Mutational Disruption of TP53: A Structural Approach to Understanding Chemoresistance
Abstract
1. Introduction
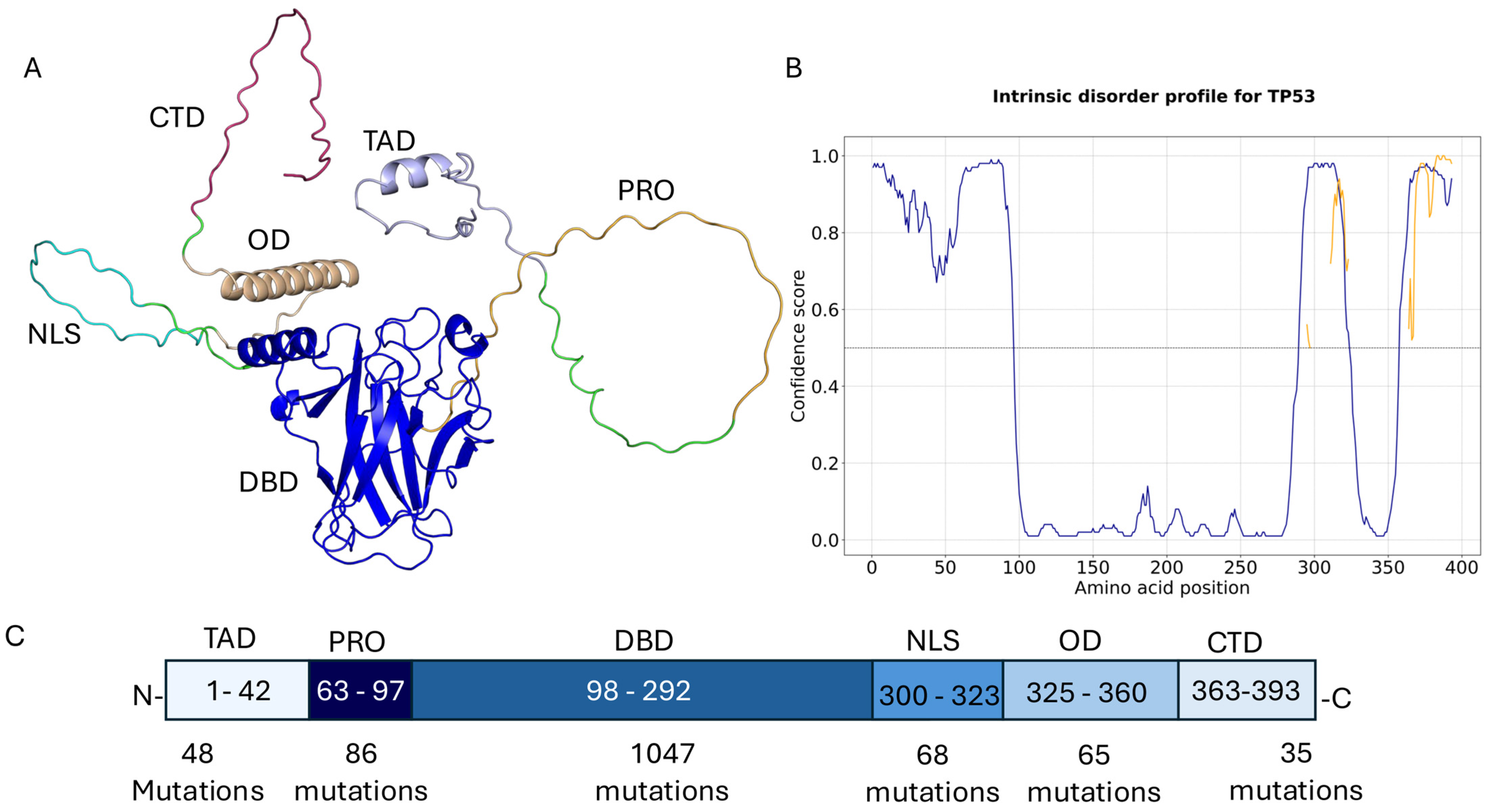
2. Results
- Very high-frequency mutations (>500 occurrences):
- High-frequency mutations (100–500 occurrences):
- Moderately high-frequency mutations (50–100 occurrences):
- Lower high-frequency mutations (20–50 occurrences):
3. Discussion
- Structure-based drug discovery targeting druggable cavities like Y220C and other destabilised cores.
