Computational Metagenomics: State of the Art
Abstract
1. Introduction
1.1. Overview of Metagenomics and Microbiome Analysis
1.2. Significance of Computational Approaches in Analyzing Microbial Communities
1.3. Recent Advancements in Computational Tools for Biological Research
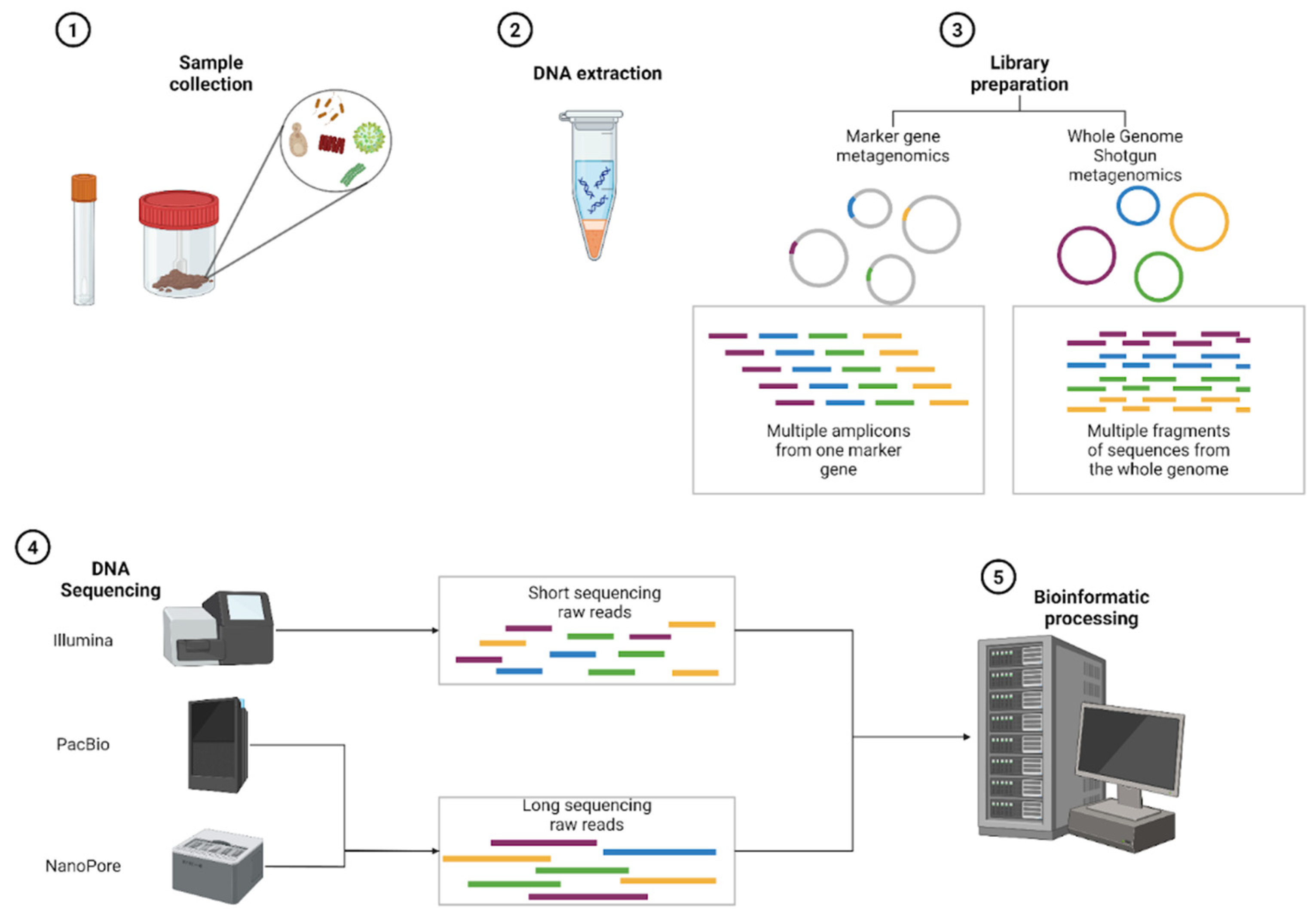
2. Computational Methods for Metagenomics and Microbiome Analysis
2.1. Method Approach and Sequencing Technologies
2.1.1. Technology Selection Framework
2.1.2. Resolution Capabilities: Species Versus Strain-Level Analysis
2.1.3. Practical Experimental Design Recommendations
2.1.4. Strategic Technology Selection
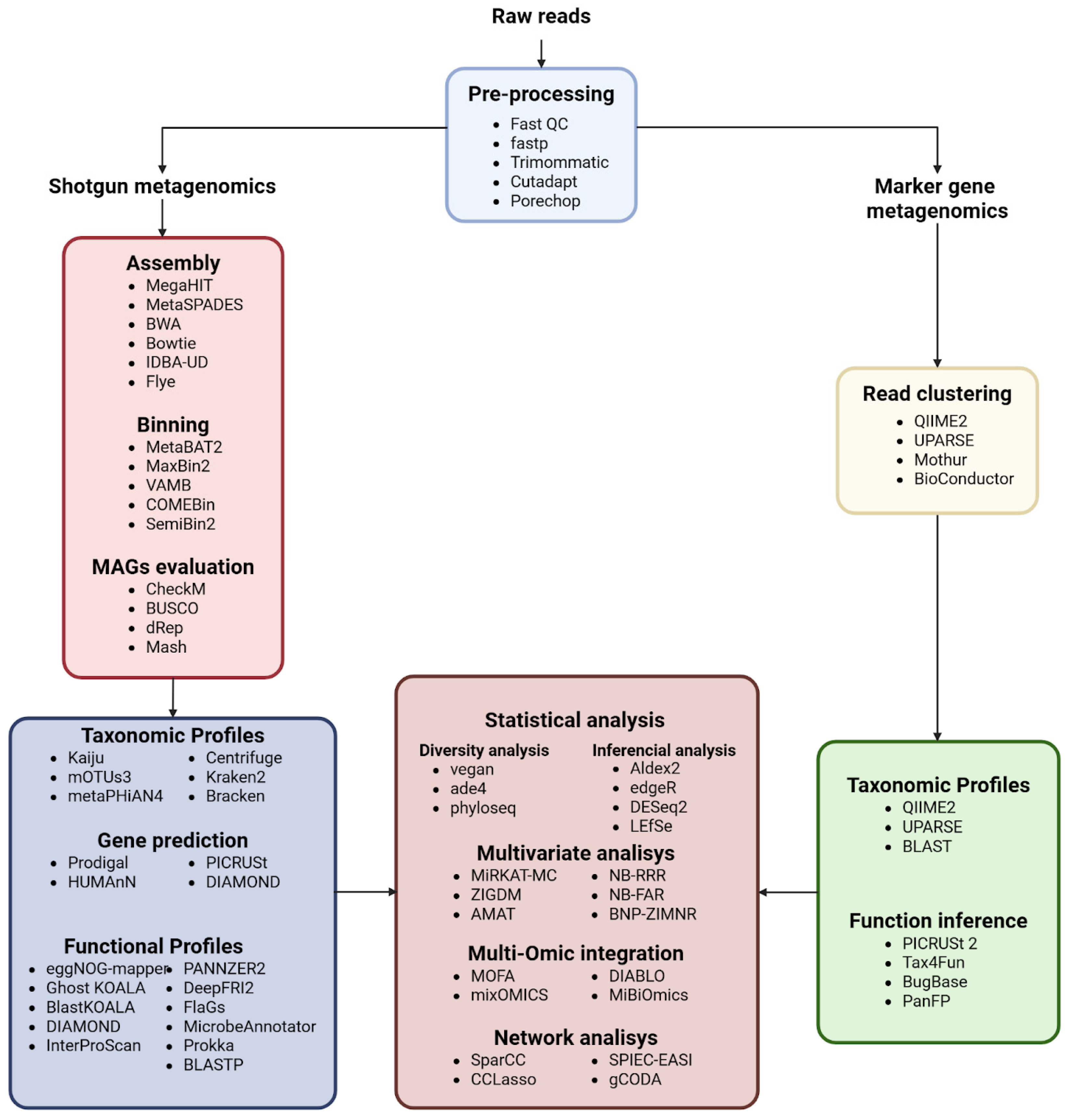
2.2. Bioinformatics Pipelines
2.2.1. Computational Paths: From Raw Data to FASTQ
2.2.2. Preprocessing and Quality Control
2.2.3. Contig/Genome Assembly Approaches
- De novo Assembly:
- Reference-guided Assembly:
2.2.4. Binning of Metagenomic Contigs and Genome Recovery
- Composition-based Methods:
- Abundance-based Methods:
- Hybrid Methods:
2.2.5. Evaluation of MAG Quality
- Completeness and Contamination:
- De-replication and Cataloging:
- Additional Metrics:
2.2.6. Reads Clustering for Taxonomic Classification
2.2.7. Taxonomic Profiles
Amplicon Metagenomics
Whole Genome Metagenomics
2.2.8. Gene Prediction and Annotation
Gene Prediction
Functional Annotation
2.2.9. Metagenomic Pipelines
2.3. Choosing an Adequate Database
2.3.1. Sequence Repositories
2.3.2. General Databases for Taxonomic Analysis
2.3.3. General Databases for Functional Analysis
2.3.4. Particular Human Microbiome
2.3.5. Specialized Questions
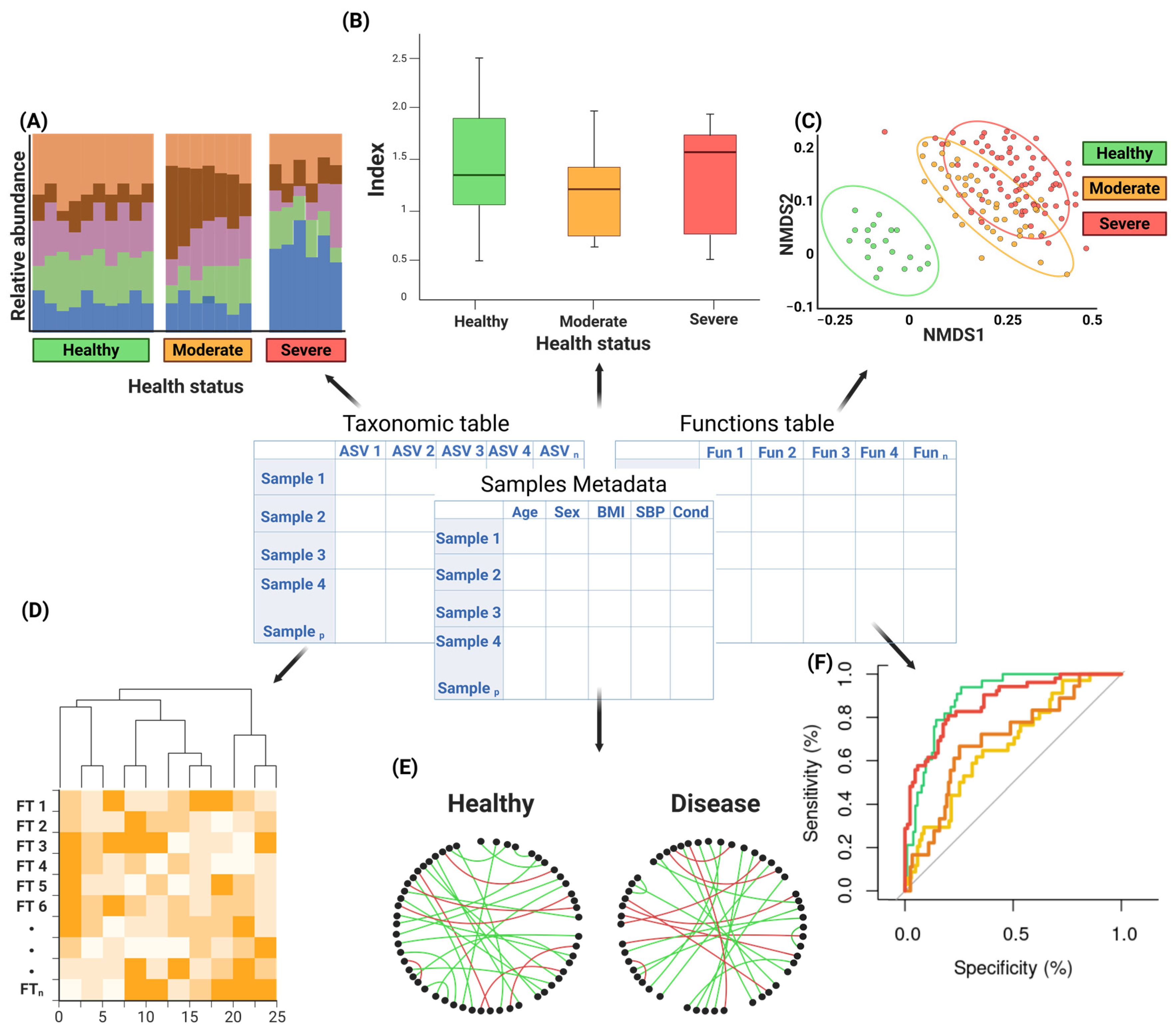
3. Downstream Analyses in Metagenomics
3.1. Descriptive Analysis
3.2. Statistical Analysis
3.2.1. Differential Abundance
3.2.2. Multivariate Analysis
3.3. Integration with Multi-Omics Data
3.4. Network and Interaction Analysis
4. AI and Machine Learning in Metagenomics
4.1. AI/ML Techniques in Microbiome Data Analysis
4.2. Current Applications of ML in Metagenomics
4.2.1. Disease Prediction
4.2.2. Identification of Microbial Signatures
4.2.3. Functional Trait Prediction
4.3. AI Tools for Metagenomics
4.4. Comparison with Traditional Tools
- Computational resources: Traditional methods often require significant computational resources, particularly for large datasets.
- Efficiency in Application: The computational profile of AI-based tools presents a critical trade-off between training and application. While the process of training deep learning models is often famously resource-intensive, requiring large datasets and significant computational power, the resulting models can be highly efficient for inference. Once trained, applying a model to classify new sequences or predict a phenotype is often computationally much faster than performing traditional, per-sample alignment-based searches against large reference databases [3].
- Pattern Recognition: Advanced algorithms identify subtle patterns and relationships that traditional tools may overlook, such as novel microbial signatures or complex microbial interactions.
- Scalability: AI tools are well-suited for managing the ever-expanding scale of metagenomic data, enabling large-scale analyses that were previously impractical.
4.5. Future Directions: Advancing AI in Metagenomics
4.5.1. Enhancing Personalized Medicine
4.5.2. Overcoming Challenges: Benchmarking, Robustness, and Interpretability
Benchmarking and External Validation
Overfitting Mitigation and Uncertainty Estimation
Interpretability, Feature Stability and Biological Validation
