Distinct Oxidative Stress Adaptations Driven by the Overexpression of miR-526b, miR-655, and COX-2 in Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Cell Viability in miRNA Overexpressing Breast Cancer
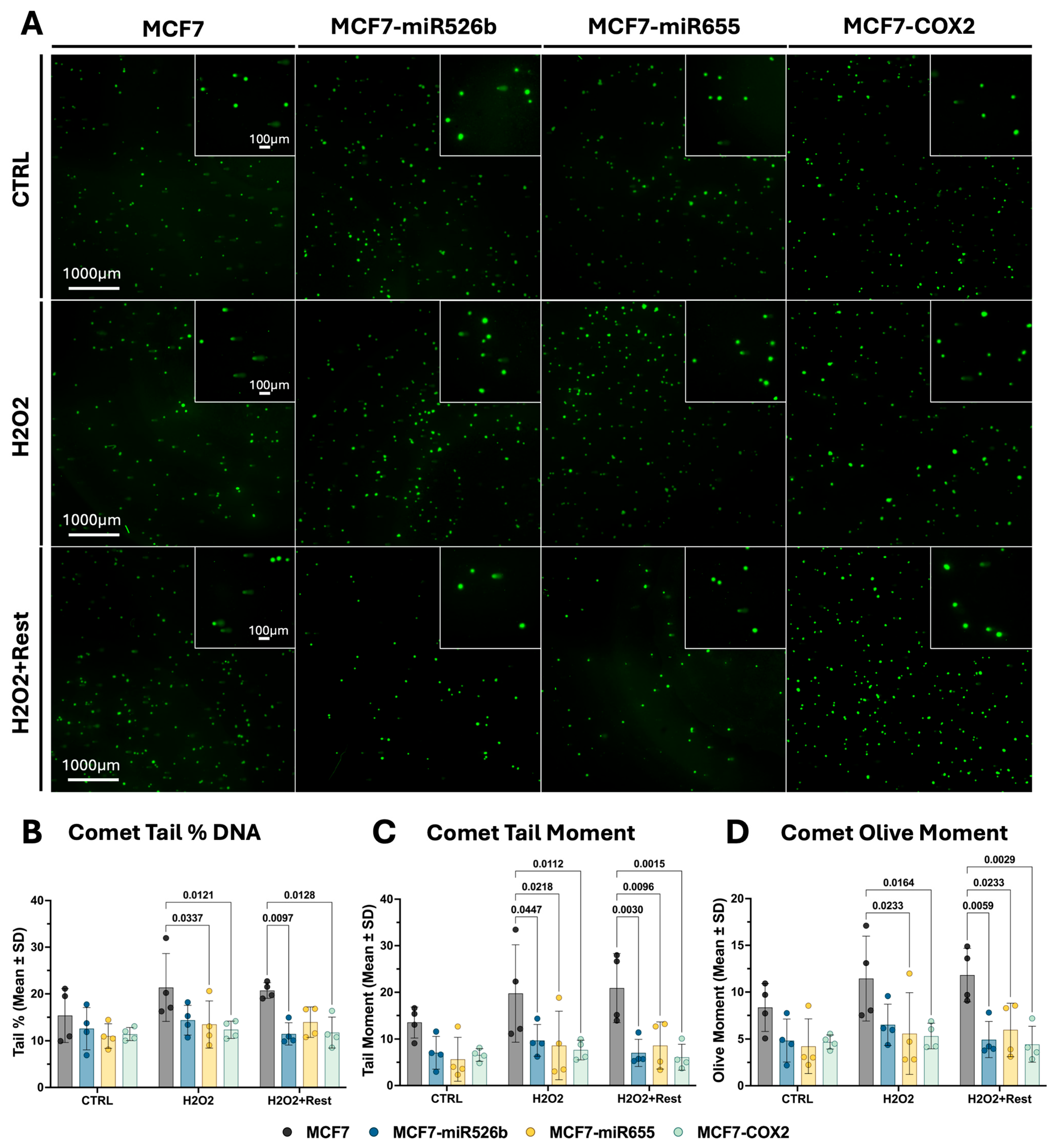
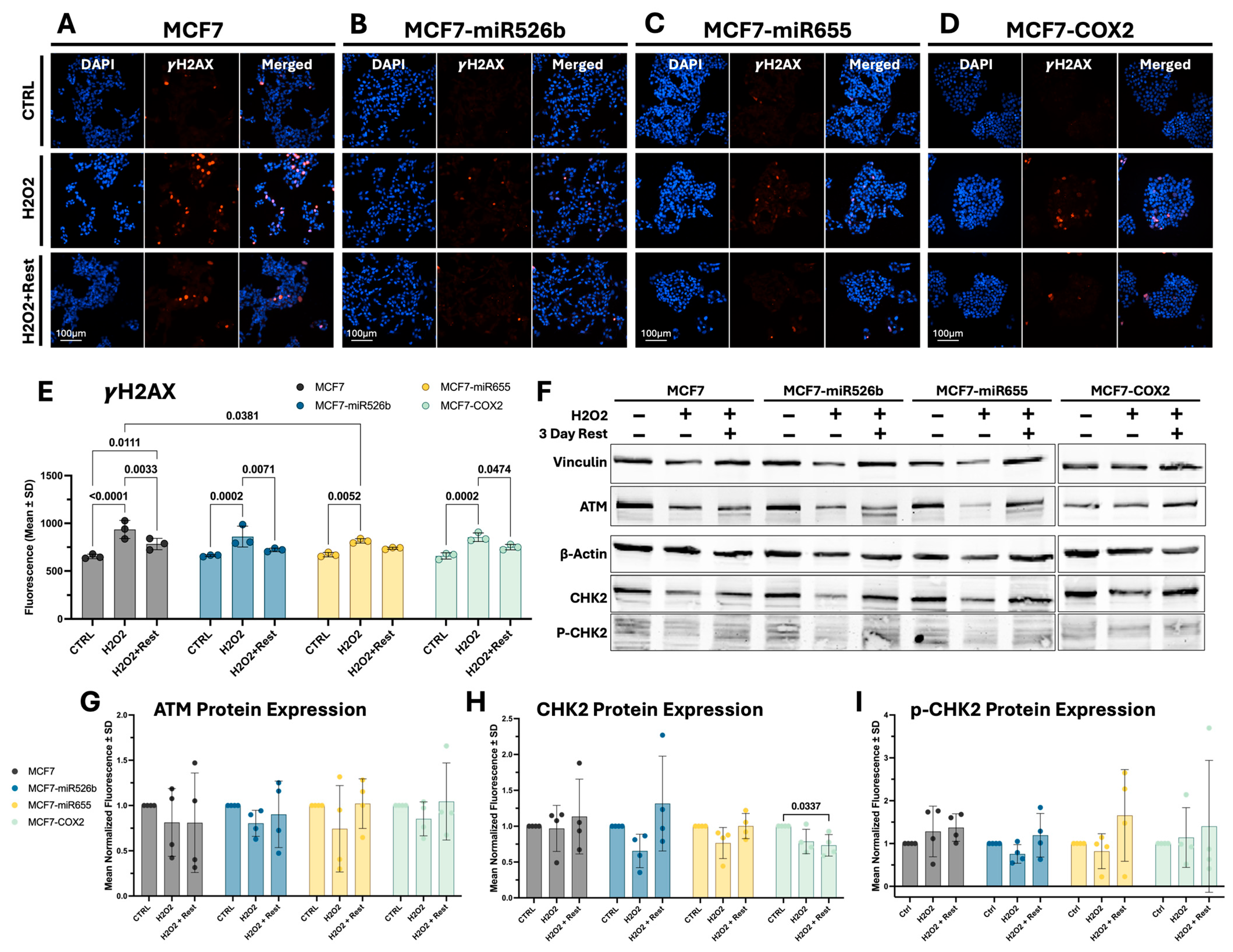
2.2. Assessing Oxidative-Stress-Induced DNA Damage and Repair
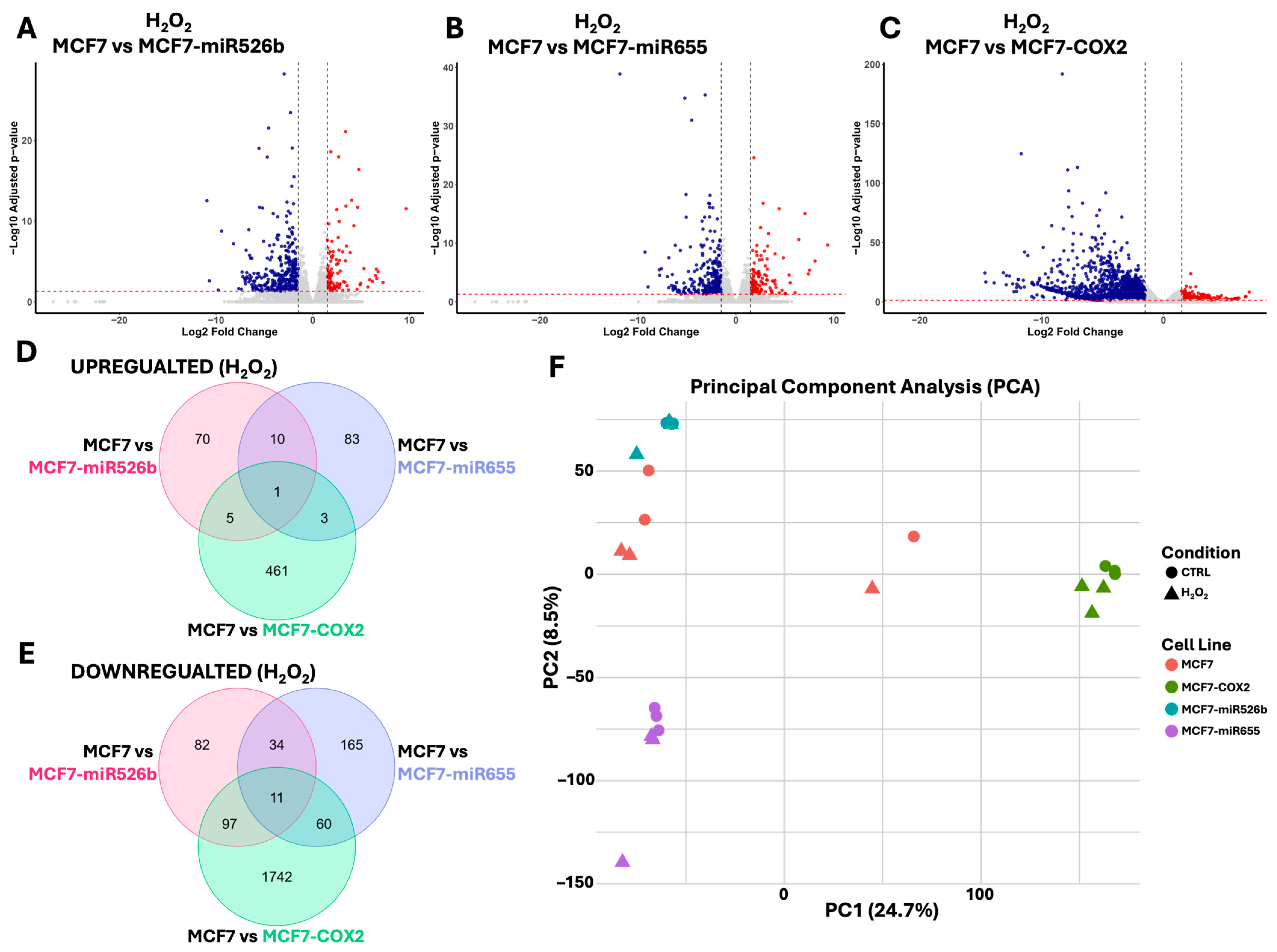
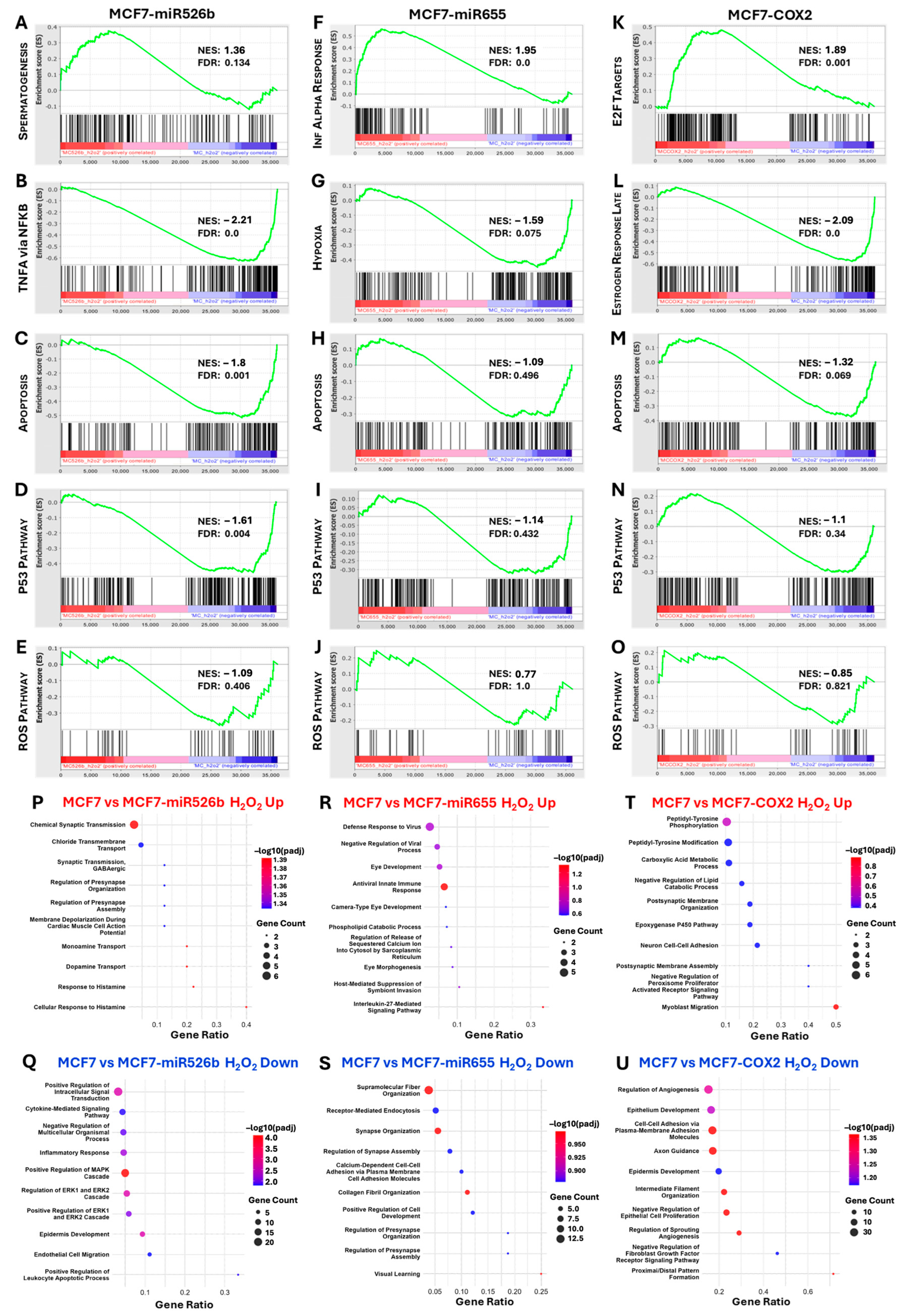
2.3. Identification of DEGs Associated with miRNA Overexpression via RNA-Sequencing
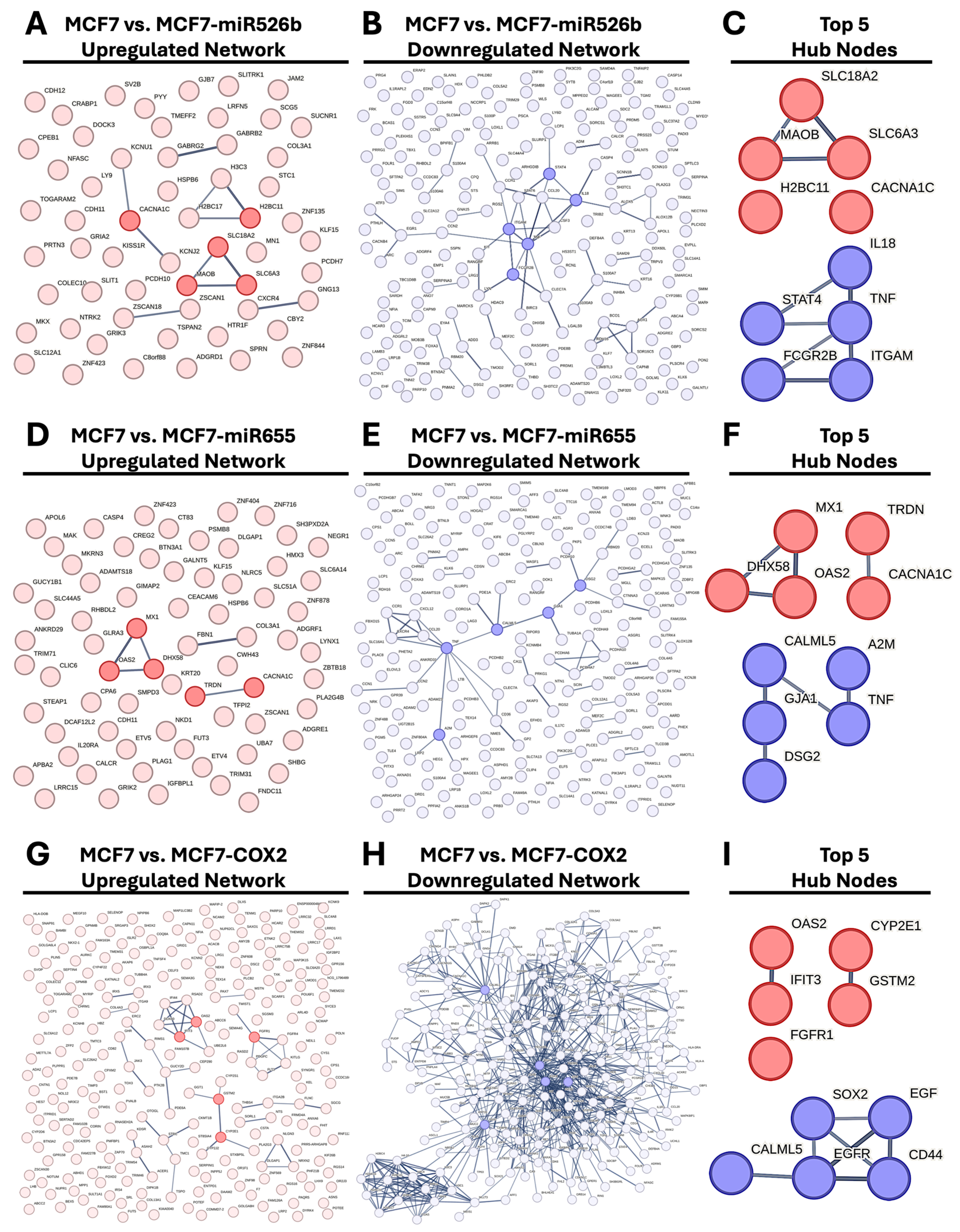
2.4. Protein–Protein Interaction (PPI) Network Construction and Hub Gene Analysis
