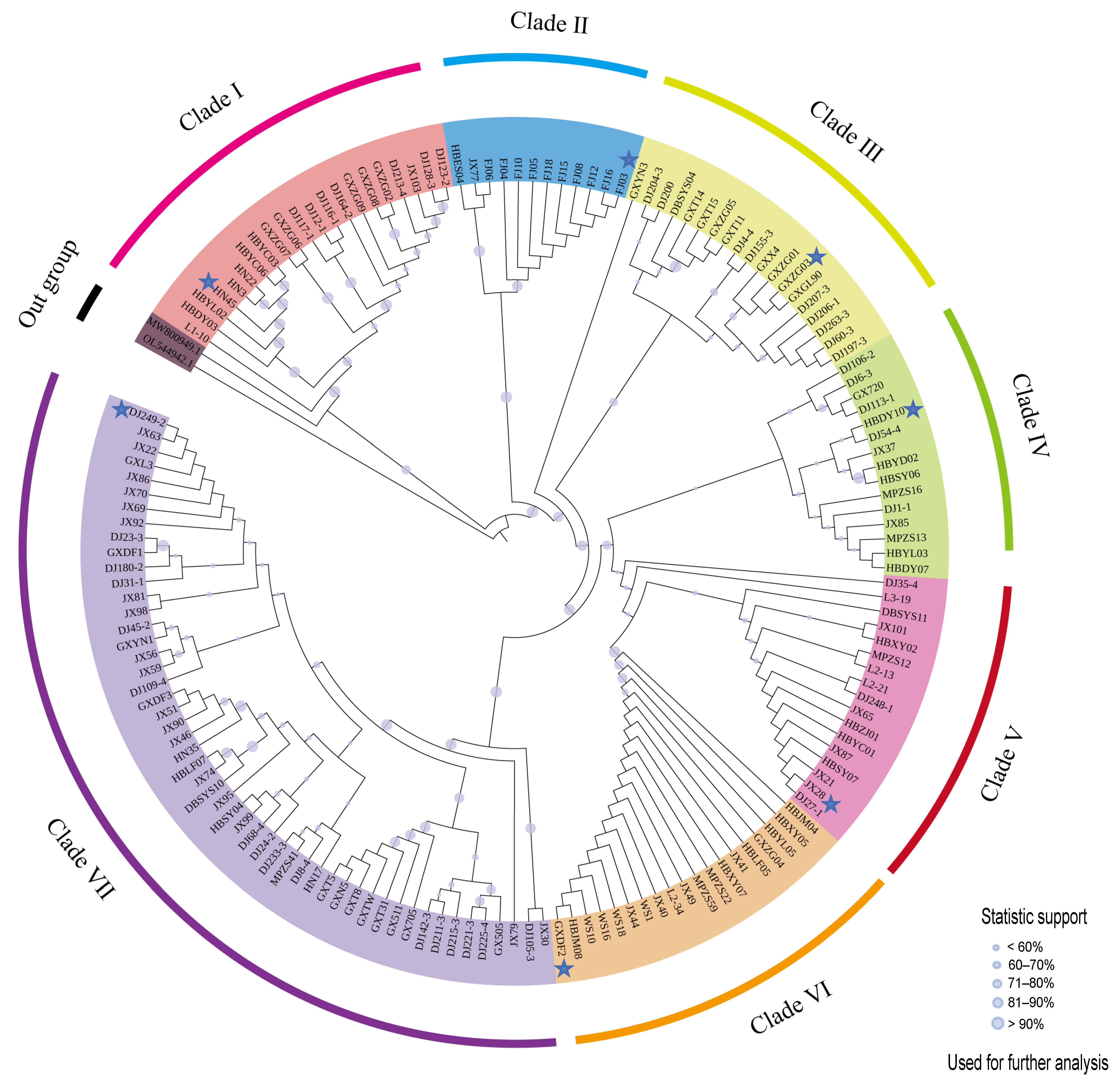
2.2. Phylogenetic Analysis Utilizing the Maximum Likelihood Method
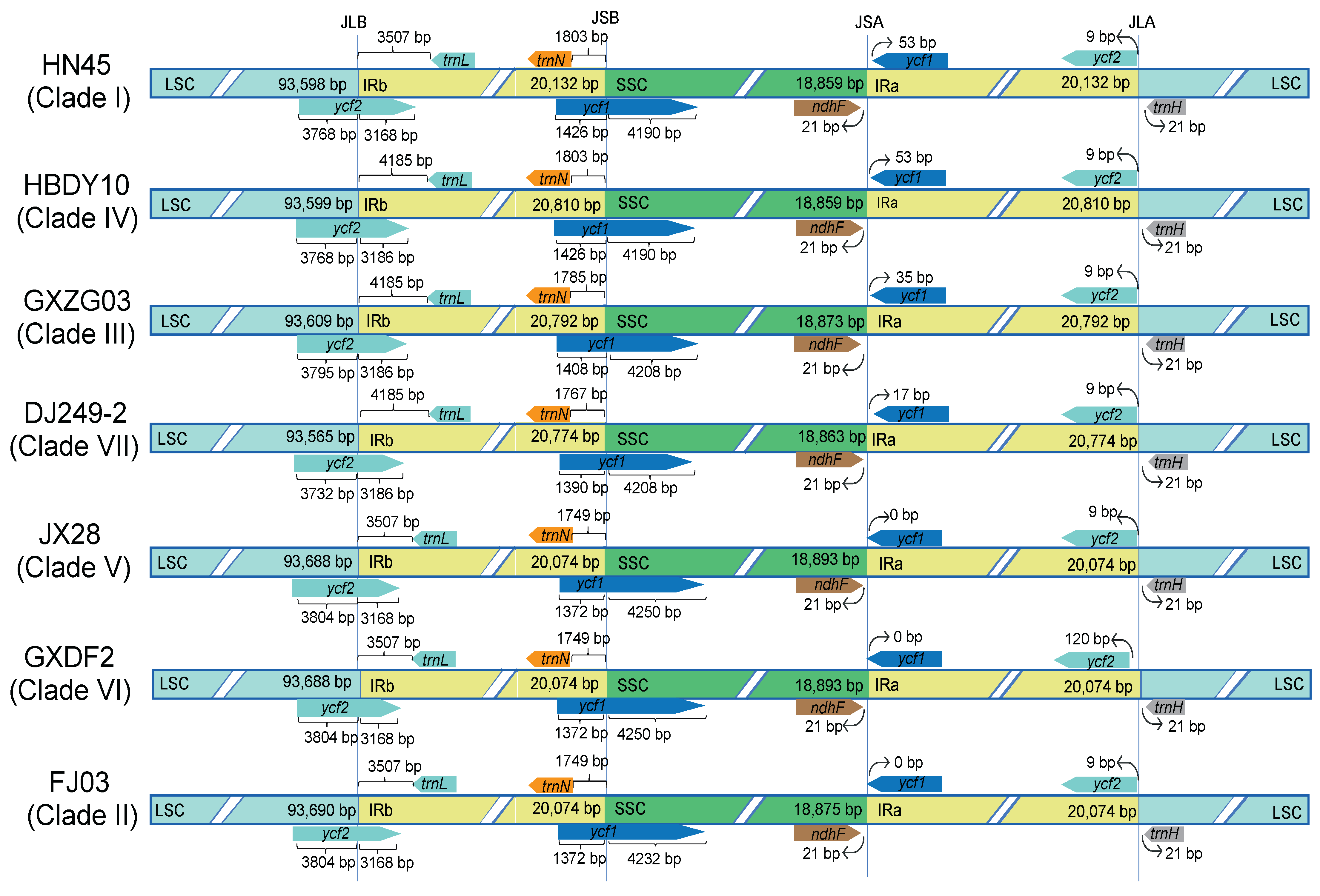
2.3. Comparative Analysis of Chloroplast Genome Boundaries in Camphor Trees
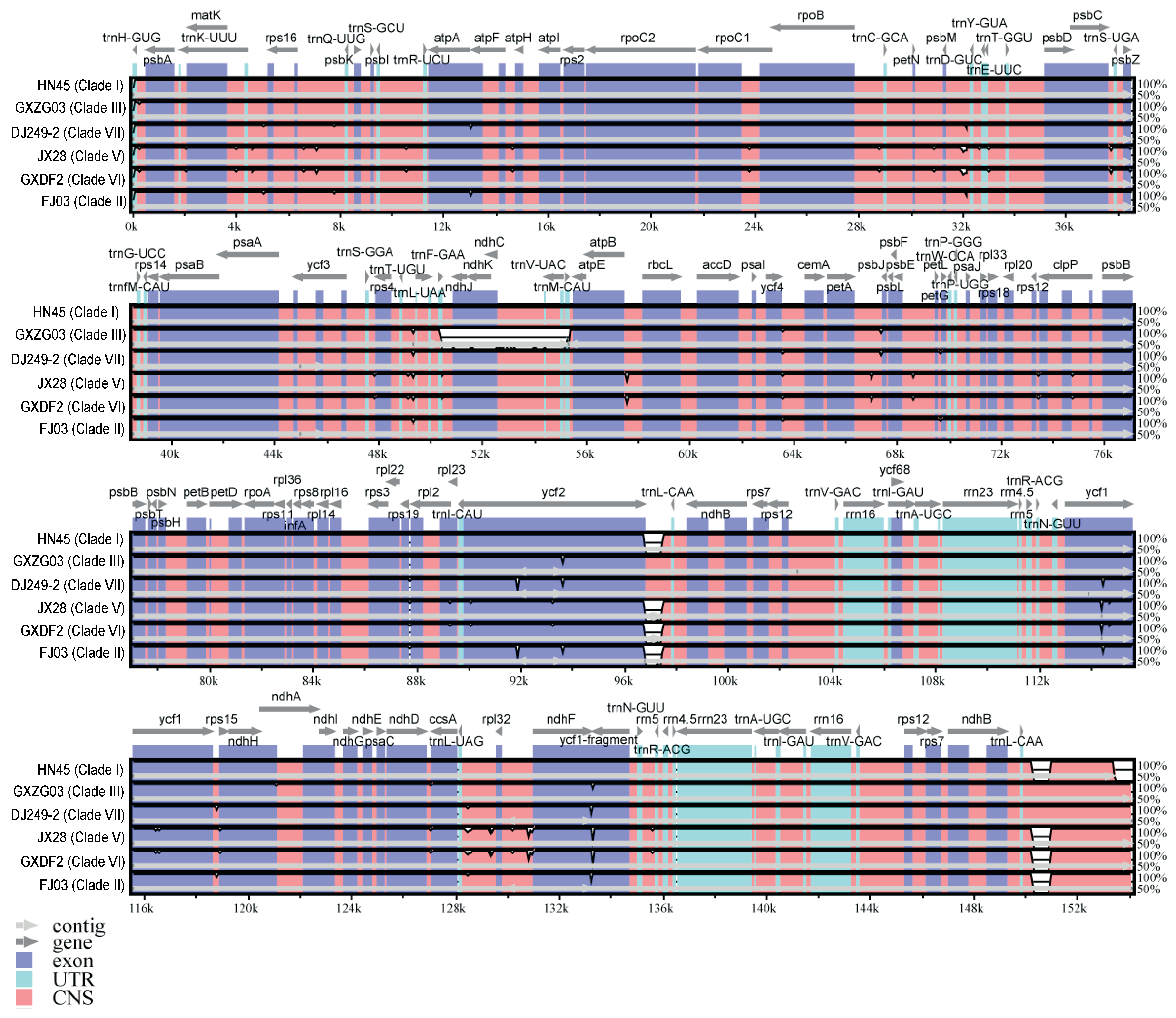
2.4. Sequence Similarity Analysis of the Seven Representative Specimens
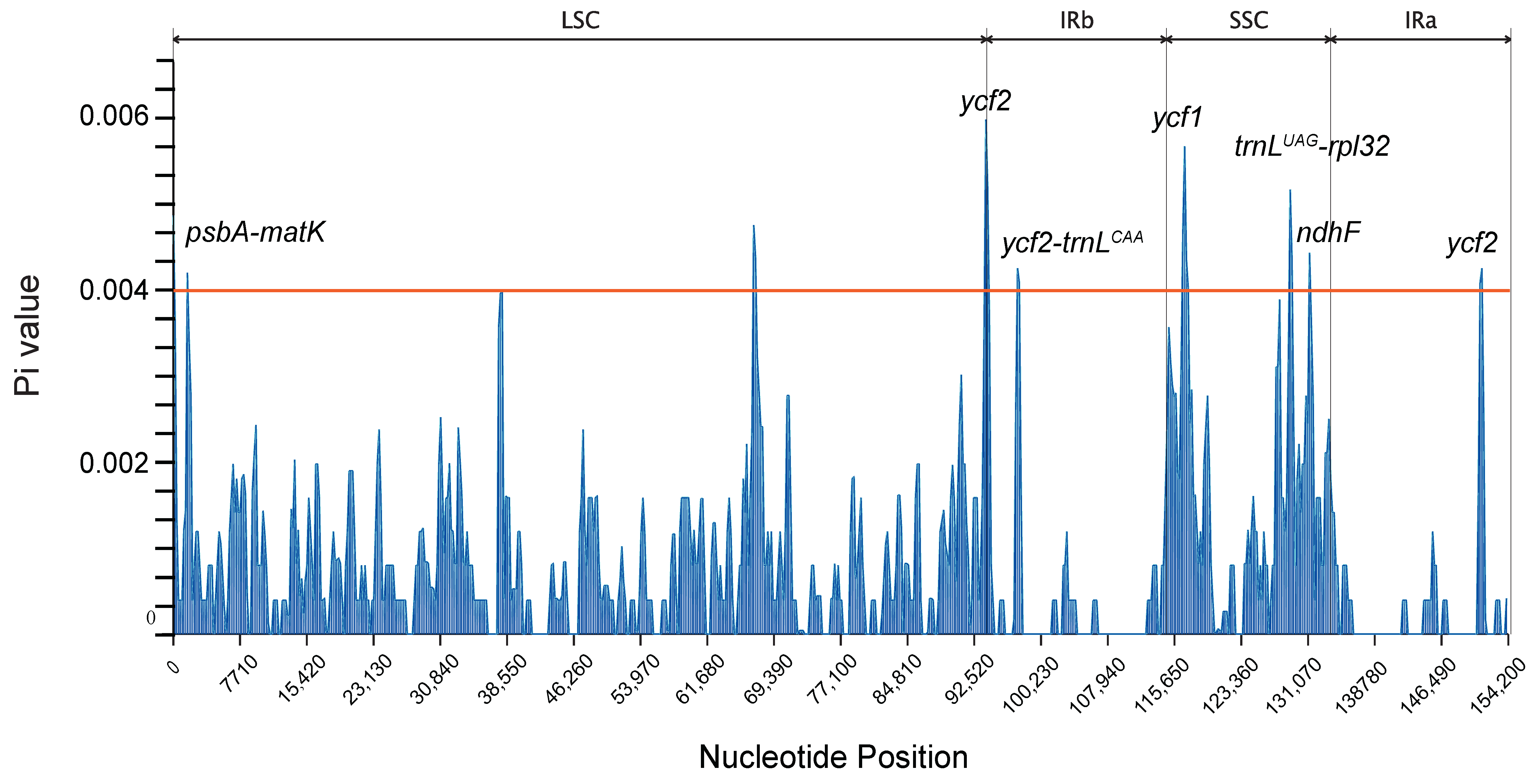
2.5. Analysis of Nucleotide Polymorphism in the Camphor Chloroplast Genome
2.6. Comparative Analysis of Chloroplast Repetitive Sequences of Camphor Tree
3. Discussion
4. Materials and Methods
4.1. Material Collection
4.2. Genome DNA Sequencing and Assembly Annotation
4.3. Construction of the Phylogenetic Tree
4.4. Chloroplast Nucleotide Polymorphism Analysis
4.5. Chloroplast Genome Structure and IR/SC Boundary Analysis
4.6. Simple Repeat Sequence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, J.D.; Jansen, R.K.; Michaels, H.J.; Chase, M.W.; Manhart, J.R. Chloroplast DNA variation and plant phylogeny. Ann. Mo. Bot. Gard. 1988, 75, 1180–1206. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Palmer, J.D.; Stein, D.B. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986, 10, 823–833. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Lu, R.-S.; Li, P.; Qiu, Y.-X. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: Comparative genomic and phylogenetic analyses. Front. Plant. Sci. 2017, 7, 2054. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Duan, X.; Zhang, R.; Guo, C.; Li, L.; Xu, G.; Shan, H.; Kong, H.; Ren, Y. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogenetics Evol. 2019, 135, 12–21. [Google Scholar] [CrossRef]