Bridging the Gap: Multidisciplinary Collaboration and Innovation
5. Applications in Human and Environmental Health
5.1. Microbiome and Disease
5.1.1. Metabolic Diseases
5.1.2. Liver Diseases
5.1.3. Pregnancy and Reproductive Disorders
5.1.4. Neurological Disorders
5.1.5. Inflammatory Skin Disorders
5.1.6. Autoimmune Diseases
- Metagenomics confirmed the depletion of key commensals and provided the genomic blueprints (via MAGs) of the community members.
- Metaproteomics complemented this by providing direct evidence of which proteins—the functional workhorses of the cell—were being produced, confirming that inflammatory and stress-response pathways were highly active [240].
- Metabolomics measured the downstream biochemical output, identifying clear shifts in bile acid metabolism and SCFA production that correlated with the functional changes observed at the gene and protein level.
5.2. Therapeutic Applications
5.2.1. Probiotics
5.2.2. Prebiotics
5.2.3. Fecal Microbiota Transplantation
5.2.4. Precision Medicine
6. Data Sharing and Open Science
7. Next Steps in Metagenomics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Lema, N.K.; Gemeda, M.T.; Woldesemayat, A.A. Recent Advances in Metagenomic Approaches, Applications, and Challenges. Curr. Microbiol. 2023, 80, 347. [Google Scholar] [CrossRef] [PubMed]
- Roy, G.; Prifti, E.; Belda, E.; Zucker, J.-D. Deep Learning Methods in Metagenomics: A Review. Microb. Genomics 2024, 10, 001231. [Google Scholar] [CrossRef]
- Wani, A.K.; Roy, P.; Kumar, V.; ul Gani Mir, T. Metagenomics and Artificial Intelligence in the Context of Human Health. Infect. Genet. Evol. 2022, 100, 105267. [Google Scholar] [CrossRef]
- Huo, D.; Wang, X. A New Era in Healthcare: The Integration of Artificial Intelligence and Microbial. Med. Nov. Technol. Devices 2024, 23, 100319. [Google Scholar] [CrossRef]
- Kumar, R.; Yadav, G.; Kuddus, M.; Ashraf, G.M.; Singh, R. Unlocking the Microbial Studies through Computational Approaches: How Far Have We Reached? Environ. Sci. Pollut. Res. 2023, 30, 48929–48947. [Google Scholar] [CrossRef]
- Bernau, C.; Riester, M.; Boulesteix, A.-L.; Parmigiani, G.; Huttenhower, C.; Waldron, L.; Trippa, L. Cross-Study Validation for the Assessment of Prediction Algorithms. Bioinformatics 2014, 30, i105–i112. [Google Scholar] [CrossRef]
- Kim, C.; Pongpanich, M.; Porntaveetus, T. Unraveling Metagenomics through Long-Read Sequencing: A Comprehensive Review. J. Transl. Med. 2024, 22, 111. [Google Scholar] [CrossRef]
- Van Uffelen, A.; Posadas, A.; Roosens, N.H.C.; Marchal, K.; De Keersmaecker, S.C.J.; Vanneste, K. Benchmarking Bacterial Taxonomic Classification Using Nanopore Metagenomics Data of Several Mock Communities. Sci. Data 2024, 11, 864. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Nesme, J.; Santos-Bay, L.; Koziol, A.; Sørensen, S.J.; Alberdi, A.; Aizpurua, O. A Comparison of Short-Read, HiFi Long-Read, and Hybrid Strategies for Genome-Resolved Metagenomics. Microbiol. Spectr. 2024, 12, e0359023. [Google Scholar] [CrossRef]
- Bertrand, D.; Chia, J.X.; Kundu, A.; Hu, B.; Lin, W.; Nagarajan, N. Comparative Analysis of Metagenomic Classifiers for Long-Read Sequencing Datasets. BMC Bioinform. 2024, 25, 22. [Google Scholar] [CrossRef]
- Samarakoon, H.; Ferguson, J.M.; Gamaarachchi, H.; Deveson, I.W.; Alkan, C. Gamaarachchi, Hasindu, Deveson Accelerated Nanopore Basecalling with SLOW5 Data Format. Bioinformatics 2023, 39, btad352. [Google Scholar] [CrossRef]
- SMRT Analysis Software. Available online: https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/ (accessed on 11 March 2025).
