The STAT Signaling Pathway in HIV-1 Infection: Roles and Dysregulation
Abstract
1. Introduction
2. The JAK/STAT Pathway
3. HIV-1 and Host STAT Pathways
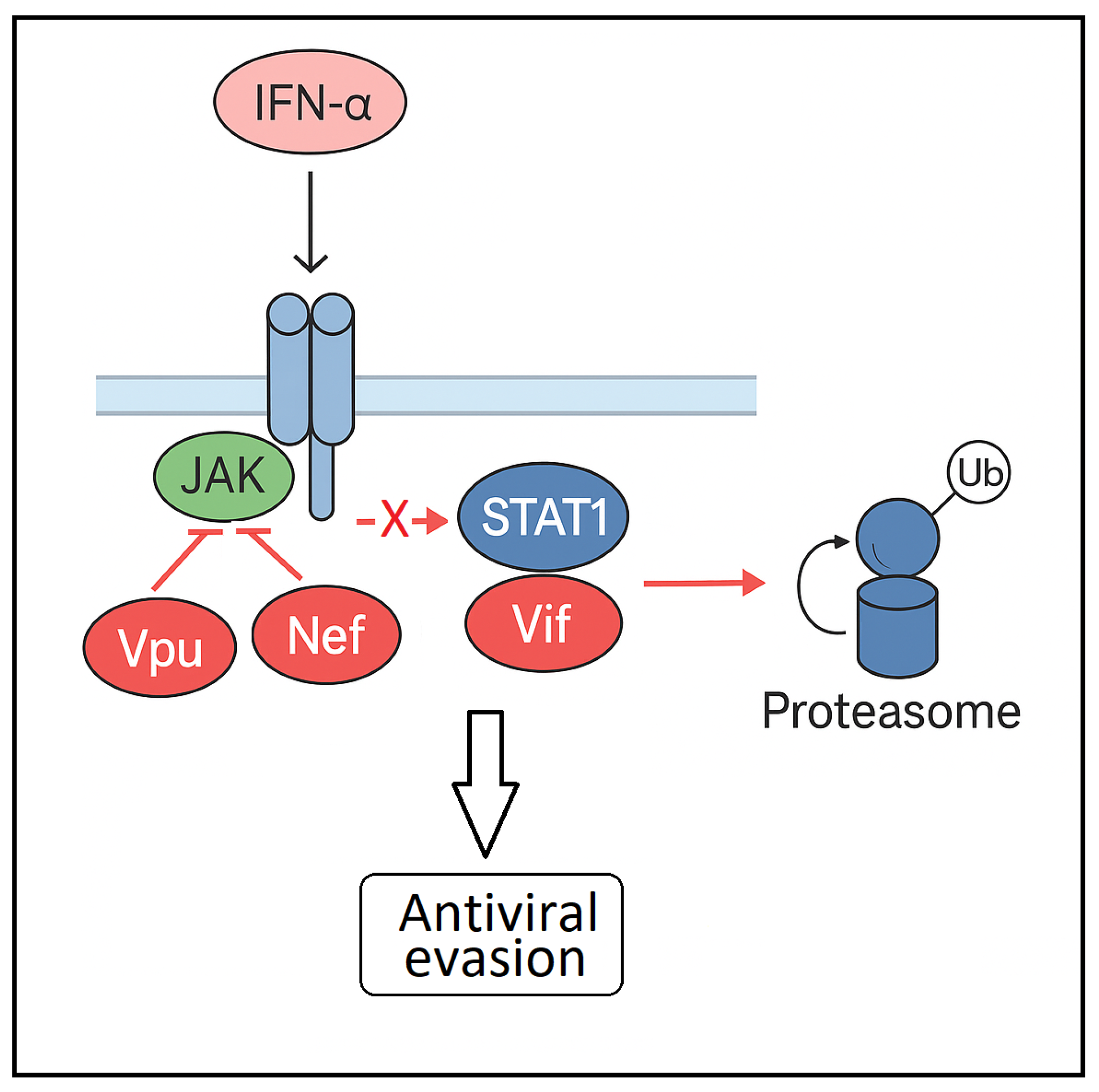
4. The Impact of HIV-1 on STAT1 Signaling and Pathway Dysregulation
4.1. HIV-1–Mediated Modulation of STAT1: Context-Dependent Mechanisms
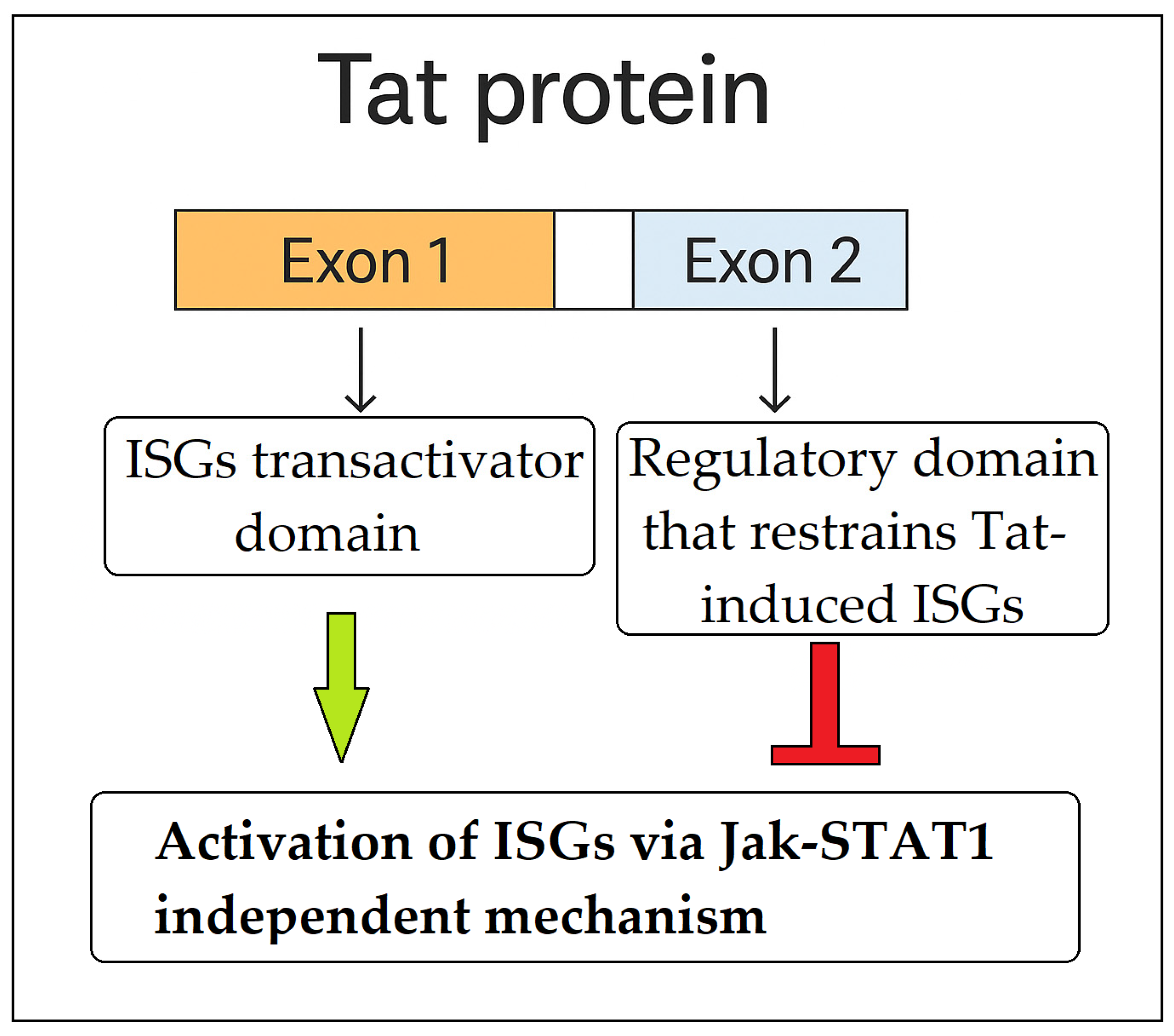
4.2. Atypical Activation of ISG and STAT1 by HIV-1 Tat Protein
5. HIV-1 and STAT3: Molecular Interactions and Pathophysiological Implications
5.1. HIV-1 gp120-Mediated Activation of the STAT3/IL-6 Axis in Dendritic Cells
5.2. Role of STAT3 in Th17 and Treg Differentiation in HIV-1 Infection
5.3. Insertional Mutagenesis and Oncogenesis
5.4. Effects of STAT3 Inhibition in HIV-1 Infected PBMCs
5.5. Conclusive Considerations on the Role of STAT3 in HIV-1 Infection
6. Indirect Evidence for STAT4 in HIV-1 Pathogenesis
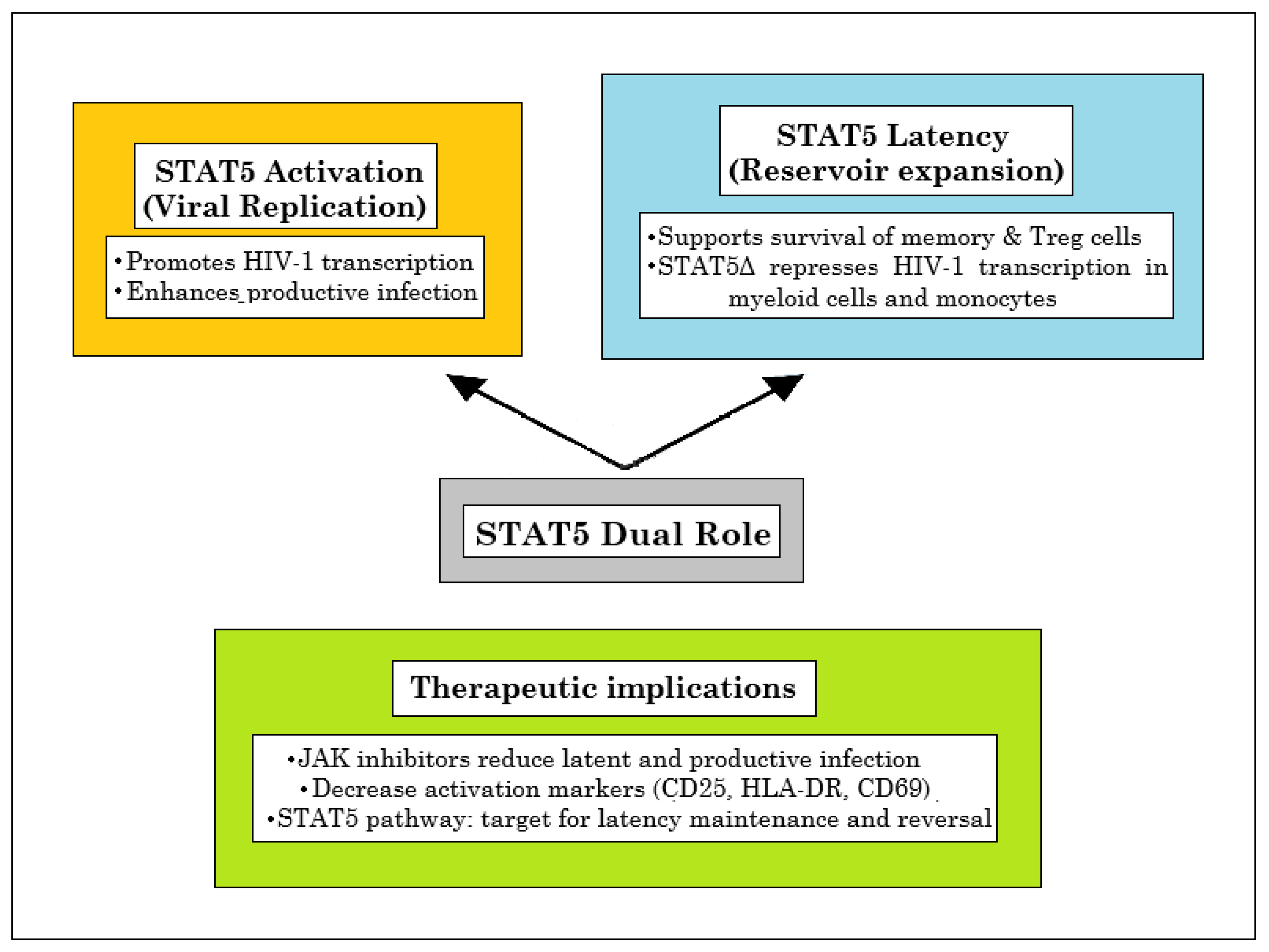
7. Dysregulation of STAT5 Signaling in HIV-1 Infection: Implications for Immune Dysfunction and Viral Persistence
7.1. Dual Roles of STAT5 Dysregulation in HIV-1 Infection: From Altered Cytokine Responsiveness to Viral Replication, and Clonal Expansion
7.2. The STAT5–CCR5 Axis: Mechanistic Insights and Implications for HIV-1 Cure Approaches
7.3. STAT5 at the Crossroads of HIV-1 Latency and Persistence
8. STAT6 as a Host Factor Manipulated by KSHV: Implications for Kaposi’s Sarcoma in HIV-1 Infection
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Bekker, L.G.; Beyrer, C.; Mgodi, N.; Lewin, S.R.; Delany-Moretlwe, S.; Taiwo, B.; Masters, M.C.; Lazarus, J.V. HIV infection. Nat. Rev. Dis. Primers 2023, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Mbonye, U.; Karn, J. The cell biology of HIV-1 latency and rebound. Retrovirology 2024, 21, 6. [Google Scholar] [CrossRef]
- Petrovas, C.; Mueller, Y.M.; Katsikis, P.D. Apoptosis of HIV-specific CD8+ T cells: An HIV evasion strategy. Cell Death Differ. 2005, 12, 859–870. [Google Scholar] [CrossRef]
- Sandstrom, T.S.; Ranganath, N.; Angel, J.B. Impairment of the Type I Interferon Response by HIV-1: Potential Targets for HIV Eradication. Cytokine Growth Factor Rev. 2017, 37, 1–16. [Google Scholar] [CrossRef]
- Scagnolari, C.; Antonelli, G. Type I interferon and HIV: Subtle balance between antiviral activity, immunopathogenesis and the microbiome. Cytokine Growth Factor Rev. 2018, 40, 19–31. [Google Scholar] [CrossRef]
- Masenga, S.K.; Mweene, B.C.; Luwaya, E.; Muchaili, L.; Chona, M.; Kirabo, A. HIV-host cell interactions. Cells 2023, 12, 1351. [Google Scholar] [CrossRef]
- Svanberg, C.; Nyström, S.; Govender, M.; Bhattacharya, P.; Che, K.F.; Ellegård, R.; Shankar, E.M.; Larsson, M. HIV-1 induction of tolerogenic dendritic cells is mediated by cellular interaction with suppressive T cells. Front. Immunol. 2022, 13, 790276. [Google Scholar] [CrossRef]
- Del Cornò, M.; Donninelli, G.; Varano, B.; Da Sacco, L.; Masotti, A.; Gessani, S. HIV-1 gp120 activates the STAT3/interleukin-6 axis in primary human monocyte-derived dendritic cells. J. Virol. 2014, 88, 11045–11055. [Google Scholar] [CrossRef]
- Tugizov, S. Human immunodeficiency virus-associated disruption of mucosal barriers and its role in HIV transmission and pathogenesis of HIV/AIDS disease. Tissue Barriers 2016, 4, e1159276. [Google Scholar]
- Estes, J.D. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol. Rev. 2013, 254, 65–77. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cavalli, A.; Cascio, A. Stat1 and its crucial role in the control of viral infections. Int. J. Mol. Sci. 2022, 23, 4095. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. The multifaced role of stat3 in cancer and its implication for anticancer therapy. Int. J. Mol. Sci. 2021, 22, 603. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. Stat4 and stat6, their role in cellular and humoral immunity and in diverse human diseases. Int. Rev. Immunol. 2024, 43, 394–418. [Google Scholar] [CrossRef] [PubMed]
- Tolomeo, M.; Meli, M.; Grimaudo, S. Stat5 and stat5 inhibitors in hematological malignancies. Anticancer Agents Med. Chem. 2019, 19, 2036–2046. [Google Scholar] [CrossRef] [PubMed]