- Zhang, X.; Rong, C.; Qin, L.; Mo, C.; Fan, L.; Yan, J.; Zhang, M. Complete chloroplast genome sequence of Malus hupehensis: Genome structure, comparative analysis, and phylogenetic relationships. Molecules 2018, 23, 2917. [Google Scholar] [CrossRef]
- Chaw, S.-M.; Chang, C.-C.; Chen, H.-L.; Li, W.-H. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 2004, 58, 424–441. [Google Scholar]
- Sun, Y.; Moore, M.J.; Zhang, S.; Soltis, P.S.; Soltis, D.E.; Zhao, T.; Meng, A.; Li, X.; Li, J.; Wang, H. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol. Phylogenetics Evol. 2016, 96, 93–101. [Google Scholar] [CrossRef]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef]
- Wu, C.-S.; Wang, T.-J.; Wu, C.-W.; Wang, Y.-N.; Chaw, S.-M. Plastome evolution in the sole hemiparasitic genus laurel dodder (Cassytha) and insights into the plastid phylogenomics of Lauraceae. Genome Biol. 2017, 9, 2604–2614. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yu, W.B.; Tan, Y.H.; Jin, J.J.; Wang, B.; Yang, J.B.; Liu, B.; Corlett, R.T. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J. Syst. Evol. 2020, 58, 423–439. [Google Scholar] [CrossRef]
- Yang, Z.; Ferguson, D.K.; Yang, Y. New insights into the plastome evolution of Lauraceae using herbariomics. BMC Plant Biol. 2023, 23, 387. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.-W.; Ge, X.-J. Plastome structure, phylogenomics, and divergence times of tribe Cinnamomeae (Lauraceae). BMC Genom. 2022, 23, 642. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, B.; Yang, Y.; Ferguson, D.K. Phylogeny and taxonomy of Cinnamomum (Lauraceae). Ecol. Evol. 2022, 12, e9378. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, H.; Yang, F.; Zhu, W.; Li, Q.; Cao, Z.; Song, Y.; Xin, P. Phylogeny of Camphora and Cinnamomum (Lauraceae) based on plastome and nuclear ribosomal DNA data. Int. J. Mol. Sci. 2025, 26, 1370. [Google Scholar] [CrossRef]
- Yang, Z.; Ferguson, D.K.; Yang, Y. Plastome phylogeny and taxonomy of Cinnamomum guizhouense (Lauraceae). Forests 2023, 14, 310. [Google Scholar] [CrossRef]
- Li, D.; Lin, H.-Y.; Wang, X.; Bi, B.; Gao, Y.; Shao, L.; Zhang, R.; Liang, Y.; Xia, Y.; Zhao, Y.-P. Genome and whole-genome resequencing of Cinnamomum camphora elucidate its dominance in subtropical urban landscapes. BMC Biol. 2023, 21, 192. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, A.; Li, Z.; Zhang, H.; Liu, L.; Wu, Z.; Li, Y.; Liu, T.; Xu, M.; Yu, F. Genetic diversity and population genetic structure of Cinnamomum camphora in South China revealed by EST-SSR markers. Forests 2019, 10, 1019. [Google Scholar] [CrossRef]
- Gong, X.; Yang, A.; Wu, Z.; Chen, C.; Li, H.; Liu, Q.; Yu, F.; Zhong, Y. Employing genome-wide SNP discovery to characterize the genetic diversity in Cinnamomum camphora using genotyping by sequencing. Forests 2021, 12, 1511. [Google Scholar] [CrossRef]
- Meng, J.; Li, M.; Guo, J.; Zhao, D.; Tao, J. Predicting suitable environments and potential occurrences for Cinnamomum camphora (Linn.) Presl. Forests 2021, 12, 1126. [Google Scholar] [CrossRef]