- Bcl2fastq2 Conversion Software v2.20 Software Guide (15051736). Available online: https://support.illumina.com/content/dam/illumina-support/documents/documentation/software_documentation/bcl2fastq/bcl2fastq2-v2-20-software-guide-15051736-03.pdf (accessed on 11 March 2025).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 February 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing Bacterial Genome Assemblies with Multiplex MinION Sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de Novo Assembler for Single-Cell and Metagenomic Sequencing Data with Highly Uneven Depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Mallawaarachchi, V.; Wickramarachchi, A.; Xue, H.; Papudeshi, B.; Grigson, S.R.; Bouras, G.; Prahl, R.E.; Kaphle, A.; Verich, A.; Talamantes-Becerra, B.; et al. Solving Genomic Puzzles: Computational Methods for Metagenomic Binning. Brief. Bioinform. 2024, 25, bbae372. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An Automated Binning Algorithm to Recover Genomes from Multiple Metagenomic Datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Nissen, J.N.; Johansen, J.; Allesøe, R.L.; Sønderby, C.K.; Armenteros, J.J.A.; Grønbech, C.H.; Jensen, L.J.; Nielsen, H.B.; Petersen, T.N.; Winther, O.; et al. Improved Metagenome Binning and Assembly Using Deep Variational Autoencoders. Nat. Biotechnol. 2021, 39, 555–560. [Google Scholar] [CrossRef]
- Wang, Z.; You, R.; Han, H.; Liu, W.; Sun, F.; Zhu, S. Effective Binning of Metagenomic Contigs Using Contrastive Multi-View Representation Learning. Nat. Commun. 2024, 15, 585. [Google Scholar] [CrossRef]
- Pan, S.; Zhao, X.-M.; Coelho, L.P. SemiBin2: Self-Supervised Contrastive Learning Leads to Better MAGs for Short- and Long-Read Sequencing. Bioinformatics 2023, 39, i21–i29. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, M.; Yang, J. Recovering Metagenome-Assembled Genomes from Shotgun Metagenomic Sequencing Data: Methods, Applications, Challenges, and Opportunities. Microbiol. Res. 2022, 260, 127023. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum Information about a Single Amplified Genome (MISAG) and a Metagenome-Assembled Genome (MIMAG) of Bacteria and Archaea. Nat. Biotechnol. 2017, 35, 725–731, Erratum in Nat Biotechnol. 2018, 36, 660. https://doi.org/10.1038/nbt0718-660a. [Google Scholar] [CrossRef]
- Kim, N.; Ma, J.; Kim, W.; Kim, J.; Belenky, P.; Lee, I. Genome-Resolved Metagenomics: A Game Changer for Microbiome Medicine. Exp. Mol. Med. 2024, 56, 1501–1512. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and Sensitive Classification of Metagenomic Sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A Practical Guide to Amplicon and Metagenomic Analysis of Microbiome Data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating Species Abundance in Metagenomics Data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Ounit, R.; Wanamaker, S.; Close, T.J.; Lonardi, S. CLARK: Fast and Accurate Classification of Metagenomic and Genomic Sequences Using Discriminative k-Mers. BMC Genom. 2015, 16, 236. [Google Scholar] [CrossRef]
- Ounit, R.; Lonardi, S. Higher Classification Sensitivity of Short Metagenomic Reads with CLARK-S. Bioinformatics 2016, 32, 3823–3825. [Google Scholar] [CrossRef]
- Meyer, F.; Lesker, T.-R.; Koslicki, D.; Fritz, A.; Gurevich, A.; Darling, A.E.; Sczyrba, A.; Bremges, A.; McHardy, A.C. Tutorial: Assessing Metagenomics Software with the CAMI Benchmarking Toolkit. Nat. Protoc. 2021, 16, 1785–1801. [Google Scholar] [CrossRef]
- Kutuzova, S.; Nielsen, M.; Piera, P.; Nissen, J.N.; Rasmussen, S. Taxometer: Improving Taxonomic Classification of Metagenomics Contigs. Nat. Commun. 2024, 15, 8357. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Milanese, A.; Mende, D.R.; Paoli, L.; Salazar, G.; Ruscheweyh, H.-J.; Cuenca, M.; Hingamp, P.; Alves, R.; Costea, P.I.; Coelho, L.P.; et al. Microbial Abundance, Activity and Population Genomic Profiling with mOTUs2. Nat. Commun. 2019, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Ruscheweyh, H.-J.; Milanese, A.; Paoli, L.; Karcher, N.; Clayssen, Q.; Keller, M.I.; Wirbel, J.; Bork, P.; Mende, D.R.; Zeller, G.; et al. Cultivation-Independent Genomes Greatly Expand Taxonomic-Profiling Capabilities of mOTUs across Various Environments. Microbiome 2022, 10, 212. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M.; et al. Extending and Improving Metagenomic Taxonomic Profiling with Uncharacterized Species Using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Bremges, A.; Belmann, P.; Janssen, S.; McHardy, A.C.; Koslicki, D. Assessing Taxonomic Metagenome Profilers with OPAL. Genome Biol. 2019, 20, 51. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Levy Karin, E.; Mirdita, M.; Söding, J. MetaEuk—Sensitive, High-Throughput Gene Discovery, and Annotation for Large-Scale Eukaryotic Metagenomics. Microbiome 2020, 8, 48. [Google Scholar] [CrossRef]
- Neely, C.J.; Hu, S.K.; Alexander, H.; Tully, B.J. The High-Throughput Gene Prediction of More than 1700 Eukaryote Genomes Using the Software Package EukMetaSanity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, Z. Introduction to Computational Metagenomics; WSPC: Singapore, 2022. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of Habitat-Specific Functional Profiles and Functional Redundancy Based on 16S rRNA Gene Sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase Predicts Organism-Level Microbiome Phenotypes. bioRxiv 2017, 133462. [Google Scholar] [CrossRef]
- Jun, S.-R.; Robeson, M.S.; Hauser, L.J.; Schadt, C.W.; Gorin, A.A. PanFP: Pangenome-Based Functional Profiles for Microbial Communities. BMC Res. Notes 2015, 8, 479. [Google Scholar] [CrossRef]
- Mongad, D.S.; Chavan, N.S.; Narwade, N.P.; Dixit, K.; Shouche, Y.S.; Dhotre, D.P. MicFunPred: A Conserved Approach to Predict Functional Profiles from 16S rRNA Gene Sequence Data. Genomics 2021, 113, 3635–3643. [Google Scholar] [CrossRef]
- Wajid, B.; Anwar, F.; Wajid, I.; Nisar, H.; Meraj, S.; Zafar, A.; Al-Shawaqfeh, M.K.; Ekti, A.R.; Khatoon, A.; Suchodolski, J.S. Music of Metagenomics-a Review of Its Applications, Analysis Pipeline, and Associated Tools. Funct. Integr. Genom. 2022, 22, 3–26. [Google Scholar] [CrossRef]
- Bharti, R.; Grimm, D.G. Current Challenges and Best-Practice Protocols for Microbiome Analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic Approaches in Microbial Ecology: An Update on Whole-Genome and Marker Gene Sequencing Analyses. Microb. Genom. 2020, 6, mgen000409. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chowdhury, D.; Zhang, Z.; Cheung, W.K.; Lu, A.; Bian, Z.; Zhang, L. A Review of Computational Tools for Generating Metagenome-Assembled Genomes from Metagenomic Sequencing Data. Comput. Struct. Biotechnol. J. 2021, 19, 6301–6314. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, C.A.; Conrad, R.E.; Konstantinidis, K.T. MicrobeAnnotator: A User-Friendly, Comprehensive Functional Annotation Pipeline for Microbial Genomes. BMC Bioinform. 2021, 22, 11. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 Enables Sensitive Protein Sequence Searching for the Analysis of Massive Data Sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Maranga, M.; Szczerbiak, P.; Bezshapkin, V.; Gligorijevic, V.; Chandler, C.; Bonneau, R.; Xavier, R.J.; Vatanen, T.; Kosciolek, T. Comprehensive Functional Annotation of Metagenomes and Microbial Genomes Using a Deep Learning-Based Method. mSystems 2023, 8, e0117822. [Google Scholar] [CrossRef]
- Saha, C.K.; Pires, R.S.; Brolin, H.; Delannoy, M.; Atkinson, G.C. Predicting Functional Associations Using Flanking Genes (FlaGs). bioRxiv 2020, 362095. [Google Scholar] [CrossRef]
- Anand, S.; Kuntal, B.K.; Mohapatra, A.; Bhatt, V.; Mande, S.S. FunGeCo: A Web-Based Tool for Estimation of Functional Potential of Bacterial Genomes and Microbiomes Using Gene Context Information. Bioinformatics 2020, 36, 2575–2577. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.; Borenstein, E. Comparative Analysis of Functional Metagenomic Annotation and the Mappability of Short Reads. PLoS ONE 2014, 9, e105776. [Google Scholar] [CrossRef] [PubMed]