- Rondanin, R.; Simoni, D.; Maccesi, M.; Romagnoli, R.; Grimaudo, S.; Pipitone, R.M.; Meli, M.; Cascio, A.; Tolomeo, M. Effects of pimozide derivatives on pStat5 in K562 cells. ChemMedChem 2017, 12, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Rondanin, R.; Simoni, D.; Romagnoli, R.; Baruchello, R.; Marchetti, P.; Costantini, C.; Fochi, S.; Padroni, G.; Grimaudo, S.; Pipitone, R.M.; et al. Inhibition of activated Stat5 in Bcr/abl expressing leukemia cells with new pimozide derivatives. Bioorganic Med. Chem. Lett. 2014, 24, 4568–4574. [Google Scholar] [CrossRef]
- Grimaudo, S.; Meli, M.; Di Cristina, A.; Ferro, A.; Pipitone, M.R.; Romagnoli, R.; Simoni, D.; Dieli, F.; Tolomeo, M. The new iodoacetamidobenzofuran derivative TR120 decreases Stat5 expression and induces antitumor effects in imatinib-sensitive and imatinib-resistant Bcr-abl-expressing leukemia cells. Anticancer Drugs 2013, 24, 384–393. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; O’Shea, J.J. Mechanisms of jak/stat signaling in immunity and disease. J. Immunol. 2015, 194, 21–27. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Holland, S.M.; Staudt, L.M. Jaks and stats in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 2013, 368, 161–170. [Google Scholar] [CrossRef]
- Banerjee, S.; Boucheron, N.; Schuster, M.; Tschurtschenthaler, G.; Sexl, V. Jak/stat signaling as a target for inflammatory and autoimmune diseases: Current and future prospects. Int. J. Mol. Sci. 2023, 24, 8123. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The jak/stat signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Shuai, K.; Liu, B. Regulation of jak-stat signalling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. [Google Scholar] [PubMed]
- Aaronson, D.S.; Horvath, C.M. A road map for those who don’t know jak-stat. Science 2002, 296, 1653–1655. [Google Scholar]
- Schindler, C.; Levy, D.E.; Decker, T. Jak-stat signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar]
- Yu, H.; Pardoll, D.; Jove, R. Stats in cancer inflammation and immunity: A leading role for stat3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Bromberg, J.F. Activation of stat proteins and growth control. BioEssays 2001, 23, 161–169. [Google Scholar] [CrossRef]
- Kaplan, M.H. Stat4: A critical regulator of inflammation in vivo. Immunol. Res. 2005, 31, 231–242. [Google Scholar] [CrossRef]
- Karpathiou, G.; Papoudou-Bai, A.; Ferrand, E.; Dumollard, J.M.; Peoc’h, M. STAT6: A Review of a Signaling Pathway Implicated in Various Diseases with a Special Emphasis in Its Usefulness in Pathology. Pathol. Res. Pract. 2021, 223, 153477. [Google Scholar] [CrossRef] [PubMed]
- Malin, S.; McManus, S.; Busslinger, M. Stat5 in B cell development and leukemia. Curr. Opin. Immunol. 2010, 22, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Naka, T.; Kubo, M. Socs proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007, 7, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G.; Overbaugh, J.; Phillips, A.; Buchbinder, S. Hiv infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. The complex dysregulations of CD4 T cell subtypes in HIV infection. Int. J. Mol. Sci. 2024, 25, 7512. [Google Scholar] [CrossRef]
- Chun, T.W.; Moir, S.; Fauci, A.S. Hiv reservoirs as obstacles and opportunities for an HIV cure. Nat. Immunol. 2015, 16, 584–589. [Google Scholar] [CrossRef]
- Siliciano, R.F.; Greene, W.C. Hiv latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. [Google Scholar] [CrossRef]
- Krebs, A.S.; Mendonça, L.M.; Zhang, P. Structural analysis of retrovirus assembly and maturation. Viruses 2021, 14, 54–65. [Google Scholar] [CrossRef]
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and function of retroviral integrase. Nat. Rev. Microbiol. 2022, 20, 20–34. [Google Scholar] [CrossRef]
- Grawenhoff, J.; Engelman, A.N. Retroviral integrase protein and intasome nucleoprotein complex structures. World J. Biol. Chem. 2017, 8, 32–44. [Google Scholar] [CrossRef]
- Tolomeo, M.; Tolomeo, F.; Cascio, A. The complex interactions between HIV-1 and human host cell genome: From molecular mechanisms to clinical practice. Int. J. Mol. Sci. 2025, 26, 3184. [Google Scholar] [CrossRef] [PubMed]
- Appay, V.; Sauce, D. Immune activation and inflammation in HIV-1 infection: Causes and consequences. J. Pathol. 2008, 214, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Nema, V. HIV-associated neurotoxicity: The interplay of host and viral proteins. Mediators Inflamm. 2021, 2021, 1267041. [Google Scholar] [CrossRef] [PubMed]
- Garg, H.; Mohl, J.; Joshi, A. HIV-1 induced bystander apoptosis. Viruses 2012, 4, 3020–3043. [Google Scholar] [CrossRef]