- Computational–experimental pipelines to screen and validate small molecules that restore TP53 folding and DNA-binding capacity.
4. Materials and Methods
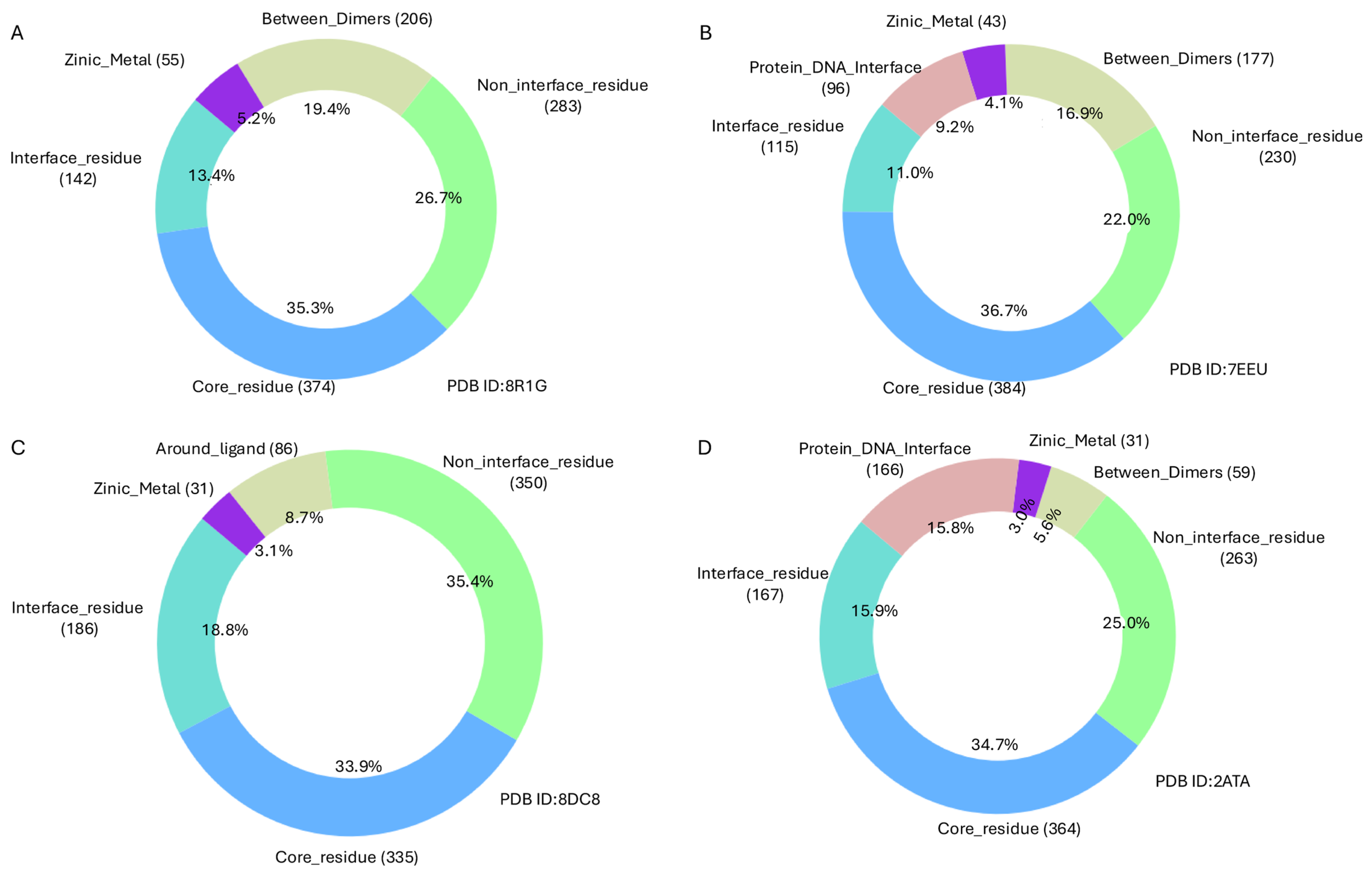
Solvent Accessibility and Residue Classification
- Core residues: SASA < 25
- Non-interface residues: 25 ≤ SASA ≤ 80
- Interface residues: SASA > 80
- 6 Å for ligand-binding sites
- 5 Å for protein-DNA interfaces, zinc ion coordination sites, and homo-/hetero-dimer interfaces
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lane, D.P. P53, Guardian of the Genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef] [PubMed]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, T.; Gleber-Netto, F.O.; Chen, Z.; McGrail, D.J.; Gomez, J.A.; Ju, W.; Gadhikar, M.A.; Ma, W.; Shen, L.; et al. Mutant P53 Gains Oncogenic Functions through a Chromosomal Instability-Induced Cytosolic DNA Response. Nat. Commun. 2024, 15, 180. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Abuetabh, Y.; Wu, H.H.; Chai, C.; Al Yousef, H.; Persad, S.; Sergi, C.M.; Leng, R. DNA Damage Response Revisited: The P53 Family and Its Regulators Provide Endless Cancer Therapy Opportunities. Exp. Mol. Med. 2022, 54, 1658–1669. [Google Scholar] [CrossRef]
- Wu, H.; Leng, R.P. MDM2 Mediates P73 Ubiquitination: A New Molecular Mechanism for Suppression of P73 Function. Oncotarget 2015, 6, 21479–21492. [Google Scholar] [CrossRef]
- Lin, J.; Chen, J.; Elenbaas, B.; Levine, A.J. Several Hydrophobic Amino Acids in the P53 Amino-Terminal Domain Are Required for Transcriptional Activation, Binding to Mdm-2 and the Adenovirus 5 E1B 55-kD Protein. Genes Dev. 1994, 8, 1235–1246. [Google Scholar] [CrossRef]
- Shieh, S.-Y.; Ikeda, M.; Taya, Y.; Prives, C. DNA Damage-Induced Phosphorylation of P53 Alleviates Inhibition by MDM2. Cell 1997, 91, 325–334. [Google Scholar] [CrossRef]
- Cabezas, M.; García-Quevedo, L.; Alonso, C.; Manubens, M.; Álvarez, Y.; Barquinero, J.F.; Ramón, Y.; Cajal, S.; Ortega, M.; Blanco, A.; et al. Polymorphisms in MDM2 and TP53 Genes and Risk of Developing Therapy-Related Myeloid Neoplasms. Sci. Rep. 2019, 9, 150. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting P53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant P53 in Cancer: From Molecular Mechanism to Therapeutic Modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. P53: 800 Million Years of Evolution and 40 Years of Discovery. Nat. Rev. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why Are There Hotspot Mutations in the TP53 Gene in Human Cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, A.O.; Yang, X.; Lintner, R.E.; McFarland, J.M.; Duby, M.; Kim, J.; Howard, T.P.; Takeda, D.Y.; Ly, S.H.; Kim, E.; et al. Mutational Processes Shape the Landscape of TP53 Mutations in Human Cancer. Nat. Genet. 2018, 50, 1381–1387. [Google Scholar] [CrossRef]
- Wang, H.; Guo, M.; Wei, H.; Chen, Y. Targeting P53 Pathways: Mechanisms, Structures and Advances in Therapy. Signal Transduct. Target. Ther. 2023, 8, 92. [Google Scholar] [CrossRef]
- Loh, S.N. Arsenic and an Old Place: Rescuing P53 Mutants in Cancer. Cancer Cell 2021, 39, 140–142. [Google Scholar] [CrossRef]
- Kitayner, M.; Rozenberg, H.; Kessler, N.; Rabinovich, D.; Shaulov, L.; Haran, T.E.; Shakked, Z. Structural Basis of DNA Recognition by P53 Tetramers. Mol. Cell 2006, 22, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor P53 and Cancer-Associated Mutants. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 2007; Volume 97, pp. 1–23. ISBN 978-0-12-006697-1. [Google Scholar]
- Tong, X.; Xu, D.; Mishra, R.K.; Jones, R.D.; Sun, L.; Schiltz, G.E.; Liao, J.; Yang, G.-Y. Identification of a Druggable Protein–Protein Interaction Site between Mutant P53 and Its Stabilizing Chaperone DNAJA1. J. Biol. Chem. 2021, 296, 100098. [Google Scholar] [CrossRef]
- Eldar, A.; Rozenberg, H.; Diskin-Posner, Y.; Rohs, R.; Shakked, Z. Structural Studies of P53 Inactivation by DNA-Contact Mutations and Its Rescue by Suppressor Mutations via Alternative Protein-DNA Interactions. Nucleic Acids Res. 2013, 41, 8748–8759. [Google Scholar] [CrossRef]
- Alsulami, A.F. Comprehensive Annotation of Mutations in Hallmark Genes Insights into Structural and Functional Implications. Comput. Biol. Med. 2025, 185, 109588. [Google Scholar] [CrossRef]
- Friedler, A.; DeDecker, B.S.; Freund, S.M.V.; Blair, C.; Rüdiger, S.; Fersht, A.R. Structural Distortion of P53 by the Mutation R249S and Its Rescue by a Designed Peptide: Implications for “Mutant Conformation”. J. Mol. Biol. 2004, 336, 187–196. [Google Scholar] [CrossRef]