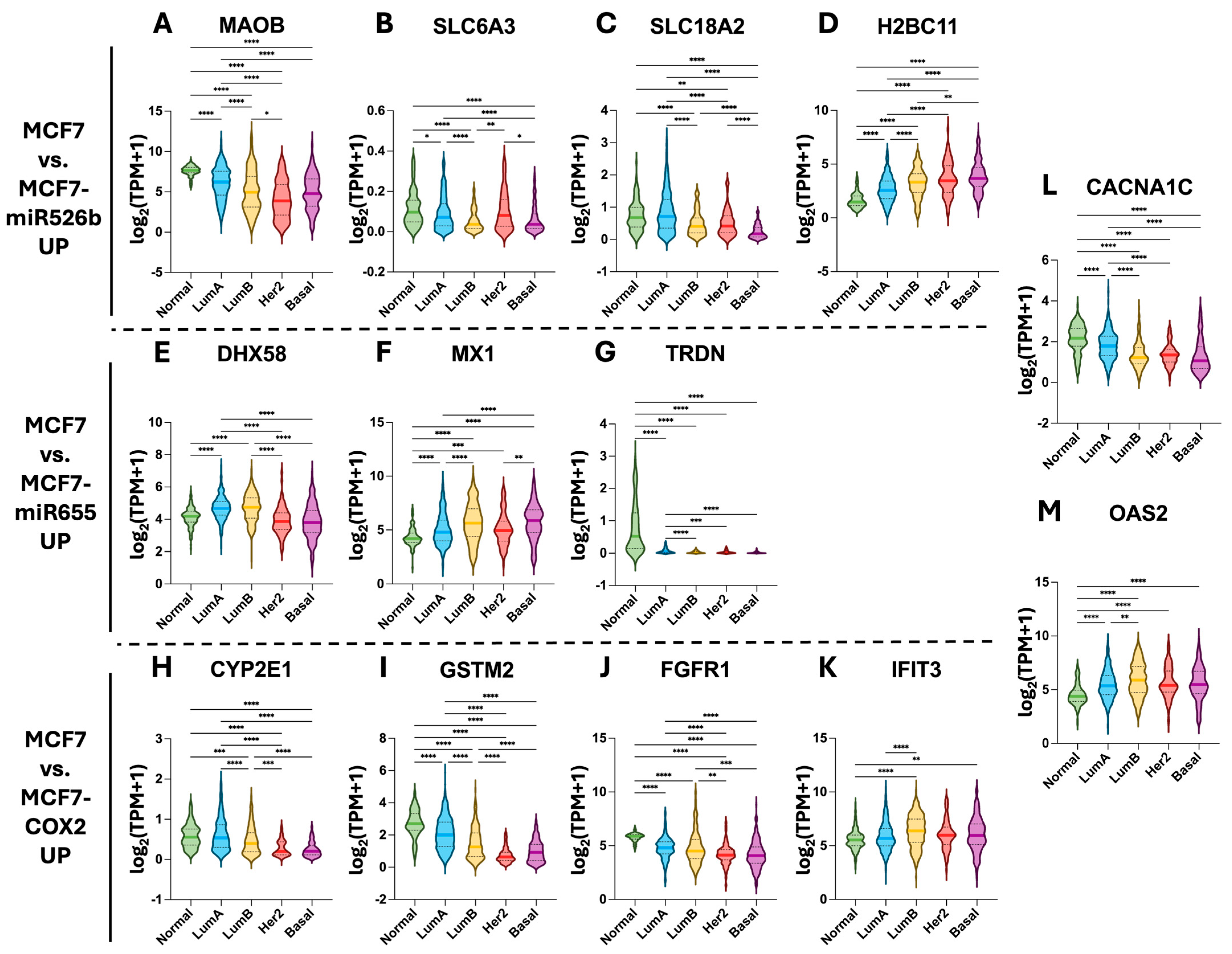
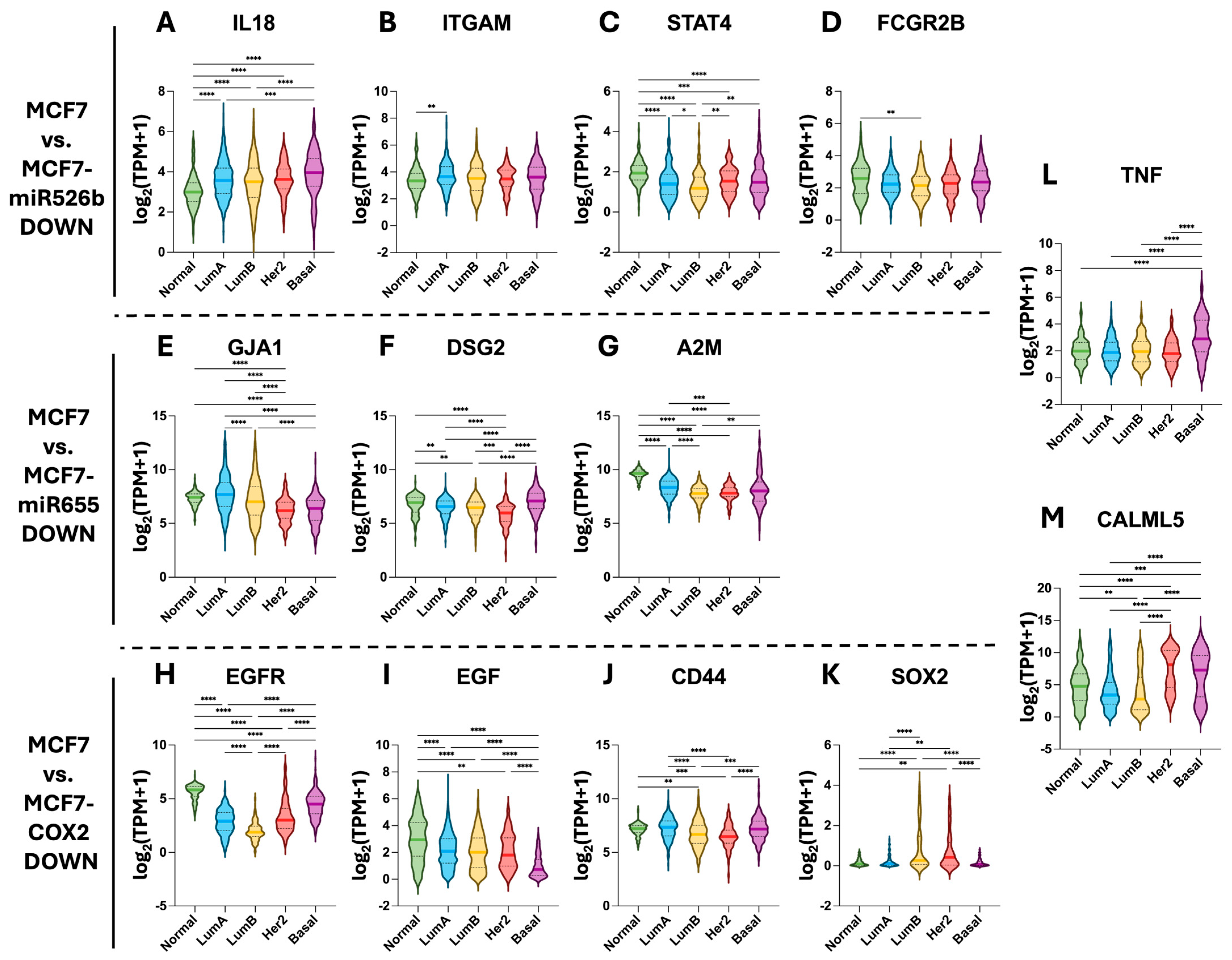
2.5. Hub Gene Expression in TCGA-BRCA Tissue Samples
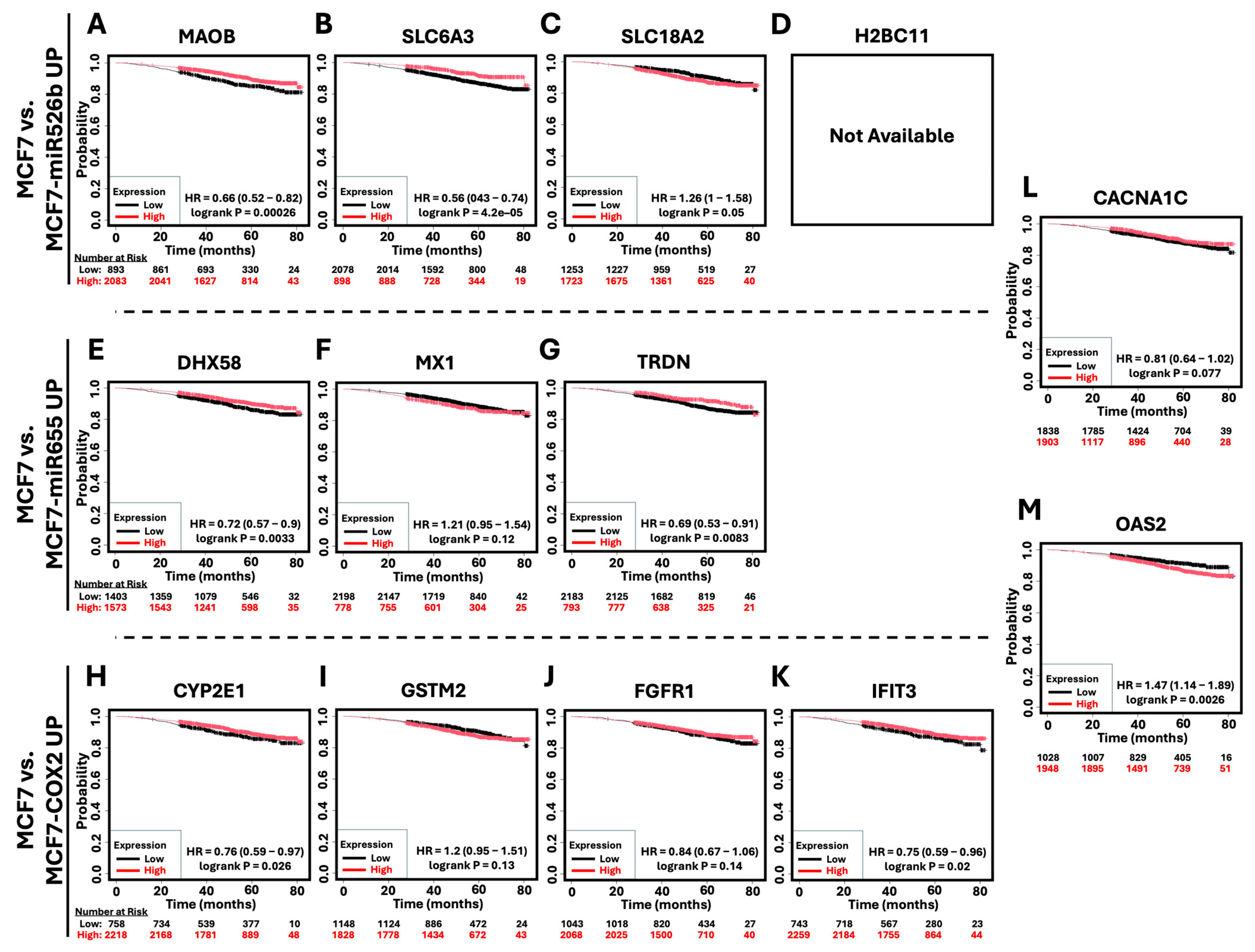
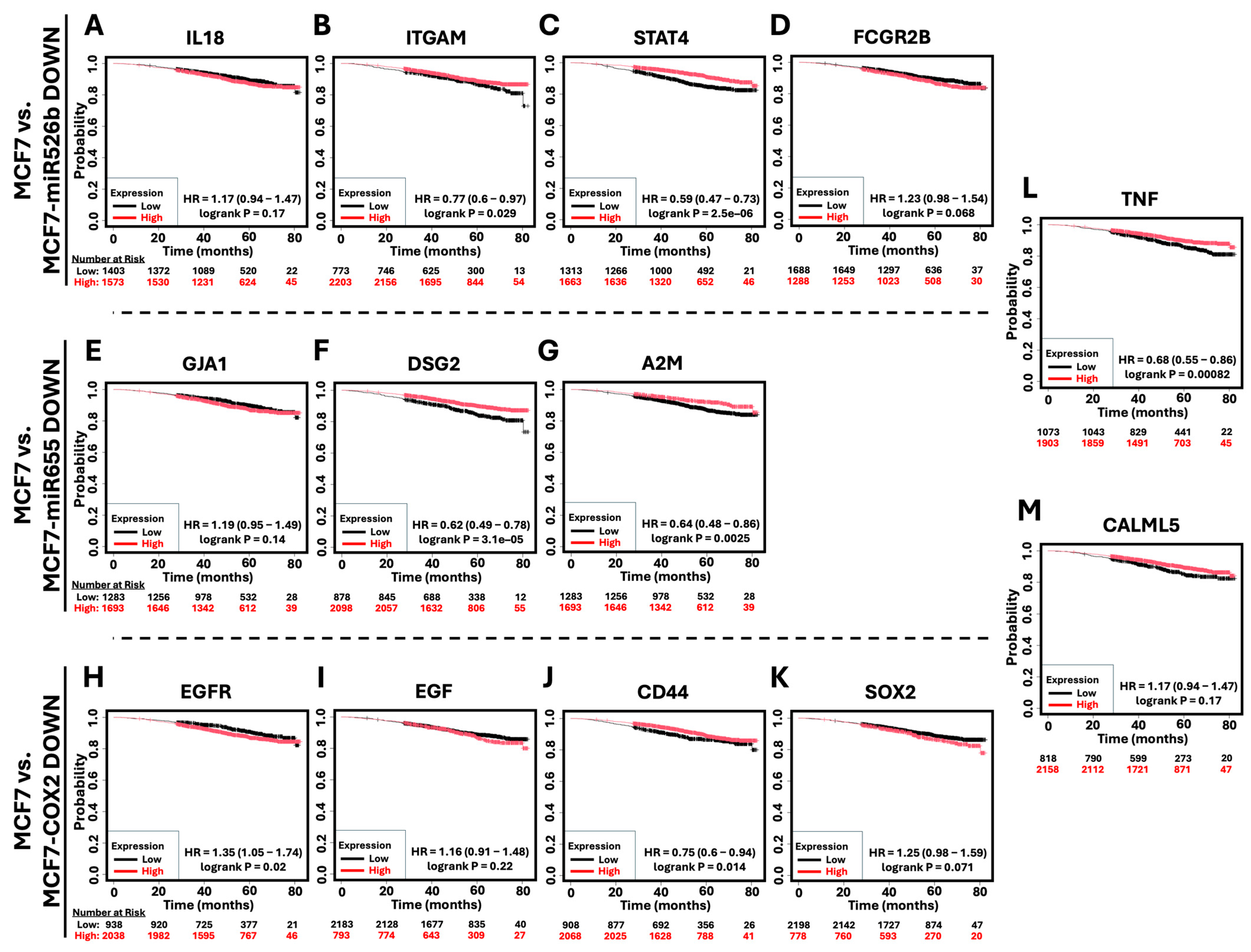
2.6. Survival Analysis of Hub Genes in Breast Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Hydrogen Peroxide Treatment
4.3. Cell Viability
4.4. RNA Extraction, cDNA Synthesis, and Real-Time qPCR
4.5. SDS-PAGE and Western Blotting
4.6. Single-Cell Gel Electrophoresis (Comet Assay)
4.7. Immunocytochemistry (ICC)
4.8. RNA-Sequencing and High-Throughput Data Pipeline