- Mouzakis, A.; Petrakis, V.; Tryfonopoulou, E.; Panopoulou, M.; Panagopoulos, P.; Chlichlia, K. Mechanisms of immune evasion in HIV-1: The role of virus-host protein interactions. Curr. Issues Mol. Biol. 2025, 47, 367. [Google Scholar] [CrossRef]
- Zimmerman, M.G.; Bowen, J.R.; McDonald, C.E.; Young, E.; Baric, R.S.; Pulendran, B.; Suthar, M.S. STAT5: A Target of Antagonism by Neurotropic Flaviviruses. J. Virol. 2019, 93, 1110–1128. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Zhang, Y.J. Interferon Independent Non-Canonical STAT Activation and Virus Induced Inflammation. Viruses 2018, 10, 196. [Google Scholar] [CrossRef]
- Roca Suarez, A.A.; Van Renne, N.; Baumert, T.F.; Lupberger, J. Viral manipulation of STAT3: Evade, exploit, and injure. PLoS Pathog. 2018, 14, e1006839. [Google Scholar] [CrossRef]
- Reece, M.D.; Song, C.; Hancock, S.C.; Pereira Ribeiro, S.; Kulpa, D.A.; Gavegnano, C. Repurposing BCL-2 and Jak 1/2 inhibitors: Cure and treatment of HIV-1 and other viral infections. Front. Immunol. 2022, 13, 1033672. [Google Scholar] [CrossRef]
- Gavegnano, C.; Brehm, J.H.; Dupuy, F.P.; Talla, A.; Ribeiro, S.P.; Kulpa, D.A.; Cameron, C.; Santos, S.; Hurwitz, S.J.; Marconi, V.C.; et al. Novel mechanisms to inhibit HIV reservoir seeding using Jak inhibitors. PLoS Pathog. 2017, 13, e1006740. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, J.; Chen, Y.; Liu, S. Targeting the STAT3 oncogenic pathway: Cancer immunotherapy and drug repurposing. Biomed. Pharmacother. 2023, 167, 115513. [Google Scholar] [CrossRef] [PubMed]
- Miklossy, G.; Hilliard, T.S.; Turkson, J. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013, 12, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Soper, A.; Kimura, I.; Nagaoka, S.; Konno, Y.; Yamamoto, K.; Koyanagi, Y.; Sato, K. Type I interferon responses by HIV-1 infection: Association with disease progression and control. Front. Immunol. 2017, 8, 1823. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Zang, T.M.; Rihn, S.J.; Zhang, F.; Kueck, T.; Alim, M.; Schoggins, J.; Rice, C.M.; Wilson, S.J.; Bieniasz, P.D. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 2016, 20, 392–405. [Google Scholar] [CrossRef]
- Chang, T.L.; Mosoian, A.; Pine, R.; Klotman, M.E.; Moore, J.P. A soluble factor(s) secreted from CD8(+) T lymphocytes inhibits human immunodeficiency virus type 1 replication through STAT1 activation. J. Virol. 2002, 76, 569–581. [Google Scholar]
- Federico, M.; Percario, Z.; Olivetta, E.; Fiorucci, G.; Muratori, C.; Micheli, A.; Romeo, G.; Affabris, E. HIV-1 Nef activates STAT1 in human monocytes/ macrophages through the release of soluble factors. Blood 2001, 98, 2752–2761. [Google Scholar] [CrossRef]
- Kohler, J.J.; Tuttle, D.L.; Coberley, C.R.; Sleasman, J.W.; Goodenow, M.M. Human Immunodeficiency virus type 1 (HIV-1) induces activation of multiple STATs in CD4+ cells of lymphocyte or monocyte/ macrophage lineages. J. Leukoc. Biol. 2003, 73, 407–416. [Google Scholar]
- Nguyen, N.V.; Tran, J.T.; Sanchez, D.J. HIV blocks type I IFN signaling through disruption of STAT1 phosphorylation. Innate Immun. 2018, 24, 490–500. [Google Scholar]
- Gargan, S.; Ahmed, S.; Mahony, R.; Bannan, C.; Napoletano, S.; O’Farrelly, C.; Borrow, P.; Bergin, C.; Stevenson, N.J. HIV-1 Promotes the degradation of components of the type 1 IFN JAK/ STAT pathway and blocks anti-viral ISG induction. EBioMedicine 2018, 30, 203–216. [Google Scholar] [CrossRef]
- Sugawara, S.; El-Diwany, R.; Cohen, L.K.; Rousseau, K.E.; Williams, C.Y.K.; Veenhuis, R.T.; Mehta, S.H.; Blankson, J.N.; Thomas, D.L.; Cox, A.L.; et al. People with HIV-1 demonstrate type 1 interferon refractoriness associated with upregulated USP18. J. Virol. 2021, 95, JVI.01777-20. [Google Scholar] [CrossRef]
- Rivera, L.E.; Kraiselburd, E.; Meléndez, L.M. Cystatin B and HIV regulate the STAT-1 signaling circuit in HIV-infected and INF-β-treated human macrophages. J. Neurovirol. 2016, 22, 666–673. [Google Scholar] [CrossRef]
- Kukkonen, S.; Martinez-Viedma, M.P.; Kim, N.; Manrique, M.; Aldovini, A. HIV-1 Tat second exon limits the extent of Tat-mediated modulation of interferon-stimulated genes in antigen presenting cells. Retrovirology 2014, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Ishihara, K.; Hibi, M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000, 19, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, J.J. STAT3 plays an important role in DNA replication by turning on WDHD1. Cell Biosci. 2021, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V. The complex role of STAT3 in viral infections. J. Immunol. Res. 2015, 2015, 272359. [Google Scholar] [CrossRef]