- Zhang, L.; Jing, Z.; Li, Z.; Liu, Y.; Fang, S. Predictive modeling of suitable habitats for Cinnamomum Camphora (L.) presl using maxent model under climate change in China. Int. J. Environ. Res. Public Health 2019, 16, 3185. [Google Scholar] [CrossRef]
- Tian, Z.; Luo, Q.; Li, Y.; Zuo, Z. Terpinene and β-pinene acting as signaling molecules to improve Cinnamomum camphora thermotolerance. Ind. Crops Prod. 2020, 154, 112641. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.-A.; Xu, M. Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef]
- Chen, C.; Zhong, Y.; Yu, F.; Xu, M. Deep sequencing identifies miRNAs and their target genes involved in the biosynthesis of terpenoids in Cinnamomum camphora. Ind. Crops Prod. 2020, 145, 111853. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, J.; Zhang, B.; Jin, X.; Zhang, H.; Jin, Z. Transcriptional analysis of metabolic pathways and regulatory mechanisms of essential oil biosynthesis in the leaves of Cinnamomum camphora (L.) Presl. Front. Genet. 2020, 11, 598714. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Han, X.; Chen, C.; Zhong, Y.; Xu, M.; Xu, L.A.; Yu, F. Integrating GC-MS and ssRNA-Seq analysis to identify long non-coding RNAs related to terpenoid biosynthesis in Cinnamomum camphora. Ind. Crops Prod. 2021, 171, 113875. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, C.; Zhan, T.; Li, L.; Liu, S.; Huang, Y.; An, W.; Zheng, X.; Huang, S. Genome-wide identification and functional characterization of the trans-isopentenyl diphosphate synthases gene family in Cinnamomum camphora. Front. Plant Sci. 2021, 12, 708697. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, F.; Chen, S.; Wang, K.; Xiang, G.; Liang, X.; An, J.; Li, K.; Liu, L. Proteomics analysis and identification of proteins related to isoprenoid biosynthesis in Cinnamomum camphora (L.) Presl. Forests 2022, 13, 1487. [Google Scholar] [CrossRef]
- Kameyama, Y.; Furumichi, J.; Li, J.; Tseng, Y.-H. Natural genetic differentiation and human-mediated gene flow: The spatiotemporal tendency observed in a long-lived Cinnamomum camphora (Lauraceae) tree. Tree Genet. Genomes 2017, 13, 38. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Liu, S.; Zhong, Y.; Wu, Y.; Li, J.; Xu, L.-A.; Xu, M. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ 2017, 5, e3820. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Jia, G.; Xin, G.; Cai, X. The complete chloroplast genome of Cinnamomum camphora (L.) Presl., a unique economic plant to China. Mitochondrial DNA Part B 2019, 4, 2511–2512. [Google Scholar] [CrossRef]
- Qiu, M.-Y.; Yang, Y.; Wang, N.; Wu, X.; Hu, Y.-L.; Zou, X.-X. The re-sequencing of complete chloroplast genome of Cinnamomum camphora (Lauraceae) from Quanzhou, China. Mitochondrial DNA Part B 2020, 5, 520–521. [Google Scholar] [CrossRef]
- Mower, J.P.; Vickrey, T.L. Structural diversity among plastid genomes of land plants. Adv. Bot. Res. 2018, 85, 263–292. [Google Scholar]
- Zhu, A.; Guo, W.; Gupta, S.; Fan, W.; Mower, J.P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016, 209, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-Y.; Yang, J.-X.; Bai, M.-Z.; Zhang, G.-Q.; Liu, Z.-J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 248. [Google Scholar] [CrossRef]