4.9. Gene Set Enrichment Analysis (GSEA)
4.10. Analysis of Enriched Biological Processes
4.11. Network and Hub Gene Analysis
4.12. Kaplan–Meier (KM) Survival Analysis
4.13. Accession of the Cancer Genome Atlas (TCGA)—Breast Cancer (BRCA) Patient Data
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | Beta Actin |
| ANOVA | Analysis of Variance |
| ATM | Ataxia Telangiectasia Mutated |
| BCRA | Breast Cancer (TCGA) |
| BP(s) | Biological Processes |
| BSA | Bovine Serum Albumin |
| CACNA1C | Calcium Voltage-Gated Channel Subunit Alpha1 C |
| CALML5 | Calmodulin Like 5 |
| CAT | Catalase |
| CD44 | Cluster of Differentiation 44 |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A (p21) |
| cDNA | Complementary DNA |
| CHK2/Chk2 | Checkpoint Kinase 2 |
| COX-2 | Cyclooxygenase-2 |
| DAPI | 4′,6-Diamidino-2-Phenylindole |
| DEG(s) | Differentially Expressed Gene(s) |
| DESeq2 | Differential Expression Sequencing 2 |
| DHX58 | DExH-Box Helicase 58 |
| DMSO | Dimethyl Sulfoxide |
| DSG2 | Desmoglein 2 |
| EGFR | Epidermal Growth Factor Receptor |
| EP4 | Prostaglandin E2 Receptor 4 |
| FDR | False Discovery Rate |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| GPX2 | Glutathione Peroxidase 2 |