- Ibba, S.V.; Fenizia, C.; Ortega, P.S.; Mercurio, V.; Saulle, I.; Lori, E.M.; Trabattoni, D.; Clerici, M.; Biasin, M. Analysing the role of STAT3 in HIV-1 infection. J. Biol. Regul. Homeost. Agents 2019, 33, 1635–1639. [Google Scholar]
- Miller, E.; Bhardwaj, N. Dendritic cell dysregulation during HIV-1 infection. Immunol. Rev. 2013, 254, 170–189. [Google Scholar] [CrossRef]
- Kanwar, B.; Favre, D.; McCune, J.M. Th17 and regulatory T cells: Implications for AIDS pathogenesis. Curr. Opin. HIV AIDS 2010, 5, 151–157. [Google Scholar] [CrossRef]
- Raffatellu, M.; Santos, R.L.; Verhoeven, D.E.; George, M.D.; Wilson, R.P.; Winter, S.E.; Godinez, I.; Sankaran, S.; Paixao, T.A.; Gordon, M.A.; et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 2008, 14, 421–428. [Google Scholar]
- Cecchinato, V.; Trindade, C.J.; Laurence, A.; Heraud, J.M.; Brenchley, J.M.; Ferrari, M.G.; Zaffiri, L.; Tryniszewska, E.; Tsai, W.P.; Vaccari, M.; et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol. 2008, 1, 279–288. [Google Scholar] [CrossRef]
- Christensen-Quick, A.; Lafferty, M.; Sun, L.; Marchionni, L.; DeVico, A.; Garzino-Demo, A. Human Th17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection. J. Virol. 2016, 90, 7833–7847. [Google Scholar] [CrossRef]
- Eyerich, K.; Foerster, S.; Rombold, S.; Seidl, H.-P.; Behrendt, H.; Hofmann, H.; Ring, J.; Traidl-Hoffmann, C. Patients with chronic mucocutaneous candidiasis exhibit reduced production of Th17-associated cytokines IL-17 and IL-22. J. Investig. Dermatol. 2008, 128, 2640–2645. [Google Scholar] [CrossRef] [PubMed]
- Elhed, A.; Unutmaz, D. Th17 cells and HIV infection. Curr. Opin. HIV AIDS 2010, 5, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Vajpayee, M.; Ali, S.A.; Mojumdar, K.; Chauhan, N.K.; Singh, R. HIV-1 disease progression associated with loss of Th17 cells in subtype ‘C’ infection. Cytokine 2012, 60, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Rocco, J.; Mellors, J.W.; Macatangay, B.J.C. Regulatory T cells: The ultimate HIV reservoir? J. Virus Erad. 2018, 4, 209–214. [Google Scholar] [CrossRef]
- Xu, L.; Kitani, A.; Fuss, I.; Strober, W. Cutting edge: Regulatory T cells induce CD4+CD25−Foxp3− T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J. Immunol. 2007, 178, 6725–6729. [Google Scholar] [CrossRef]
- Osorio, F.; LeibundGut-Landmann, S.; Lochner, M.; Lahl, K.; Sparwasser, T.; Eberl, G.; Reis e Sousa, C. DC activated via dectin-1 convert Treg into IL-17 producers. Eur. J. Immunol. 2008, 38, 3274–3281. [Google Scholar] [CrossRef]
- Koenen, H.J.; Smeets, R.L.; Vink, P.M.; van Rijssen, E.; Boots, A.M.; Joosten, I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood 2008, 112, 2340–2352. [Google Scholar] [CrossRef]
- Beriou, G.; Costantino, C.M.; Ashley, C.W.; Yang, L.; Kuchroo, V.K.; Baecher-Allan, C.; Hafler, D.A. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood 2009, 113, 4240–4249. [Google Scholar] [CrossRef]
- Yang, X.O.; Panopoulos, A.D.; Nurieva, R.; Chang, S.H.; Wang, D.; Watowich, S.S.; Dong, C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007, 282, 9358–9363. [Google Scholar] [CrossRef]
- Tripathi, S.K.; Chen, Z.; Larjo, A.; Kanduri, K.; Nousiainen, K.; Äijo, T.; Ricaño-Ponce, I.; Hrdlickova, B.; Tuomela, S.; Laajala, E.; et al. Genome-wide analysis of STAT3-mediated transcription during early human Th17 cell differentiation. Cell Rep. 2017, 19, 1888–1901. [Google Scholar] [CrossRef]
- Egwuagu, C.E. STAT3 in CD4⁺ T helper cell differentiation and inflammatory diseases. Cytokine 2009, 47, 149–156. [Google Scholar] [CrossRef]
- Schmetterer, K.G.; Neunkirchner, A.; Wojta-Stremayr, D.; Leitner, J.; Steinberger, P.; Pickl, W.F. STAT3 governs hyporesponsiveness and granzyme B-dependent suppressive capacity in human CD4⁺ T cells. FASEB J. 2015, 29, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Yero, A.; Bouassa, R.M.; Ancuta, P.; Estaquier, J.; Jenabian, M.A. Immuno-metabolic control of the balance between Th17-polarized and regulatory T cells during HIV infection. Cytokine Growth Factor Rev. 2023, 69, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Renault, C.; Veyrenche, N.; Mennechet, F.; Bedin, A.S.; Routy, J.P.; Van de Perre, P.; Reynes, J.; Tuaillon, E. Th17 CD4⁺ T cell as a preferential target for HIV reservoirs. Front. Immunol. 2022, 13, 822576. [Google Scholar] [CrossRef] [PubMed]