- Kojoma, M.; Kurihara, K.; Yamada, K.; Sekita, S.; Satake, M.; Iida, O. Genetic identification of cinnamon (Cinnamomum spp.) based on the trnL-trnF chloroplast DNA. Planta Medica 2002, 68, 94–96. [Google Scholar] [CrossRef]
- Chanderbali, A.S.; Van Der Werff, H.; Renner, S.S. Phylogeny and historical biogeography of Lauraceae: Evidence from the chloroplast and nuclear genomes. Ann. Mo. Bot. Gard. 2001, 88, 104–134. [Google Scholar] [CrossRef]
- Xia, L.; Wang, H.; Zhao, X.; Obel, H.O.; Yu, X.; Lou, Q.; Chen, J.; Cheng, C. Chloroplast pan-genomes and comparative transcriptomics reveal genetic variation and temperature adaptation in the cucumber. Int. J. Mol. Sci. 2023, 24, 8943. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; DePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Krumsiek, J.; Arnold, R.; Rattei, T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007, 23, 1026–1028. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Nystuen, A. Lasergene 5.0.1. Biotech Softw. Internet Rep. 2001, 2, 239–244. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
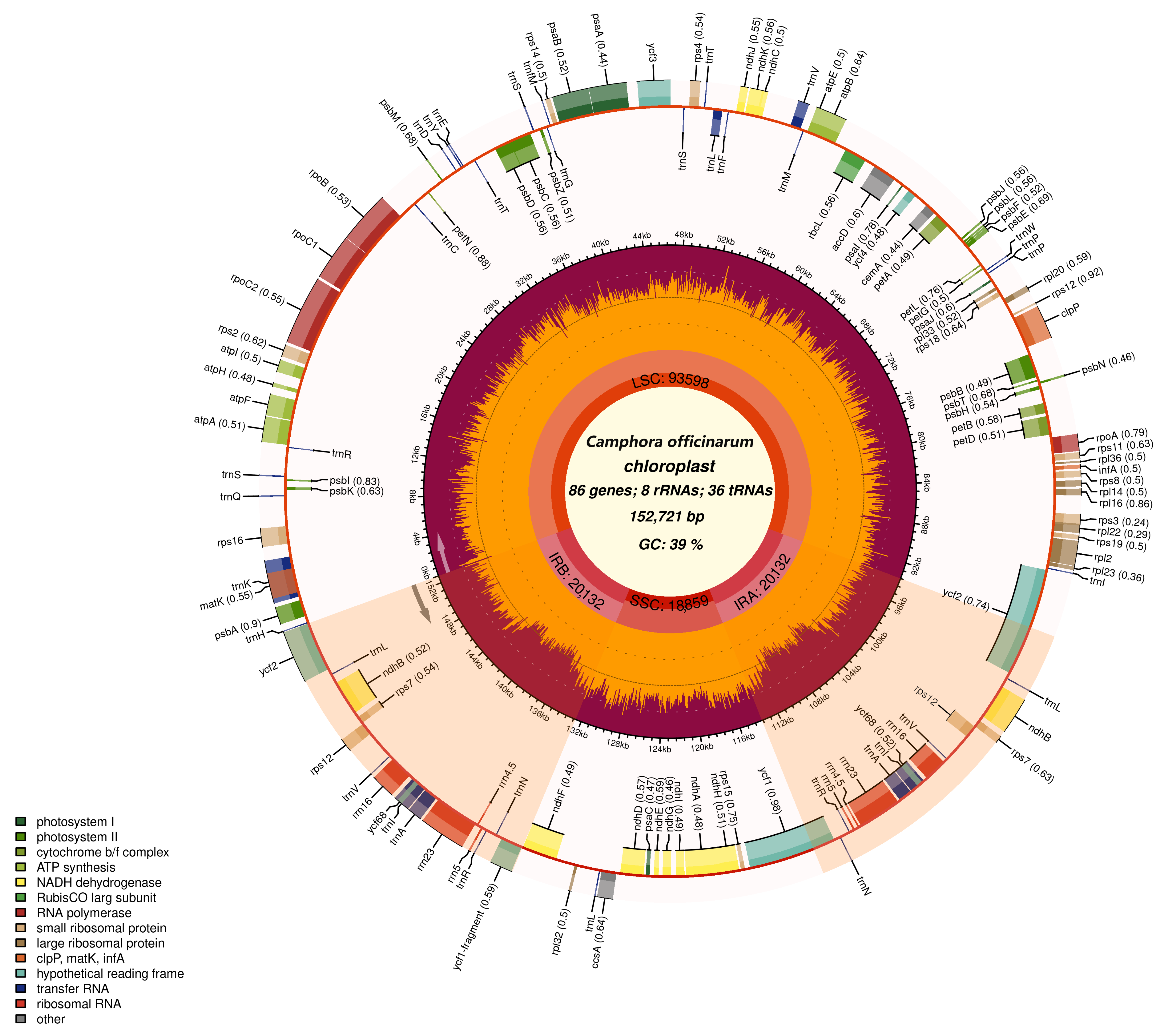

| Category | Gene Functions | Genes |
|---|---|---|
| Photosynthesis | Photosynthem I | psaA, psaB, psaC, psaI, psaJ |
| Photosynthem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| NADH dehydrogenase | ndhA, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| Large subunit Rubisco | rbcL | |