- Mirete, S.; Sánchez-Costa, M.; Díaz-Rullo, J.; González de Figueras, C.; Martínez-Rodríguez, P.; González-Pastor, J.E. Metagenome-Assembled Genomes (MAGs): Advances, Challenges, and Ecological Insights. Microorganisms 2025, 13, 985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, J.; Wang, H.; Yang, C.; Huang, Y.; Yue, Z.; Chen, Y.; Han, L.; Yin, K.; Lyu, A.; et al. Exploring High-Quality Microbial Genomes by Assembling Short-Reads with Long-Range Connectivity. Nat. Commun. 2024, 15, 4631. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An Algorithm for Hybrid Assembly of Short and Long Reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Bertrand, D.; Shaw, J.; Kalathiyappan, M.; Ng, A.H.Q.; Kumar, M.S.; Li, C.; Dvornicic, M.; Soldo, J.P.; Koh, J.Y.; Tong, C.; et al. Hybrid Metagenomic Assembly Enables High-Resolution Analysis of Resistance Determinants and Mobile Elements in Human Microbiomes. Nat. Biotechnol. 2019, 37, 937–944. [Google Scholar] [CrossRef]
- Kajitani, R.; Noguchi, H.; Gotoh, Y.; Ogura, Y.; Yoshimura, D.; Okuno, M.; Toyoda, A.; Kuwahara, T.; Hayashi, T.; Itoh, T. MetaPlatanus: A Metagenome Assembler That Combines Long-Range Sequence Links and Species-Specific Features. Nucleic Acids Res. 2021, 49, e130. [Google Scholar] [CrossRef]
- Jung, S. Advances in Functional Analysis of the Microbiome: Integrating Metabolic Modeling, Metabolite Prediction, and Pathway Inference with Next-Generation Sequencing Data. J. Microbiol. 2025, 63, e2411006. [Google Scholar] [CrossRef]
- Marizzoni, M.; Gurry, T.; Provasi, S.; Greub, G.; Lopizzo, N.; Ribaldi, F.; Festari, C.; Mazzelli, M.; Mombelli, E.; Salvatore, M.; et al. Comparison of Bioinformatics Pipelines and Operating Systems for the Analyses of 16S rRNA Gene Amplicon Sequences in Human Fecal Samples. Front. Microbiol. 2020, 11, 1262. [Google Scholar] [CrossRef]
- Shaw, J.; Yu, Y.W. Rapid Species-Level Metagenome Profiling and Containment Estimation with Sylph. Nat. Biotechnol. 2025, 43, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.H.; Siddle, K.J.; Park, D.J.; Sabeti, P.C. Benchmarking Metagenomics Tools for Taxonomic Classification. Cell 2019, 178, 779–794. [Google Scholar] [CrossRef]
- Alonso-Reyes, D.G.; Albarracin, V.H. Arche: An Advanced Flexible Tool for High-Throuput Annotation of Functions on Microbial Contigs. bioRxiv 2024. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A Flexible Pipeline for Genome-Resolved Metagenomic Data Analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Kultima, J.R.; Coelho, L.P.; Forslund, K.; Huerta-Cepas, J.; Li, S.S.; Driessen, M.; Voigt, A.Y.; Zeller, G.; Sunagawa, S.; Bork, P. MOCAT2: A Metagenomic Assembly, Annotation and Profiling Framework. Bioinformatics 2016, 32, 2520–2523. [Google Scholar] [CrossRef] [PubMed]
- Narayanasamy, S.; Jarosz, Y.; Muller, E.E.L.; Heintz-Buschart, A.; Herold, M.; Kaysen, A.; Laczny, C.C.; Pinel, N.; May, P.; Wilmes, P. IMP: A Pipeline for Reproducible Reference-Independent Integrated Metagenomic and Metatranscriptomic Analyses. Genome Biol. 2016, 17, 260. [Google Scholar] [CrossRef]
- Köster, J.; Rahmann, S. Snakemake—A Scalable Bioinformatics Workflow Engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow Enables Reproducible Computational Workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Minot, S.S.; Garb, B.; Roldan, A.; Tang, A.S.; Oskotsky, T.T.; Rosenthal, C.; Hoffman, N.G.; Sirota, M.; Golob, J.L. MaLiAmPi Enables Generalizable and Taxonomy-Independent Microbiome Features from Technically Diverse 16S-Based Microbiome Studies. Cell Rep. Methods 2023, 3, 100639. [Google Scholar] [CrossRef]
- Kieser, S.; Brown, J.; Zdobnov, E.M.; Trajkovski, M.; McCue, L.A. ATLAS: A Snakemake Workflow for Assembly, Annotation, and Genomic Binning of Metagenome Sequence Data. BMC Bioinform. 2020, 21, 257. [Google Scholar] [CrossRef] [PubMed]
- Tadrent, N.; Dedeine, F.; Hervé, V. SnakeMAGs: A Simple, Efficient, Flexible and Scalable Workflow to Reconstruct Prokaryotic Genomes from Metagenomes. F1000Research 2022, 11, 1522. [Google Scholar] [CrossRef] [PubMed]
- Krapohl, J.; Pickett, B.E. SnakeWRAP: A Snakemake Workflow to Facilitate Automated Processing of Metagenomic Data through the metaWRAP Pipeline. F1000Research 2022, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A Review of Methods and Databases for Metagenomic Classification and Assembly. Brief. Bioinform. 2017, 20, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Chorlton, S.D. Ten Common Issues with Reference Sequence Databases and How to Mitigate Them. Front. Bioinforma. 2024, 4, 1278228. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Godoy-Lozano, E.E.; Raggi, L.; Segovia, L.; Merino, E.; Gutiérrez-Rios, R.M.; Juarez, K.; Licea-Navarro, A.F.; Pardo-Lopez, L.; Sanchez-Flores, A. Author Correction: Analysis of Sequencing Strategies and Tools for Taxonomic Annotation: Defining Standards for Progressive Metagenomics. Sci. Rep. 2020, 10, 4259. [Google Scholar] [CrossRef]
- Wright, R.J.; Comeau, A.M.; Langille, M.G.I. From Defaults to Databases: Parameter and Database Choice Dramatically Impact the Performance of Metagenomic Taxonomic Classification Tools. Microb. Genom. 2023, 9, 000949. [Google Scholar] [CrossRef]
- Xu, R.; Rajeev, S.; Salvador, L.C.M. The Selection of Software and Database for Metagenomics Sequence Analysis Impacts the Outcome of Microbial Profiling and Pathogen Detection. PLoS ONE 2023, 18, e0284031. [Google Scholar] [CrossRef]
- Zheng, B.; Xu, J.; Zhang, Y.; Qin, J.; Yuan, D.; Fan, T.; Wu, W.; Chen, Y.; Jiang, Y. MBCN: A Novel Reference Database for Effcient Metagenomic Analysis of Human Gut Microbiome. Heliyon 2024, 10, e37422. [Google Scholar] [CrossRef]
- Martins, I.B.; Miguel Silva, J.; Almeida, J.R. A Comprehensive Study of Databases to Assess the Reliability of Metagenomic Tools. In Proceedings of the 2024 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Natal, Brazil, 27–29 August 2024; pp. 1–6. [Google Scholar]
- Cummins, C.; Ahamed, A.; Aslam, R.; Burgin, J.; Devraj, R.; Edbali, O.; Gupta, D.; Harrison, P.W.; Haseeb, M.; Holt, S.; et al. The European Nucleotide Archive in 2021. Nucleic Acids Res. 2022, 50, D106–D110. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2022, 50, D161–D164. [Google Scholar] [CrossRef]
- Katz, K.; Shutov, O.; Lapoint, R.; Kimelman, M.; Brister, J.R.; O’Sullivan, C. The Sequence Read Archive: A Decade More of Explosive Growth. Nucleic Acids Res. 2022, 50, D387–D390. [Google Scholar] [CrossRef]
- Ogasawara, O.; Kodama, Y.; Mashima, J.; Kosuge, T.; Fujisawa, T. DDBJ Database Updates and Computational Infrastructure Enhancement. Nucleic Acids Res. 2020, 48, D45–D50. [Google Scholar] [CrossRef]
- Arita, M.; Karsch-Mizrachi, I.; Cochrane, G.; The International Nucleotide Sequence Database Collaboration. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2021, 49, D121–D124. [Google Scholar] [CrossRef]
- Federhen, S.; Clark, K.; Barrett, T.; Parkinson, H.; Ostell, J.; Kodama, Y.; Mashima, J.; Nakamura, Y.; Cochrane, G.; Karsch-Mizrachi, I. Toward Richer Metadata for Microbial Sequences: Replacing Strain-Level NCBI Taxonomy Taxids with BioProject, BioSample and Assembly Records. Stand. Genom. Sci. 2014, 9, 1275–1277. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Henrik Nilsson, R.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE Database for Molecular Identification of Fungi—Recent Updates and Future Perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Hernández-Plaza, A.; Szklarczyk, D.; Botas, J.; Cantalapiedra, C.P.; Giner-Lamia, J.; Mende, D.R.; Kirsch, R.; Rattei, T.; Letunic, I.; Jensen, L.J.; et al. eggNOG 6.0: Enabling Comparative Genomics across 12 535 Organisms. Nucleic Acids Res. 2022, 51, D389–D394. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Wolf, Y.I.; Makarova, K.S.; Vera Alvarez, R.; Landsman, D.; Koonin, E.V. COG Database Update: Focus on Microbial Diversity, Model Organisms, and Widespread Pathogens. Nucleic Acids Res. 2021, 49, D274–D281. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc Database of Metabolic Pathways and Enzymes—A 2019 Update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, K.; Li, S.; Zhang, X.; Zhao, Q.; Zhao, X.; Liu, Z.; Cheng, H.; Liu, Z.-X.; Li, X. gutMEGA: A Database of the Human Gut MEtaGenome Atlas. Brief. Bioinform. 2021, 22, bbaa082. [Google Scholar] [CrossRef]