- Valverde-Villegas, J.M.; Matte, M.C.; de Medeiros, R.M.; Chies, J.A. New insights about Treg and Th17 cells in HIV infection and disease progression. J. Immunol. Res. 2015, 2015, 647916. [Google Scholar] [CrossRef]
- Fernandes, J.R.; Berthoud, T.K.; Kumar, A.; Angel, J.B. IL-23 signaling in Th17 cells is inhibited by HIV infection and is not restored by HAART: Implications for persistent immune activation. PLoS ONE 2017, 12, e0186823. [Google Scholar] [CrossRef]
- Rist, M.; Kaku, M.; Coffin, J.M. In vitro HIV DNA integration in STAT3 drives T cell persistence-A model of HIV-associated T cell lymphoma. PLoS Pathog. 2025, 21, e1013087. [Google Scholar] [CrossRef]
- Adoro, S.; Cubillos-Ruiz, J.R.; Chen, X.; Deruaz, M.; Vrbanac, V.D.; Song, M.; Park, S.; Murooka, T.T.; Dudek, T.E.; Luster, A.D.; et al. IL-21 induces antiviral microRNA-29 in CD4 T cells to limit HIV-1 infection. Nat. Commun. 2015, 6, 7562. [Google Scholar] [CrossRef]
- Vanpouille, C.; Introini, A.; Morris, S.; Margolis, L.; Daar, E.S.; Dube, M.P.; Little, S.J.; Smith, D.M.; Lisco, A.; Gianella, S. Distinct cytokine/chemokine network in semen and blood characterize different stages of HIV infection. AIDS 2016, 30, 193–201. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Lahesmaa, R.; Vahedi, G.; Laurence, A.; Kanno, Y. Genomic views of STAT function in CD4⁺ T helper cell differentiation. Nat. Rev. Immunol. 2011, 11, 239–250. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, Q.; Li, X.; Liu, Y.; Shen, Q.; Yang, M.; Wang, C.; Li, N.; Cao, X. Cytoplasmic STAT4 promotes antiviral type I IFN production by blocking CHIP-mediated degradation of RIG-I. J. Immunol. 2016, 196, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.D.; Smith, J.A.; Lichtinger, M.; Wang, L.; Su, L. Activation of the Signal Transducer and Activator of Transcription 1 signaling pathway in thymocytes from HIV-1-infected human thymus. AIDS 2003, 17, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, C.; Camorali, L.; Lorini, A.L.; Ghezzi, S.; Vicenzi, E.; Lazzarin, A.; Poli, G. Constitutive activation of STATs upon in vivo human immunodeficiency virus infection. Blood 1999, 94, 4202–4209. [Google Scholar] [CrossRef]
- Lee, A.W.; Sharp, E.R.; O’Mahony, A.; Rosenberg, M.G.; Israelski, D.M.; Nolan, G.P.; Nixon, D.F. Single-cell, phosphoepitope-specific analysis demonstrates cell type- and pathway-specific dysregulation of Jak/STAT and MAPK signaling associated with in vivo human immunodeficiency virus type 1 infection. J. Virol. 2008, 82, 3702–3712. [Google Scholar] [CrossRef]
- Lawless, V.A.; Zhang, S.; Ozes, O.N.; Bruns, H.A.; Oldham, I.; Hoey, T.; Grusby, M.J.; Kaplan, M.H. Stat4 regulates multiple components of IFN-gamma-inducing signaling pathways. J. Immunol. 2000, 165, 6803–6808. [Google Scholar] [CrossRef]
- Roff, S.R.; Noon-Song, E.N.; Yamamoto, J.K. The significance of interferon-γ in HIV-1 pathogenesis, therapy, and prophylaxis. Front. Immunol. 2014, 4, 498. [Google Scholar] [CrossRef]
- Miller, R.C.; Schlaepfer, E.; Baenziger, S.; Crameri, R.; Zeller, S.; Byland, R.; Audigé, A.; Nadal, D.; Speck, R.F. HIV interferes with SOCS-1 and -3 expression levels driving immune activation. Eur. J. Immunol. 2011, 41, 1058–1069. [Google Scholar] [CrossRef]
- Tamiya, T.; Kashiwagi, I.; Takahashi, R.; Yasukawa, H.; Yoshimura, A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: Regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 980–985. [Google Scholar] [CrossRef]
- Xia, Y.; Xie, Y.; Zhang, H.; Liu, L. STAT4 gene polymorphisms in human diseases. Front. Immunol. 2024, 15, 1479418. [Google Scholar] [CrossRef]
- Madera-Salcedo, I.K.; Ramírez-Sánchez, A.L.; Rodríguez-Rodríguez, N.; García-Quintero, R.; Rubio, R.M.; Morales-Montes de Oca, G.; Dávalos, E.; Cuervo, R.; Furuzawa-Carballeda, J.; Alcocer-Varela, J.; et al. Down-regulation-resistant STAT4 risk haplotype contributes to lupus nephritis through CD4⁺ T cell interferon-γ production. Arthritis Rheumatol. 2023, 75, 961–972. [Google Scholar] [CrossRef]