| Transcription and Translation | Ribosomal protein, LSU | rpl14, rpl16, rpl2 *, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Ribosomal protein, SSU | rps11, rps12 *, rps14, rps15, rps16, rps18, rps19, rps2, rps3, rps4, rps7 *, rps8 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Ribosomal RNAs | rrn16 *, rrn23 *, rrn4.5 *, rrn5 * | |
| Transfer RNAs | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-UCC, trnfM-CAU, trnH-GUG, trnI-CAU, trnK-UUU, trnL-UAA, trnL-UAG, trnA-UGC, trnP-GGG, trnP-UGG, trnQ-UUG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-UAC, trnW-CCA, trnY-GUA, trnI-GAU *, trnL-CAA *, trnM-CAU *, trnN-GUU *, trnR-ACG *, trnV-GAC * | |
| Maturase | matK | |
| Biosynthesis | Protease | clpP |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis | ccsA | |
| Unknown function | Unknown group | ycf1 *, ycf2 *, ycf3, ycf4 |
| Samples | Total Length | LSC Length (GC Content) | SSC Length (GC Content) | IR Length (GC Content) | Total GC Content |
|---|---|---|---|---|---|
| HN45 (Clade I) | 152,729 bp | 93,687 bp (38.0%) | 18,894 bp (33.9%) | 40,148 bp (44.4%) | 39.2% |
| FJ03 (Clade II) | 152,620 bp | 93,565 bp (37.9%) | 18,863 bp (33.8%) | 40,192 bp (44.4%) | 39.1% |
| GXZG03 (Clade III) | 154,066 bp | 93,609 bp (37.9%) | 18,873 bp (33.8%) | 41,584 bp (44.3%) | 39.1% |
| HBDY10 (Clade IV) | 152,729 bp | 93,687 bp (38.0%) | 18,894 bp (33.9%) | 40,148 bp (44.4%) | 39.2% |
| JX28 (Clade V) | 154,078 bp | 93,599 bp (37.9%) | 18,859 bp (33.8%) | 41,620 bp (44.3%) | 39.1% |
| GXDF2 (Clade VI) | 152,729 bp | 93,688 bp (38.04%) | 18,893 bp (33.9%) | 40,148 bp (44.4%) | 39.2% |