- Almeida, A.; Nayfach, S.; Boland, M.; Strozzi, F.; Beracochea, M.; Shi, Z.J.; Pollard, K.S.; Sakharova, E.; Parks, D.H.; Hugenholtz, P.; et al. A Unified Catalog of 204,938 Reference Genomes from the Human Gut Microbiome. Nat. Biotechnol. 2021, 39, 105–114. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.-H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A Web Accessible Resource for Investigating Oral Microbe Taxonomic and Genomic Information. Database 2010, 2010, baq013. [Google Scholar] [CrossRef]
- Agostinetto, G.; Bozzi, D.; Porro, D.; Casiraghi, M.; Labra, M.; Bruno, A. SKIOME Project: A Curated Collection of Skin Microbiome Datasets Enriched with Study-Related Metadata. Database 2022, 2022, baac033. [Google Scholar] [CrossRef] [PubMed]
- Fettweis, J.M.; Serrano, M.G.; Sheth, N.U.; Mayer, C.M.; Glascock, A.L.; Brooks, J.P.; Jefferson, K.K.; Buck, G.A.; Vaginal Microbiome Consortium (additional members). Species-Level Classification of the Vaginal Microbiome. BMC Genom. 2012, 13 (Suppl. S8), S17. [Google Scholar] [CrossRef] [PubMed]
- Baltoumas, F.A.; Karatzas, E.; Liu, S.; Ovchinnikov, S.; Sofianatos, Y.; Chen, I.-M.; Kyrpides, N.C.; Pavlopoulos, G.A. NMPFamsDB: A Database of Novel Protein Families from Microbial Metagenomes and Metatranscriptomes. Nucleic Acids Res. 2024, 52, D502–D512. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A Database for Classification of Antimicrobial Drug, Biocide and Metal Resistance Determinants in Metagenomic Sequence Data. Nucleic Acids Res. 2019, 48, D561–D569. [Google Scholar] [CrossRef] [PubMed]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved Annotation of Antibiotic Resistance Determinants Reveals Microbial Resistomes Cluster by Ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef]
- Nam, N.N.; Do, H.D.K.; Loan Trinh, K.T.; Lee, N.Y. Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions. Foods 2023, 12, 2140. [Google Scholar] [CrossRef]
- Konopiński, M.K. Shannon Diversity Index: A Call to Replace the Original Shannon’s Formula with Unbiased Estimator in the Population Genetics Studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric Estimation of the Number of Classes in a Population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Faith, D.P. Conservation Evaluation and Phylogenetic Diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Calle, M.L. Statistical Analysis of Metagenomics Data. Genomics Inform. 2019, 17, e6. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.; Bose, A.; Mitra, S. Metagenomics Data Visualization Using R. In Metagenomic Data Analysis; Mitra, S., Ed.; Springer: New York, NY, USA, 2023; pp. 359–392. ISBN 978-1-0716-3072-3. [Google Scholar]
- Hauptfeld, E.; Pappas, N.; van Iwaarden, S.; Snoek, B.L.; Aldas-Vargas, A.; Dutilh, B.E.; von Meijenfeldt, F.A.B. Integrating Taxonomic Signals from MAGs and Contigs Improves Read Annotation and Taxonomic Profiling of Metagenomes. Nat. Commun. 2024, 15, 3373. [Google Scholar] [CrossRef] [PubMed]
- Greenacre, M.; Primicerio, R. Multivariate Analysis of Ecological Data; Fundación BBVA: Bilbao, Spain, 2013; ISBN 978-84-92937-50-9. [Google Scholar]
- Oksanen, J. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. 2015. Available online: https://john-quensen.com/wp-content/uploads/2018/10/Oksanen-Jari-vegantutor.pdf (accessed on 14 September 2025).
- Thioulouse, J.; Dray, S.; Dufour, A.-B.; Siberchicot, A.; Jombart, T.; Pavoine, S. Multivariate Analysis of Ecological Data with Ade4; Springer: New York, NY, USA, 2018; ISBN 978-1-4939-8848-8. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Khleborodova, A.; Gamboa-Tuz, S.D.; Ramos, M.; Segata, N.; Waldron, L.; Oh, S. Lefser: Implementation of Metagenomic Biomarker Discovery Tool, LEfSe, in R. Bioinformatics 2024, 40, btae707. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the Analysis of High-Throughput Sequencing Datasets: Characterizing RNA-Seq, 16S rRNA Gene Sequencing and Selective Growth Experiments by Compositional Data Analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D. Multigroup Analysis of Compositions of Microbiomes with Covariate Adjustments and Repeated Measures. Nat. Methods 2024, 21, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome Differential Abundance Methods Produce Different Results across 38 Datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Aldirawi, H.; Morales, F.G. Univariate and Multivariate Statistical Analysis of Microbiome Data: An Overview. Appl. Microbiol. 2023, 3, 322–338. [Google Scholar] [CrossRef]
- Lutz, K.C.; Jiang, S.; Neugent, M.L.; De Nisco, N.J.; Zhan, X.; Li, Q. A Survey of Statistical Methods for Microbiome Data Analysis. Front. Appl. Math. Stat. 2022, 8, 884810. [Google Scholar] [CrossRef]
- Jiang, Z.; He, M.; Chen, J.; Zhao, N.; Zhan, X. MiRKAT-MC: A Distance-Based Microbiome Kernel Association Test with Multi-Categorical Outcomes. Front. Genet. 2022, 13, 841764. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Chen, G. Zero-Inflated Generalized Dirichlet Multinomial Regression Model for Microbiome Compositional Data Analysis. Bioinformatics 2019, 20, 698–713. [Google Scholar] [CrossRef]
- Mishra, A.K.; Müller, C.L. Negative Binomial Factor Regression with Application to Microbiome Data Analysis. Stat. Med. 2022, 41, 2786–2803. [Google Scholar] [CrossRef]
- Banerjee, K.; Chen, J.; Zhan, X. Adaptive and Powerful Microbiome Multivariate Association Analysis via Feature Selection. NAR Genom. Bioinform. 2022, 4, lqab120. [Google Scholar] [CrossRef] [PubMed]
- Shuler, K.; Verbanic, S.; Chen, I.A.; Lee, J. A Bayesian Nonparametric Analysis for Zero-Inflated Multivariate Count Data with Application to Microbiome Study. J. R. Stat. Soc. Ser. C Appl. Stat. 2021, 70, 961–979. [Google Scholar] [CrossRef] [PubMed]
- Arıkan, M.; Muth, T. Integrated Multi-Omics Analyses of Microbial Communities: A Review of the Current State and Future Directions. Mol. Omics 2023, 19, 607–623. [Google Scholar] [CrossRef]
- Muller, E.; Shiryan, I.; Borenstein, E. Multi-Omic Integration of Microbiome Data for Identifying Disease-Associated Modules. Nat. Commun. 2024, 15, 2621. [Google Scholar] [CrossRef]
- Wu, J.; Singleton, S.S.; Bhuiyan, U.; Krammer, L.; Mazumder, R. Multi-Omics Approaches to Studying Gastrointestinal Microbiome in the Context of Precision Medicine and Machine Learning. Front. Mol. Biosci. 2024, 10, 1337373. [Google Scholar] [CrossRef] [PubMed]
- Athieniti, E.; Spyrou, G.M. A Guide to Multi-Omics Data Collection and Integration for Translational Medicine. Comput. Struct. Biotechnol. J. 2022, 21, 134–149. [Google Scholar] [CrossRef]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-Omics Data Integration Considerations and Study Design for Biological Systems and Disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Westerhuis, J.A. A Beginner’s Guide to Integrating Multi-Omics Data from Microbial Communities. Biochemist 2022, 44, 23–29. [Google Scholar] [CrossRef]
- Flores, J.E.; Claborne, D.M.; Weller, Z.D.; Webb-Robertson, B.-J.M.; Waters, K.M.; Bramer, L.M. Missing Data in Multi-Omics Integration: Recent Advances through Artificial Intelligence. Front. Artif. Intell. 2023, 6, 1098308. [Google Scholar] [CrossRef]
- Hayes, C.N.; Nakahara, H.; Ono, A.; Tsuge, M.; Oka, S. From Omics to Multi-Omics: A Review of Advantages and Tradeoffs. Genes 2024, 15, 1551. [Google Scholar] [CrossRef]
- Yu, Y.; Mai, Y.; Zheng, Y.; Shi, L. Assessing and Mitigating Batch Effects in Large-Scale Omics Studies. Genome Biol. 2024, 25, 254. [Google Scholar] [CrossRef] [PubMed]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis—A Framework for Unsupervised Integration of Multi-omics Data Sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Lê Cao, K.-A. DIABLO: An Integrative Approach for Identifying Key Molecular Drivers from Multi-Omics Assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef] [PubMed]