- Lamana, A.; López-Santalla, M.; Castillo-González, R.; Ortiz, A.M.; Martín, J.; García-Vicuña, R.; González-Álvaro, I. The minor allele of rs7574865 in the STAT4 gene is associated with increased mRNA and protein expression. PLoS ONE 2015, 10, e0142683. [Google Scholar] [CrossRef]
- Hagberg, N.; Joelsson, M.; Leonard, D.; Reid, S.; Eloranta, M.L.; Mo, J.; Nilsson, M.K.; Syvänen, A.C.; Bryceson, Y.T.; Rönnblom, L. The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann. Rheum. Dis. 2018, 77, 1070–1077. [Google Scholar] [CrossRef]
- Buzzelli, A.A.; McWilliams, I.L.; Shin, B.; Bryars, M.T.; Harrington, L.E. Intrinsic STAT4 expression controls effector CD4 T cell migration and Th17 pathogenicity. J. Immunol. 2023, 210, 1667–1676. [Google Scholar] [CrossRef]
- Kryworuchko, M.; Pasquier, V.; Keller, H.; David, D.; Goujard, C.; Gilquin, J.; Viard, J.P.; Joussemet, M.; Delfraissy, J.F.; Theze, J. Defective interleukin-2-dependent STAT5 signalling in CD8 T lymphocytes from HIV-positive patients: Restoration by antiretroviral therapy. AIDS 2004, 18, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Warby, T.J.; Crowe, S.M.; Jaworowski, A. Human immunodeficiency virus type 1 infection inhibits granulocyte-macrophage colony-stimulating factor-induced activation of STAT5A in human monocyte-derived macrophages. J. Virol. 2003, 77, 12630–12638. [Google Scholar] [CrossRef] [PubMed]
- Selliah, N.; Zhang, M.; DeSimone, D.; Kim, H.; Brunner, M.; Ittenbach, R.F.; Rui, H.; Cron, R.Q.; Finkel, T.H. The gammac-cytokine regulated transcription factor, STAT5, increases HIV-1 production in primary CD4 T cells. Virology 2006, 344, 283–291. [Google Scholar] [CrossRef][Green Version]
- Della Chiara, G.; Crotti, A.; Liboi, E.; Giacca, M.; Poli, G.; Lusic, M. Negative regulation of HIV-1 transcription by a heterodimeric NF-κB1/p50 and C-terminally truncated STAT5 complex. J. Mol. Biol. 2011, 410, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Crotti, A.; Lusic, M.; Lupo, R.; Lievens, P.M.; Liboi, E.; Della Chiara, G.; Tinelli, M.; Lazzarin, A.; Patterson, B.K.; Giacca, M.; et al. Naturally occurring C-terminally truncated STAT5 is a negative regulator of HIV-1 expression. Blood 2007, 109, 5380–5389. [Google Scholar] [CrossRef]
- Landires, I.; Bugault, F.; Lambotte, O.; de Truchis, P.; Slama, L.; Danckaert, A.; Delfraissy, J.F.; Thèze, J.; Chakrabarti, L.A. HIV infection perturbs interleukin-7 signaling at the step of STAT5 nuclear relocalization. AIDS 2011, 25, 1843–1853. [Google Scholar] [CrossRef]
- Cesana, D.; Santoni de Sio, F.R.; Rudilosso, L.; Gallina, P.; Calabria, A.; Beretta, S.; Merelli, I.; Bruzzesi, E.; Passerini, L.; Nozza, S.; et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat. Commun. 2017, 8, 498. [Google Scholar] [CrossRef]
- Christian, M.L.; Dapp, M.J.; Scharffenberger, S.C.; Jones, H.; Song, C.; Frenkel, L.M.; Krumm, A.; Mullins, J.I.; Rawlings, D.J. CRISPR/Cas9-mediated insertion of HIV long terminal repeat within BACH2 promotes expansion of T regulatory-like cells. J. Immunol. 2022, 208, 1700–1710. [Google Scholar] [CrossRef]
- Ikeda, T.; Shibata, J.; Yoshimura, K.; Koito, A.; Matsushita, S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 2007, 195, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Yeh, Y.J.; Varabyou, A.; Collora, J.A.; Sherrill-Mix, S.; Talbot, C.C., Jr.; Mehta, S.; Albrecht, K.; Hao, H.; Zhang, H.; et al. Single-cell transcriptional landscapes reveal HIV-1-driven aberrant host gene transcription as a potential therapeutic target. Sci. Transl. Med. 2020, 12, eaaz0802. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Peppa, D.; Hill, A.L.; Gálvez, C.; Salgado, M.; Pace, M.; McCoy, L.E.; Griffith, S.A.; Thornhill, J.; Alrubayyi, A. Evidence for HIV-1 cure after CCR5Δ32/Δ32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption: A case report. Lancet HIV 2020, 7, e340–e347. [Google Scholar] [CrossRef] [PubMed]
- Hütter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müßig, A.; Allers, K.; Schneider, T.; Homann, J.; Kücherer, C.; Blau, O. Long-term remission and HIV cure in a patient with a CCR5-delta32/Delta32 mutation after stem cell transplantation. N. Engl. J. Med. 2009, 360, 692–698. [Google Scholar] [CrossRef]
- Wang, X.; Peng, J.; He, J. CCR5 is a prognostic biomarker and an immune regulator for triple negative breast cancer. Aging 2021, 13, 23810–23830. [Google Scholar] [CrossRef]