- Walerych, D.; Napoli, M.; Collavin, L.; Del Sal, G. The Rebel Angel: Mutant P53 as the Driving Oncogene in Breast Cancer. Carcinogenesis 2012, 33, 2007–2017. [Google Scholar] [CrossRef]
- Nishikawa, S.; Iwakuma, T. Drugs Targeting P53 Mutations with FDA Approval and in Clinical Trials. Cancers 2023, 15, 429. [Google Scholar] [CrossRef]
- Bourgeois, B.; Spreitzer, E.; Platero-Rochart, D.; Paar, M.; Zhou, Q.; Usluer, S.; De Keizer, P.L.J.; Burgering, B.M.T.; Sánchez-Murcia, P.A.; Madl, T. The Disordered P53 Transactivation Domain Is the Target of FOXO4 and the Senolytic Compound FOXO4-DRI. Nat. Commun. 2025, 16, 5672. [Google Scholar] [CrossRef]
- Venot, C.; Maratrat, M.; Dureuil, C.; Conseiller, E.; Bracco, L.; Debussche, L. The Requirement for the P53 Proline-Rich Functional Domain for Mediation of Apoptosis Is Correlated with Specific PIG3 Gene Transactivation and with Transcriptional Repression. EMBO J. 1998, 17, 4668–4679. [Google Scholar] [CrossRef]
- Clore, G.M.; Omichinski, J.G.; Sakaguchi, K.; Zambrano, N.; Sakamoto, H.; Appella, E.; Gronenborn, A.M. High-Resolution Structure of the Oligomerization Domain of P53 by Multidimensional NMR. Science 1994, 265, 386–391, Erratum in Science 1995, 267, 1515–1516. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.D.; Galbraith, M.D.; Andrysik, Z.; Espinosa, J.M. Mechanisms of Transcriptional Regulation by P53. Cell Death Differ. 2018, 25, 133–143. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, H.; Wiesmüller, L.; Chen, M. Canonical and Non-Canonical Functions of P53 Isoforms: Potentiating the Complexity of Tumor Development and Therapy Resistance. Cell Death Dis. 2024, 15, 412. [Google Scholar] [CrossRef]
- Hassin, O.; Nataraj, N.B.; Shreberk-Shaked, M.; Aylon, Y.; Yaeger, R.; Fontemaggi, G.; Mukherjee, S.; Maddalena, M.; Avioz, A.; Iancu, O.; et al. Different Hotspot P53 Mutants Exert Distinct Phenotypes and Predict Outcome of Colorectal Cancer Patients. Nat. Commun. 2022, 13, 2800. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-H.; Tran, T.T.M.; Tsai Chang, J.; Huang, Y.-T.; Nguyen, A.T.; Chang, I.Y.; Chen, Y.-T.; Hsieh, H.-W.; Juang, Y.-L.; Chang, P.M.-H.; et al. Utilizing TP53 Hotspot Mutations as Effective Predictors of Gemcitabine Treatment Outcome in Non-Small-Cell Lung Cancer. Cell Death Discov. 2025, 11, 26, Correction in Cell Death Discov. 2025, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M. TP53 Mutations in Cancer: Molecular Features and Therapeutic Opportunities (Review). Int. J. Mol. Med. 2024, 55, 7. [Google Scholar] [CrossRef]