- Zoppi, J.; Guillaume, J.-F.; Neunlist, M.; Chaffron, S. MiBiOmics: An Interactive Web Application for Multi-Omics Data Exploration and Integration. BMC Bioinform. 2021, 22, 6. [Google Scholar] [CrossRef]
- Hawinkel, S.; Bijnens, L.; Cao, K.-A.L.; Thas, O. Model-Based Joint Visualization of Multiple Compositional Omics Datasets. NAR Genom. Bioinform. 2020, 2, lqaa050. [Google Scholar] [CrossRef]
- Chicco, D.; Cumbo, F.; Angione, C. Ten Quick Tips for Avoiding Pitfalls in Multi-Omics Data Integration Analyses. PLoS Comput. Biol. 2023, 19, e1011224. [Google Scholar] [CrossRef]
- Fabbrini, M.; Scicchitano, D.; Candela, M.; Turroni, S.; Rampelli, S. Connect the Dots: Sketching out Microbiome Interactions through Networking Approaches. Microbiome Res. Rep. 2023, 2, 25. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Fang, H.; Huang, C.; Zhao, H.; Deng, M. CCLasso: Correlation Inference for Compositional Data through Lasso. Bioinformatics 2015, 31, 3172–3180. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Fang, H.; Huang, C.; Zhao, H.; Deng, M. gCoda: Conditional Dependence Network Inference for Compositional Data. J. Comput. Biol. 2017, 24, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network Analysis Methods for Studying Microbial Communities: A Mini Review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef]
- Rios Garza, D.; Gonze, D.; Zafeiropoulos, H.; Liu, B.; Faust, K. Metabolic Models of Human Gut Microbiota: Advances and Challenges. Cell Syst. 2023, 14, 109–121. [Google Scholar] [CrossRef]
- Selber-Hnatiw, S.; Sultana, T.; Tse, W.; Abdollahi, N.; Abdullah, S.; Al Rahbani, J.; Alazar, D.; Alrumhein, N.J.; Aprikian, S.; Arshad, R.; et al. Metabolic Networks of the Human Gut Microbiota. Microbiology 2020, 166, 96–119. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, K.; Kamada, N. Metabolic Network of the Gut Microbiota in Inflammatory Bowel Disease. Inflamm. Regen. 2024, 44, 11. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, K.T.; Hynson, N.A. Networks as Tools for Defining Emergent Properties of Microbiomes and Their Stability. Microbiome 2024, 12, 184. [Google Scholar] [CrossRef]
- Oña, L.; Shreekar, S.K.; Kost, C. Disentangling Microbial Interaction Networks. Trends Microbiol. 2025, 33, 619–634. [Google Scholar] [CrossRef]
- Lange, E.; Kranert, L.; Krüger, J.; Benndorf, D.; Heyer, R. Microbiome Modeling: A Beginner’s Guide. Front. Microbiol. 2024, 15, 1368377. [Google Scholar] [CrossRef]
- Mohseni, P.; Ghorbani, A. Exploring the Synergy of Artificial Intelligence in Microbiology: Advancements, Challenges, and Future Prospects. Comput. Struct. Biotechnol. Rep. 2024, 1, 100005. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Passerini, A.; Waldron, L.; Segata, N. Machine Learning for Microbiologists. Nat. Rev. Microbiol. 2024, 22, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Lorusso, E.; Fosso, B.; Pesole, G. A Comprehensive Overview of Microbiome Data in the Light of Machine Learning Applications: Categorization, Accessibility, and Future Directions. Front. Microbiol. 2024, 15, 1343572. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rahman, H.; Ottom, M.A.; Dinov, I.D. Machine Learning-Based Colorectal Cancer Prediction Using Global Dietary Data. BMC Cancer 2023, 23, 144. [Google Scholar] [CrossRef]
- Zeng, T.; Yu, X.; Chen, Z. Applying Artificial Intelligence in the Microbiome for Gastrointestinal Diseases: A Review. J. Gastroenterol. Hepatol. 2021, 36, 832–840. [Google Scholar] [CrossRef]
- Reiman, D.; Metwally, A.; Sun, J.; Dai, Y. Meta-Signer: Metagenomic Signature Identifier Based Onrank Aggregation of Features. F1000Research 2021, 10, 194. [Google Scholar] [CrossRef]
- Oh, M.; Zhang, L. DeepMicro: Deep Representation Learning for Disease Prediction Based on Microbiome Data. Sci. Rep. 2020, 10, 6026. [Google Scholar] [CrossRef]
- Yang, F.; Zou, Q. mAML: An Automated Machine Learning Pipeline with a Microbiome Repository for Human Disease Classification. Database J. Biol. Databases Curation 2020, 2020, baaa050. [Google Scholar] [CrossRef] [PubMed]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A Deep Learning Approach for Predicting Antibiotic Resistance Genes from Metagenomic Data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef]
- Deneke, C.; Rentzsch, R.; Renard, B.Y. PaPrBaG: A Machine Learning Approach for the Detection of Novel Pathogens from NGS Data. Sci. Rep. 2017, 7, 39194. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; López-Molina, V.M.; Bakir-Gungor, B.; Frohme, M.; Karaduzovic-Hadziabdic, K.; Klammsteiner, T.; Ibrahimi, E.; Lahti, L.; Loncar-Turukalo, T.; Dhamo, X.; et al. A Toolbox of Machine Learning Software to Support Microbiome Analysis. Front. Microbiol. 2023, 14, 1250806. [Google Scholar] [CrossRef]
- Tian, Q.; Zhang, P.; Zhai, Y.; Wang, Y.; Zou, Q. Application and Comparison of Machine Learning and Database-Based Methods in Taxonomic Classification of High-Throughput Sequencing Data. Genome Biol. Evol. 2024, 16, evae102. [Google Scholar] [CrossRef]
- Papoutsoglou, G.; Tarazona, S.; Lopes, M.B.; Klammsteiner, T.; Ibrahimi, E.; Eckenberger, J.; Novielli, P.; Tonda, A.; Simeon, A.; Shigdel, R.; et al. Machine Learning Approaches in Microbiome Research: Challenges and Best Practices. Front. Microbiol. 2023, 14, 1261889. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That Are Specific for Colorectal Cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Wirbel, J.; Zych, K.; Essex, M.; Karcher, N.; Kartal, E.; Salazar, G.; Bork, P.; Sunagawa, S.; Zeller, G. Microbiome Meta-Analysis and Cross-Disease Comparison Enabled by the SIAMCAT Machine Learning Toolbox. Genome Biol. 2021, 22, 93. [Google Scholar] [CrossRef]
- Topçuoğlu, B.D.; Lesniak, N.A.; Ruffin, M.T.; Wiens, J.; Schloss, P.D. A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems. mBio 2020, 11, e00434-20. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, E.G.; Adjei, K.P.; Andersen, C.W.; Hetle Aspheim, J.C.; Battistin, C.; Bulso, N.; Christensen, H.M.; Cretois, B.; Cubero, R.; Davidovich, I.A.; et al. Insights into the Quantification and Reporting of Model-Related Uncertainty across Different Disciplines. iScience 2022, 25, 105512. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar] [CrossRef]
- Novakovsky, G.; Fornes, O.; Saraswat, M.; Mostafavi, S.; Wasserman, W.W. ExplaiNN: Interpretable and Transparent Neural Networks for Genomics. Genome Biol. 2023, 24, 154. [Google Scholar] [CrossRef]
- van Hilten, A.; Katz, S.; Saccenti, E.; Niessen, W.J.; Roshchupkin, G.V. Designing Interpretable Deep Learning Applications for Functional Genomics: A Quantitative Analysis. Brief. Bioinform. 2024, 25, bbae449. [Google Scholar] [CrossRef]
- Karwowska, Z.; Aasmets, O.; Metspalu, M.; Metspalu, A.; Milani, L.; Esko, T.; Kosciolek, T.; Org, E.; Estonian Biobank Research Team. Effects of Data Transformation and Model Selection on Feature Importance in Microbiome Classification Data. Microbiome 2025, 13, 2. [Google Scholar] [CrossRef]
- Lee, Y.; Cappellato, M.; Di Camillo, B. Machine Learning-Based Feature Selection to Search Stable Microbial Biomarkers: Application to Inflammatory Bowel Disease. GigaScience 2022, 12, giad083. [Google Scholar] [CrossRef]
- Rynazal, R.; Fujisawa, K.; Shiroma, H.; Salim, F.; Mizutani, S.; Shiba, S.; Yachida, S.; Yamada, T. Leveraging Explainable AI for Gut Microbiome-Based Colorectal Cancer Classification. Genome Biol. 2023, 24, 21. [Google Scholar] [CrossRef] [PubMed]
- Manos, J. The Human Microbiome in Disease and Pathology. APMIS 2022, 130, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Rutayisire, E.; Huang, K.; Liu, Y.; Tao, F. The Mode of Delivery Affects the Diversity and Colonization Pattern of the Gut Microbiota during the First Year of Infants’ Life: A Systematic Review. BMC Gastroenterol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Guaraldi, F.; Salvatori, G. Effect of Breast and Formula Feeding on Gut Microbiota Shaping in Newborns. Front. Cell. Infect. Microbiol. 2012, 2, 94. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, H.; Qiu, P. Aging Progression of Human Gut Microbiota. BMC Microbiol. 2019, 19, 236. [Google Scholar] [CrossRef]