- Hemmatazad, H.; Berger, M.D. CCR5 is a potential therapeutic target for cancer. Expert Opin. Ther. Targets 2021, 25, 311–327. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, S.; Sun, Y.; Chen, C.; Yang, S.; Pei, G.; Lin, M.; Yu, J.; Liu, X.; Wang, H.; et al. CCR5 and inflammatory storm. Ageing Res. Rev. 2024, 96, 102286. [Google Scholar] [CrossRef]
- Zeng, Z.; Lan, T.; Wei, Y.; Wei, X. CCL5/CCR5 axis in human diseases and related treatments. Genes Dis. 2022, 9, 12–27. [Google Scholar] [CrossRef]
- Deng, H.K.; Liu, R.; Choe, S.; Unutmaz, D.; Burkhart, M.; Di Marzio, P.; Marmon, S.; Sutton, R.E.; Hill, C.M.; Davis, C.B.; et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996, 381, 661–666. [Google Scholar] [CrossRef]
- Wierda, R.J.; van den Elsen, P.J. Genetic and epigenetic regulation of CCR5 transcription. Biology 2012, 1, 869–879. [Google Scholar] [CrossRef]
- Mummidi, S.; VanCompernolle, S.E.; Kalkonde, M.; Camargo, J.F.; Kulkarni, H.; Bellinger, A.S.; Bonello, G.; Tagoh, H.; Ahuja, S.S.; Unutmaz, D.; et al. Production of specific mRNA transcripts, usage of an alternate promoter, and octamer-binding transcription factors influence the surface expression levels of the HIV coreceptor CCR5 on primary T cells. J. Immunol. 2007, 178, 5668–5681. [Google Scholar] [CrossRef] [PubMed]
- Manganaro, L.; Hong, P.; Hernandez, M.M.; Argyle, D.; Mulder, L.C.F.; Potla, U.; Diaz-Griffero, F.; Lee, B.; Fernandez-Sesma, A.; Simon, V. IL-15 regulates susceptibility of CD4(+) T cells to HIV infection. Proc. Natl. Acad. Sci. USA 2018, 115, E9659–E9667. [Google Scholar] [CrossRef]
- Skariah, N.; James, O.J.; Swamy, M. Signalling mechanisms driving homeostatic and inflammatory effects of interleukin-15 on tissue lymphocytes. Discov. Immunol. 2024, 3, kyae002. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yukselten, Y.; Nuwagaba, J.; Sutton, R.E. JAK/STAT signaling pathway affects CCR5 expression in human CD4+ T cells. Sci. Adv. 2024, 10, eadl0368. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Chen, Y.; Su, B.; Yang, X.; Zhang, Q.; Song, T.; Wu, H.; Liu, C.; Liu, L.; Zhang, T. Alterations of CCR2 and CX3CR1 on three monocyte subsets during HIV-1/Treponema pallidum coinfection. Front. Med. 2020, 7, 272. [Google Scholar] [CrossRef]
- Covino, D.A.; Sabbatucci, M.; Fantuzzi, L. The CCL2/CCR2 axis in the pathogenesis of HIV-1 infection: A new cellular target for therapy? Curr. Drug Targets 2016, 17, 76–110. [Google Scholar] [CrossRef]
- Kruize, Z.; Kootstra, N.A. The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Microbiol. 2019, 10, 2828. [Google Scholar] [CrossRef]
- de Armas, L.R.; Gavegnano, C.; Pallikkuth, S.; Pahwa, S.; Schinazi, R.F.; Rinaldi, T.; Banga, R.; Rinaldi, J.C.; Bosque, A.; Stevenson, M.; et al. The Effect of JAK1/2 Inhibitors on HIV Reservoir Using Primary Lymphoid Cell Model of HIV Latency. Front. Immunol. 2021, 12, 720697. [Google Scholar] [CrossRef]
- Venkatachari, N.J.; Zerbato, J.M.; Jain, S.; Mota, T.; Shao, W.; Halper-Stromberg, A.; Mellors, J.W.; Valente, S.T. Temporal transcriptional response to latency reversing agents identifies specific factors regulating HIV-1 viral transcriptional switch. Retrovirology 2015, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Janssens, J.; Kim, P.; Kim, S.J.; Wedrychowski, A.; Kadiyala, G.N.; Hunt, P.W.; Deeks, S.G.; Wong, J.K.; Yukl, S.A. Mechanisms and efficacy of small molecule latency-promoting agents to inhibit HIV reactivation ex vivo. JCI Insight 2024, 9, e183084. [Google Scholar] [CrossRef] [PubMed]
- Goenka, S.; Kaplan, M.H. Transcriptional regulation by STAT6. Immunol. Res. 2011, 50, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Zha, B. The roles of STAT6 in regulating B cell fate, activation, and function. Immunol. Lett. 2021, 233, 87–91. [Google Scholar] [CrossRef]
- Chen, H.; Sun, H.; You, F.; Sun, W.; Zhou, X.; Chen, L.; Yang, J.; Wang, Y.; Tang, H.; Guan, Y.; et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 2011, 147, 436–446. [Google Scholar] [CrossRef]
- Chiang, H.S.; Liu, H.M. The molecular basis of viral inhibition of IRF- and STAT-dependent immune responses. Front. Immunol. 2018, 9, 3086. [Google Scholar] [CrossRef]
- Mahjoor, M.; Mahmoudvand, G.; Farokhi, S.; Saghabashi, A.; Rostami, S.F.A.; Modarressi, M.H. Double-edged sword of JAK/STAT signaling pathway in viral infections: Novel insights into virotherapy. Cell Commun. Signal. 2023, 21, 272. [Google Scholar] [CrossRef]