| GSEA | Gene Set Enrichment Analysis |
| GSR | Glutathione-Disulfide Reductase |
| H2O2 | Hydrogen Peroxide |
| HTSeq | High-Throughput Sequencing |
| IC50 | Half-Maximal Inhibitory Concentration |
| IFN-γ | Interferon Gamma |
| IL18 | Interleukin 18 |
| KM | Kaplan–Meier |
| MAOB | Monoamine Oxidase B |
| MAPK | Mitogen-Activated Protein Kinase |
| MCF7 | Michigan Cancer Foundation 7 |
| miRNA | MicroRNA |
| MSigDB | Molecular Signatures Database |
| NES | Normalized Enrichment Score |
| NIR | Near-Infrared |
| OAS2 | 2′-5′-Oligoadenylate Synthetase 2 |
| PAM50 | Prediction Analysis of Microarray 50 |
| PARP | Poly (ADP-ribose) Polymerase |
| PBS(T) | Phosphate-Buffered Saline (Tween-20) |
| PI3K | Phosphoinositide 3-kinase |
| PUMA | P53 Upregulated Modulator of Apoptosis |
| qRT-PCR | Quantitative Real-Time Polymerase Chain Reaction |
| RNA-seq | RNA-sequencing |
| ROS | Reactive Oxygen Species |
| RPLP5 | Ribosomal Protein L5 |
| RPMI | Roswell Park Memorial Institute |
| SDS-PAGE | Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis |
| SLC6A3 | Solute Carrier Family 6 Member 3 |
| SO | Super Oxide |
| SOX2 | SRY-Box Transcription Factor 2 |
| STAR | Spliced Transcripts Alignment to a Reference |