- Beam, A.; Clinger, E.; Hao, L. Effect of Diet and Dietary Components on the Composition of the Gut Microbiota. Nutrients 2021, 13, 2795. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, C.; Morrow, A.L.; Lagomarcino, A.J.; Altaye, M.; Taft, D.H.; Yu, Z.; Newburg, D.S.; Ward, D.V.; Schibler, K.R. Early Empiric Antibiotic Use in Preterm Infants Is Associated with Lower Bacterial Diversity and Higher Relative Abundance of Enterobacter. J. Pediatr. 2014, 165, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the Gut Microbiota in Disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Graves, D.T.; Corrêa, J.D.; Silva, T.A. The Oral Microbiota Is Modified by Systemic Diseases. J. Dent. Res. 2019, 98, 148–156. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Salvatore, F. The Role of the Gut Microbiome in the Healthy Adult Status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Liu, B.-N.; Liu, X.-T.; Liang, Z.-H.; Wang, J.-H. Gut Microbiota in Obesity. World J. Gastroenterol. 2021, 27, 3837–3850. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between Body Mass Index and Firmicutes/Bacteroidetes Ratio in an Adult Ukrainian Population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A Core Gut Microbiome in Obese and Lean Twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Bielka, W.; Przezak, A.; Pawlik, A. The Role of the Gut Microbiota in the Pathogenesis of Diabetes. Int. J. Mol. Sci. 2022, 23, 480. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human Genetics Shape the Gut Microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Hamamah, S.; Iatcu, O.C.; Covasa, M. Dietary Influences on Gut Microbiota and Their Role in Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD). Nutrients 2024, 17, 143. [Google Scholar] [CrossRef]
- Solé, C.; Guilly, S.; Da Silva, K.; Llopis, M.; Le-Chatelier, E.; Huelin, P.; Carol, M.; Moreira, R.; Fabrellas, N.; De Prada, G.; et al. Alterations in Gut Microbiome in Cirrhosis as Assessed by Quantitative Metagenomics: Relationship with Acute-on-Chronic Liver Failure and Prognosis. Gastroenterology 2021, 160, 206–218.e13. [Google Scholar] [CrossRef]
- Gorczyca, K.; Obuchowska, A.; Kimber-Trojnar, Ż.; Wierzchowska-Opoka, M.; Leszczyńska-Gorzelak, B. Changes in the Gut Microbiome and Pathologies in Pregnancy. Int. J. Environ. Res. Public Health 2022, 19, 9961. [Google Scholar] [CrossRef] [PubMed]
- Yurtdaş, G.; Akdevelioğlu, Y. A New Approach to Polycystic Ovary Syndrome: The Gut Microbiota. J. Am. Coll. Nutr. 2020, 39, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Jiang, Y.; Xi, H.; Chen, L.; Feng, X. Exploration of the Relationship Between Gut Microbiota and Polycystic Ovary Syndrome (PCOS): A Review. Geburtshilfe Frauenheilkd. 2020, 80, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Hsiao, E.Y. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef]
- Finegold, S.M.; Molitoris, D.; Song, Y.; Liu, C.; Vaisanen, M.; Bolte, E.; McTeague, M.; Sandler, R.; Wexler, H.; Marlowe, E.M.; et al. Gastrointestinal Microflora Studies in Late-Onset Autism. Clin. Infect. Dis. 2002, 35, S6–S16. [Google Scholar] [CrossRef] [PubMed]
- Parracho, H.M.; Bingham, M.O.; Gibson, G.R.; McCartney, A.L. Differences between the Gut Microflora of Children with Autistic Spectrum Disorders and That of Healthy Children. J. Med. Microbiol. 2005, 54, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Liu, J.; Zhang, H.; Shan, C.; Guo, Y.; Gong, X.; Cui, M.; Li, X.; Tang, M. Correlation between the Gut Microbiome and Neurodegenerative Diseases: A Review of Metagenomics Evidence. Neural Regen. Res. 2024, 19, 833–845. [Google Scholar] [CrossRef]
- Thye, A.Y.-K.; Bah, Y.-R.; Law, J.W.-F.; Tan, L.T.-H.; He, Y.-W.; Wong, S.-H.; Thurairajasingam, S.; Chan, K.-G.; Lee, L.-H.; Letchumanan, V. Gut–Skin Axis: Unravelling the Connection between the Gut Microbiome and Psoriasis. Biomedicines 2022, 10, 1037. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Lee, E.; Park, Y.M.; Hong, S.-J. Microbiome in the Gut-Skin Axis in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2018, 10, 354. [Google Scholar] [CrossRef]
- Mahmud, M.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Akter, S.; Hasan, M.R.; Acharjee, M.; Hossain, M.S.; et al. Impact of Gut Microbiome on Skin Health: Gut-Skin Axis Observed through the Lenses of Therapeutics and Skin Diseases. Gut Microbes 2022, 14, 2096995. [Google Scholar] [CrossRef] [PubMed]
- Widhiati, S.; Purnomosari, D.; Wibawa, T.; Soebono, H. The Role of Gut Microbiome in Inflammatory Skin Disorders: A Systematic Review. Dermatol. Rep. 2021, 14, 9188. [Google Scholar] [CrossRef]
- Aldars-García, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Armengaud, J. Metaproteomics to Understand How Microbiota Function: The Crystal Ball Predicts a Promising Future. Environ. Microbiol. 2023, 25, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Sanders, M.E.; Szajewska, H.; Cohen, H.; Eliakim, R.; Herrera-deGuise, C.; Karakan, T.; Merenstein, D.; Piscoya, A.; Ramakrishna, B.; et al. World Gastroenterology Organisation Global Guidelines: Probiotics and Prebiotics. J. Clin. Gastroenterol. 2024, 58, 533–553. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Guarino, A.; Guandalini, S.; Lo Vecchio, A. Probiotics for Prevention and Treatment of Diarrhea. J. Clin. Gastroenterol. 2015, 49, S37–S45. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Ma, Y.; Yan, B.; Pei, W.; Wu, Q.; Ding, C.; Huang, C. The Promotion Mechanism of Prebiotics for Probiotics: A Review. Front. Nutr. 2022, 9, 1000517. [Google Scholar] [CrossRef]
- Florowska, A.; Krygier, K.; Florowski, T.; Dłużewska, E. Prebiotics as Functional Food Ingredients Preventing Diet-Related Diseases. Food Funct. 2016, 7, 2147–2155. [Google Scholar] [CrossRef]
- Khoruts, A.; Sadowsky, M.J. Understanding the Mechanisms of Faecal Microbiota Transplantation. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 508–516. [Google Scholar] [CrossRef]
- Koenigsknecht, M.J.; Young, V.B. Faecal Microbiota Transplantation for the Treatment of Recurrent Clostridium Difficile Infection: Current Promise and Future Needs. Curr. Opin. Gastroenterol. 2013, 29, 628–632. [Google Scholar] [CrossRef]
- Yadegar, A.; Bar-Yoseph, H.; Monaghan, T.M.; Pakpour, S.; Severino, A.; Kuijper, E.J.; Smits, W.K.; Terveer, E.M.; Neupane, S.; Nabavi-Rad, A.; et al. Fecal Microbiota Transplantation: Current Challenges and Future Landscapes. Clin. Microbiol. Rev. 2024, 37, e0006022. [Google Scholar] [CrossRef]
- Zmora, N.; Zeevi, D.; Korem, T.; Segal, E.; Elinav, E. Taking It Personally: Personalized Utilization of the Human Microbiome in Health and Disease. Cell Host Microbe 2016, 19, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Ahamed, A.; Burgin, J.; Cummins, C.; Devraj, R.; Gueye, K.; Gupta, D.; Gupta, V.; Haseeb, M.; Ihsan, M.; et al. The European Nucleotide Archive in 2023. Nucleic Acids Res. 2024, 52, D92–D97. [Google Scholar] [CrossRef]
- Thakur, M.; Bateman, A.; Brooksbank, C.; Freeberg, M.; Harrison, M.; Hartley, M.; Keane, T.; Kleywegt, G.; Leach, A.; Levchenko, M.; et al. EMBL’s European Bioinformatics Institute (EMBL-EBI) in 2022. Nucleic Acids Res. 2023, 51, D9–D17. [Google Scholar] [CrossRef] [PubMed]
- Kyrpides, N.C.; Eloe-Fadrosh, E.A.; Ivanova, N.N. Microbiome Data Science: Understanding Our Microbial Planet. Trends Microbiol. 2016, 24, 425–427. [Google Scholar] [CrossRef]
- Sengupta, P.; Sivabalan, S.K.M.; Mahesh, A.; Palanikumar, I.; Kuppa Baskaran, D.K.; Raman, K. Big Data for a Small World: A Review on Databases and Resources for Studying Microbiomes. J. Indian Inst. Sci. 2023, 103, 891–907. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Kottmann, R.; Field, D.; Knight, R.; Cole, J.R.; Amaral-Zettler, L.; Gilbert, J.A.; Karsch-Mizrachi, I.; Johnston, A.; Cochrane, G.; et al. Minimum Information about a Marker Gene Sequence (MIMARKS) and Minimum Information about Any (x) Sequence (MIxS) Specifications. Nat. Biotechnol. 2011, 29, 415–420. [Google Scholar] [CrossRef]
- MINSEQE FGED Society—MINSEQE. Available online: https://www.fged.org/projects/minseqe (accessed on 26 March 2025).