- Su, Y.; Zhao, J.; Fu, H.; Liu, Z.; Du, P.; Zheng, J.; Wu, J.; Zhang, J. TP53R175H Mutation Promotes Breast Cancer Cell Proliferation through CORO1A–P38 MAPK Pathway Regulation. Biochem. Pharmacol. 2024, 221, 116047. [Google Scholar] [CrossRef] [PubMed]
- Allende-Vega, N.; Villalba, M. Metabolic Stress Controls Mutant P53 R248Q Stability in Acute Myeloid Leukemia Cells. Sci. Rep. 2019, 9, 5637. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.S.; Tiong, K.H.; Choo, H.L.; Fei-Lei Chung, F.; Hii, L.-W.; Tan, S.H.; Yap, I.K.; Pani, S.; Khor, N.T.; Wong, S.F.; et al. Mutant P53-R273H Mediates Cancer Cell Survival and Anoikis Resistance through AKT-Dependent Suppression of BCL2-Modifying Factor (BMF). Cell Death Dis. 2015, 6, e1826. [Google Scholar] [CrossRef]
- Li, J.; Yang, L.; Gaur, S.; Zhang, K.; Wu, X.; Yuan, Y.-C.; Li, H.; Hu, S.; Weng, Y.; Yen, Y. Mutants TP53 p.R273H and p.R273C but Not p.R273G Enhance Cancer Cell Malignancy. Hum. Mutat. 2014, 35, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Coillie, S.V.; Fang, J.-Y.; Xu, J. Gain of Function of Mutant P53: R282W on the Peak? Oncogenesis 2016, 5, e196. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data Bank (RCSB.Org): Delivery of Experimentally-Determined PDB Structures alongside One Million Computed Structure Models of Proteins from Artificial Intelligence/Machine Learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Jones, D.T.; Cozzetto, D. DISOPRED3: Precise Disordered Region Predictions with Annotated Protein-Binding Activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Guiley, K.Z.; Shokat, K.M. A Small Molecule Reacts with the P53 Somatic Mutant Y220C to Rescue Wild-Type Thermal Stability. Cancer Discov. 2023, 13, 56–69. [Google Scholar] [CrossRef]
- Sandate, C.R.; Chakraborty, D.; Kater, L.; Kempf, G.; Thomä, N.H. Structural Insights into Viral Hijacking of P53 by E6 and E6AP. bioRxiv 2023. [Google Scholar] [CrossRef]
- Xing, Y.F.; Lu, M. Human P53 Core Domain with Hot Spot Mutation R282W in Complex with the Natural CDKN1A(P21) P53-Response Element and Arsenic: 7eeu. 2022. Available online: https://www.rcsb.org/structure/7EEU (accessed on 16 August 2025).