| DJ249-2 (Clade VII) | 153,976 bp | 93,565 bp (37.9%) | 18,863 bp (33.8%) | 41,548 bp (44.3%) | 39.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, C.; Jiang, Y.; Zhang, Q.; Yao, J.; Lian, H.; Wang, M.; Xie, P.; Chen, Y.; Cai, Y. Plastid Phylogenomics of Camphora officinarum Nees: Unraveling Genetic Diversity and Geographic Differentiation in East Asian Subtropical Forests. Int. J. Mol. Sci. 2025, 26, 9229. https://doi.org/10.3390/ijms26189229
Hou C, Jiang Y, Zhang Q, Yao J, Lian H, Wang M, Xie P, Chen Y, Cai Y. Plastid Phylogenomics of Camphora officinarum Nees: Unraveling Genetic Diversity and Geographic Differentiation in East Asian Subtropical Forests. International Journal of Molecular Sciences. 2025; 26(18):9229. https://doi.org/10.3390/ijms26189229
Chicago/Turabian StyleHou, Chen, Yingchao Jiang, Qian Zhang, Jun Yao, Huiming Lian, Minghuai Wang, Peiwu Xie, Yiqun Chen, and Yanling Cai. 2025. "Plastid Phylogenomics of Camphora officinarum Nees: Unraveling Genetic Diversity and Geographic Differentiation in East Asian Subtropical Forests" International Journal of Molecular Sciences 26, no. 18: 9229. https://doi.org/10.3390/ijms26189229
APA StyleHou, C., Jiang, Y., Zhang, Q., Yao, J., Lian, H., Wang, M., Xie, P., Chen, Y., & Cai, Y. (2025). Plastid Phylogenomics of Camphora officinarum Nees: Unraveling Genetic Diversity and Geographic Differentiation in East Asian Subtropical Forests. International Journal of Molecular Sciences, 26(18), 9229. https://doi.org/10.3390/ijms26189229