- Dorst, M.; Zeevenhooven, N.; Wilding, R.; Mende, D.; Brandt, B.W.; Zaura, E.; Hoekstra, A.; Sheraton, V.M. FAIR Compliant Database Development for Human Microbiome Data Samples. Front. Cell. Infect. Microbiol. 2024, 14, 1384809. [Google Scholar] [CrossRef]
- Ling, M.; Szarvas, J.; Kurmauskaitė, V.; Kiseliovas, V.; Žilionis, R.; Avot, B.; Munk, P.; Aarestrup, F.M. High Throughput Single Cell Metagenomic Sequencing with Semi-Permeable Capsules: Unraveling Microbial Diversity at the Single-Cell Level in Sewage and Fecal Microbiomes. Front. Microbiol. 2025, 15, 1516656. [Google Scholar] [CrossRef]
- Hosokawa, M.; Nishikawa, Y. Tools for Microbial Single-Cell Genomics for Obtaining Uncultured Microbial Genomes. Biophys. Rev. 2023, 16, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.A.; Watson-Haigh, N.S.; Burton, R.A.; Sheppard, A.E. A Gentle Introduction to Pangenomics. Brief. Bioinform. 2024, 25, bbae588. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.H.; Ju, Y.K.; Lee, H.J.; Park, S.M.; Suh, J.W.; Kim, J.Y.; Sohn, J.W.; Yoon, Y.K. Clinical Performance of Real-Time Nanopore Metagenomic Sequencing for Rapid Identification of Bacterial Pathogens in Cerebrospinal Fluid: A Pilot Study. Sci. Rep. 2025, 15, 3493. [Google Scholar] [CrossRef] [PubMed]
- Oehler, J.B.; Burns, K.; Warner, J.; Schmitz, U. Long-Read Sequencing for the Rapid Response to Infectious Diseases Outbreaks. Curr. Clin. Microbiol. Rep. 2025, 12, 10. [Google Scholar] [CrossRef]
| Technology | Taxonomic Resolution | Typical Input DNA | Typical Sequencing Depth | Main Strengths | Primary Limitations |
|---|---|---|---|---|---|
| 16S rRNA (Illumina) | Genus → Species * | 5–50 ng | 10–100 K reads/sample | Standardized workflows, extensive databases, cost-effective | Limited to prokaryotes, poor species resolution, no functional data |
| Shotgun SRS (Illumina) | Species → Strain ** | 100–500 ng | 5–50 M reads/sample | High accuracy, established tools, functional profiling | Poor repetitive region assembly, reference-dependent |
| PacBio HiFi | Species → Strain | 1–10 μg HMW DNA | 0.5–5 M reads/sample | Very high accuracy, excellent MAG recovery, complete genomes | Higher cost, shorter reads than ONT |
| ONT MinION/GridION | Species → Strain | 1–10 μg HMW DNA | 0.5–5 M reads/sample | Ultra-long reads, real-time sequencing, structural variants | Moderate accuracy in homopolymers, specialized workflows |
| Tool | Approach | Advantages | Limitations |
|---|---|---|---|
| MEGAHIT | De Bruijn Graph Ultra-fast assembly | Extremely low memory consumption; suitable for large complex metagenomes | May yield lower completeness in complex communities; shorter contigs compared to metaSPAdes |
| metaSPAdes | De Bruijn Graph Produces longer contigs | High fraction of reads assembled; optimized for metagenomic data | Higher mis-assembly rate; more resource intensive; longer runtime |
| BWA/Bowtie2 | Reference-guided alignment | High accuracy for known taxa; improved resolution for well-characterized genomes | Not suitable for novel or poorly represented organisms; requires high-quality reference databases |
| IDBA-UD | Iterative De Bruijn Graph | Handles uneven sequencing depths well; good for complex communities | Slower than MEGAHIT; higher memory requirements than succinct graph approaches |
| Flye | Overlap-layout-consensus (long-read) | Excellent for long-read data; produces highly contiguous assemblies; good error correction | Primarily designed for long reads; computationally intensive for large datasets |
| Tool | Approach | Advantages | Limitations |
|---|---|---|---|
| MetaBAT2 | Hybrid clustering using coverage and k-mer profiles | Widely used and well-validated; effective use of abundance patterns; good performance on diverse datasets | Requires contigs ≥ 1500 bp for optimal performance; struggles with low-abundance species |
| MaxBin2 | Expectation-maximization with abundance and composition | Effective for low-abundance species; robust statistical framework; handles uneven coverage well | Less effective on very complex communities; sensitive to parameter settings |
| VAMB | Deep learning with variational autoencoder | Robust across different contig length thresholds; learns complex feature representations; good scalability | Requires substantial training data; black-box approach limits interpretability |
| COMEBin | Contrastive multi-view representation learning with Leiden clustering | Demonstrates higher recovery of near-complete bins; uses advanced community detection; data augmentation improves robustness | Emerging method with limited field validation; computationally intensive; requires expertise to optimize |
| SemiBin2 | Semi-supervised deep learning with Siamese networks | Leverages both labeled and unlabeled data; good performance across diverse environments; user-friendly | Requires some reference genomes for training; newer tool with growing but limited validation |
| Process | Tool | Dataset | Memory Use | Running Time | Source |
|---|---|---|---|---|---|
| Taxonomic profiling | UPARSE | 4.7 Million sequences | Not specified | 1 h | Marizzoni et al., 2020 [84] |
| Bioconductor | ~8 h | ||||
| QIIME2 | 3 h | ||||
| Mothur | 9 h | ||||
| mOTUs 3 | 50 randomly selected human gut metagenomes | ~15 GB | ~4 min | Shaw & Yu, 2025 [85] | |
| MetaPhIAn 4 | ~18 GB | ~6 min | |||
| Centrifuge | 5.7 million sequences | 20 Gb | 7 min | Ye et al., 2019a [86] | |
| Kraken2 | 36 GB | 1 min | |||
| Bracken | <1 GB | <1 min | |||
| CLARK | 80 GB | 2 min | |||
| Functional Profiles | PICRUSt2 | Human HMP dataset (Not specified) | ~15 GB | ~45 min | Mongad et al., 2021 [63] |
| MicFunPred | ~6 GB | ~30 min | |||
| Tax4Fun2 | ~4 GB | ~15 min | |||
| eggNOG-mapper v2 | 4296 coding sequences for Escherichia coli K-12 | 6 GB | ~26 min | Alonso-Reyes & Albarracin, 2024 [87] | |
| GhostKOALA | 3.8 Million sequences | Web service | ~6 min | Kanehisa et al., 2016 [71] | |
| BlastKOALA | 2.6 Million sequences | Web service | ~41 min | ||
| DIAMOND | ~1.7 million sequences | ~14 GB | 8 min | Buchfink et al., 2021 [72] | |
| MMSeqs2 | 11 GB | 53 min | |||
| BLASTP | Web service | 46 days | |||
| MicrobeAnnotator | 100 E. coli genomes | ~19.4 GB/genome | 3.7 h/genome | Ruiz-Perez et al., 2021 [70] | |
| InterProScan | ~4 GB/genome | ~2.7 min/genome | |||
| Prokka | 204 MB/genome | ~2.5 min/genome |
| AI/ML Tool | Application |
|---|---|
| Meta-Signer (Reiman et al., 2021 [186]) | Feature ranking through ensemble learning and metagenome signature identification |
| DeepMicro (Oh & Zhang, 2020 [187]) | Deep representation learning for infection/disease prediction using microbiome data |
| mAML (F. Yang & Zou, 2020 [188]) | Automated human disease classification through reproducible models |
| DeepARG (Arango-Argoty et al., 2018 [189]) | Utilizes deep learning to predict novel antibiotic resistance genes |
| PaPrBaG (Deneke et al., 2017 [190]) | Pathogenicity prediction, reliable even at low genomic coverage |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pita-Galeana, M.A.; Ruhle, M.; López-Vázquez, L.; de Anda-Jáuregui, G.; Hernández-Lemus, E. Computational Metagenomics: State of the Art. Int. J. Mol. Sci. 2025, 26, 9206. https://doi.org/10.3390/ijms26189206
Pita-Galeana MA, Ruhle M, López-Vázquez L, de Anda-Jáuregui G, Hernández-Lemus E. Computational Metagenomics: State of the Art. International Journal of Molecular Sciences. 2025; 26(18):9206. https://doi.org/10.3390/ijms26189206
Chicago/Turabian StylePita-Galeana, Marco Antonio, Martin Ruhle, Lucía López-Vázquez, Guillermo de Anda-Jáuregui, and Enrique Hernández-Lemus. 2025. "Computational Metagenomics: State of the Art" International Journal of Molecular Sciences 26, no. 18: 9206. https://doi.org/10.3390/ijms26189206
APA StylePita-Galeana, M. A., Ruhle, M., López-Vázquez, L., de Anda-Jáuregui, G., & Hernández-Lemus, E. (2025). Computational Metagenomics: State of the Art. International Journal of Molecular Sciences, 26(18), 9206. https://doi.org/10.3390/ijms26189206