- Pires, D.E.V.; Ascher, D.B.; Blundell, T.L. MCSM: Predicting the Effects of Mutations in Proteins Using Graph-Based Signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Laimer, J.; Hofer, H.; Fritz, M.; Wegenkittl, S.; Lackner, P. MAESTRO-Multi Agent Stability Prediction upon Point Mutations. BMC Bioinform. 2015, 16, 116. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX Web Server: An Online Force Field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulyte, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate Proteome-Wide Missense Variant Effect Prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Guo, S.-B.; Meng, Y.; Lin, L.; Zhou, Z.-Z.; Li, H.-L.; Tian, X.-P.; Huang, W.-J. Artificial Intelligence Alphafold Model for Molecular Biology and Drug Discovery: A Machine-Learning-Driven Informatics Investigation. Mol. Cancer 2024, 23, 223. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Faezov, B.; Dunbrack, R.L. PDBrenum: A Webserver and Program Providing Protein Data Bank Files Renumbered According to Their UniProt Sequences. PLoS ONE 2021, 16, e0253411. [Google Scholar] [CrossRef]
- Mitternacht, S. FreeSASA: An Open Source C Library for Solvent Accessible Surface Area Calculations. F1000Research 2016, 5, 189. [Google Scholar] [CrossRef]
- Alsulami, A.F. Mut-Map: Comprehensive Computational Pipeline for Structural Mapping and Analysis of Cancer-Associated Mutations. Brief. Bioinform. 2024, 25, bbae514. [Google Scholar] [CrossRef] [PubMed]
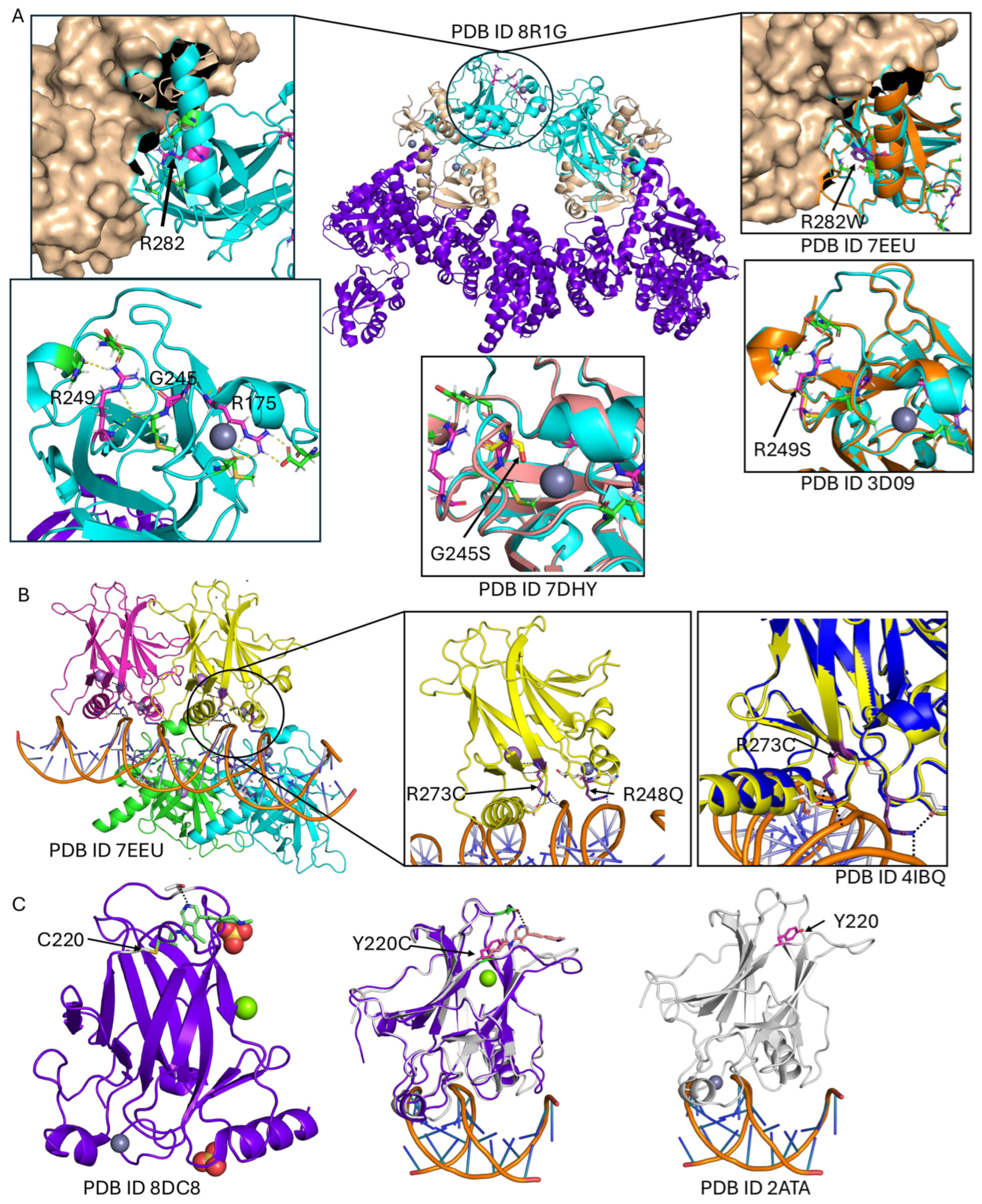

| Mutations | Frequency | PDB | Locations | mCSM | Maestro | FoldX | AlphaMissense |