- Zeng, Y.; Zhang, X.; Huang, Z.; Cheng, L.; Yao, S.; Qin, D.; Chen, X.; Tang, Q.; Lv, Z.; Zhang, L.; et al. Intracellular Tat of human immunodeficiency virus type 1 activates lytic cycle replication of Kaposi’s sarcoma-associated herpesvirus: Role of JAK/STAT signaling. J. Virol. 2007, 81, 2401–2417, Erratum in: J. Virol. 2021, 95, e00234-21. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, C.; Wei, F.; Gao, S.; Zhang, L.; Li, Y.; Feng, Y.; Tong, Y.; Xu, J.; Wang, B.; et al. Nuclear localization and cleavage of STAT6 is induced by Kaposi’s sarcoma-associated herpesvirus for viral latency. PLoS Pathog. 2017, 13, e1006124. [Google Scholar] [CrossRef]
- Marconi, V.C.; Moser, C.; Gavegnano, C.; Deeks, S.G.; Lederman, M.M.; Overton, E.T.; Tsibris, A.; Hunt, P.W.; Kantor, A.; Sekaly, R.P.; et al. Randomized trial of ruxolitinib in antiretroviral-treated adults with human immunodeficiency virus. Clin. Infect. Dis. 2022, 74, 95–104. [Google Scholar] [CrossRef]
- Hurwitz, S.J.; Tao, S.; Gavegnano, C.; Jiang, Y.; Tressler, R.L.; Tsibris, A.; Del Rio, C.; Overton, E.T.; Lederman, M.M.; Kantor, A.; et al. Pharmacokinetics of ruxolitinib in HIV suppressed individuals on antiretroviral agent therapy from the ACTG A5336 study. J. Clin. Pharmacol. 2021, 61, 1555–1566. [Google Scholar] [CrossRef]


| Protein | Antiviral Mechanism Against HIV-1 | References |
|---|---|---|
| APOBEC3G | Cytidine deamination to uracil (hypermutation); inhibition of reverse transcriptase. | [53] |
| Tetherin (BST2) | Blocks release of HIV-1 virions from cell surface | [53] |
| SAMHD1 | Depletes dNTPs, restricting reverse transcription | [53] |
| MX2 (MXB) | Inhibits nuclear import of HIV-1 pre-integration complex | [53] |
| GBP5 | Inhibits HIV-1 envelope processing and infectivity | [53] |
| Schlafen 11 (SLFN11) | Inhibits HIV-1 protein synthesis by tRNA restriction | [53] |
| TRIM56 | Enhances ISG induction, inhibits late HIV-1 gene expression | [54] |
| IDO1 | Depletes tryptophan, suppressing HIV-1 replication | [54] |
| IRF-1 | Transcription factor, suppresses HIV-1 LTR-driven gene expression | [54] |
| ISG15 | Ubiquitin-like modifier, modulates immune signaling and restricts HIV-1 | [12] |
| Aspect | Description | Cell Types Involved | References |
|---|---|---|---|
| STAT5 in HIV-1 infected cells | Increased STAT5 phosphorylation following HIV-1 exposure in vitro. | CD4+ T cells, monocytes. | [57,94] |
| Altered monocyte/macrophage function | Impaired GM-CSF-induced STAT5 phosphorylation and enhanced MAPK signaling contribute to defective antigen presentation. | Monocytes and macrophages. | [95] |
| Impaired cytokine responsiveness | Reduced STAT5 phosphorylation in response to IL-2 (CD8+ T cells) and GM-CSF (macrophages). | CD8+ T cells, macrophages. | [105,106] |
| Role in viral replication | Full-length STAT5, activated by IL-2, IL-7, or IL-15, enhances HIV-1 LTR transcription and viral protein production (e.g., p24+ cells). | CD4+ T cells. | [107] |
| STAT5Δ (truncated isoform) | Constitutively active; binds the HIV-1 LTR and inhibits viral transcription by blocking RNA polymerase II recruitment. | Myeloid cells, monocytes. | [94,108,109] |
| Disrupted IL-7 signaling | Hyperphosphorylation of STAT5 at S726 and Y694, but defective nuclear translocation; correlates with elevated HLA-DR expression. | CD4+ T cells. | [110] |
| Insertional activation of STAT5B | HIV-1 integration in STAT5B and BACH2 driving clonal expansion. | Treg cells, central memory T cells. | [111,112,113,114] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tolomeo, M.; Cascio, A. The STAT Signaling Pathway in HIV-1 Infection: Roles and Dysregulation. Int. J. Mol. Sci. 2025, 26, 9123. https://doi.org/10.3390/ijms26189123
Tolomeo M, Cascio A. The STAT Signaling Pathway in HIV-1 Infection: Roles and Dysregulation. International Journal of Molecular Sciences. 2025; 26(18):9123. https://doi.org/10.3390/ijms26189123
Chicago/Turabian StyleTolomeo, Manlio, and Antonio Cascio. 2025. "The STAT Signaling Pathway in HIV-1 Infection: Roles and Dysregulation" International Journal of Molecular Sciences 26, no. 18: 9123. https://doi.org/10.3390/ijms26189123
APA StyleTolomeo, M., & Cascio, A. (2025). The STAT Signaling Pathway in HIV-1 Infection: Roles and Dysregulation. International Journal of Molecular Sciences, 26(18), 9123. https://doi.org/10.3390/ijms26189123