|---|---|---|---|---|---|---|---|
| R175H | 2513 | 8R1G | around ZN metal | −1.07 | 0.3237 | 11.13 | 0.98 |
| R248Q | 1692 | 7EEU | protein DNA interface | −0.85 | −0.24 | −0.98 | 0.99 |
| R273H | 1521 | 7EEU | protein DNA interface | −1.96 | 0.44 | 1.22 | 0.98 |
| R273C | 1492 | 7EEU | protein DNA interface | −2.02 | −0.11 | 1.32 | 0.99 |
| R248W | 1308 | 7EEU | protein DNA interface | −1.277 | −0.11 | 0.01 | 0.99 |
| R282W | 1165 | 8R1G | between dimers interface | −1.41 | 1.02 | 9.07 | 0.94 |
| G245S | 809 | 8R1G | around ZN metal | −1.536 | 0.69 | 4.03 | 0.97 |
| Y220C | 761 | 2ATA | ligand binding site | −3.524 | 3.27 | 5.11 | 0.97 |
| R249S | 646 | 8R1G | non-interface | −1.859 | 1.85 | 2.72 | 0.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsulami, A.F. Mutational Disruption of TP53: A Structural Approach to Understanding Chemoresistance. Int. J. Mol. Sci. 2025, 26, 9135. https://doi.org/10.3390/ijms26189135
Alsulami AF. Mutational Disruption of TP53: A Structural Approach to Understanding Chemoresistance. International Journal of Molecular Sciences. 2025; 26(18):9135. https://doi.org/10.3390/ijms26189135
Chicago/Turabian StyleAlsulami, Ali F. 2025. "Mutational Disruption of TP53: A Structural Approach to Understanding Chemoresistance" International Journal of Molecular Sciences 26, no. 18: 9135. https://doi.org/10.3390/ijms26189135
APA StyleAlsulami, A. F. (2025). Mutational Disruption of TP53: A Structural Approach to Understanding Chemoresistance. International Journal of Molecular Sciences, 26(18), 9135. https://doi.org/10.3390/ijms26189135