| TCGA | The Cancer Genome Atlas |
| TPM | Transcripts Per Million |
| Tuba1 | Tubulin Alpha 1 |
| γ-H2AX | (Serine 139) Phosphorylated H2A Histone Family Member X |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Gandhi, S.; Yan, L.; Tokumaru, Y.; Wu, R.; Yamada, A.; Matsuyama, R.; Endo, I.; Takabe, K. Abundance of reactive oxygen species (ROS) is associated with tumor aggressiveness, immune response, and worse survival in breast cancer. Breast Cancer Res. Treat. 2022, 194, 231–241. [Google Scholar] [CrossRef]
- Zou, J.; Jiang, C.; Hu, Q.; Jia, X.; Wang, S.; Wan, S.; Mao, Y.; Zhang, D.; Zhang, P.; Dai, B.; et al. Tumor microenvironment-responsive engineered hybrid nanomedicine for photodynamic-immunotherapy via multi-pronged amplification of reactive oxygen species. Nat. Commun. 2025, 16, 424. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Zheng, Z.; Su, J.; Bao, X.; Wang, H.; Bian, C.; Zhao, Q.; Jiang, X. Mechanisms and applications of radiation-induced oxidative stress in regulating cancer immunotherapy. Front. Immunol. 2023, 14, 1247268. [Google Scholar] [CrossRef]
- van der Zanden, S.Y.; Qiao, X.; Neefjes, J. New insights into the activities and toxicities of the old anticancer drug doxorubicin. FEBS J. 2021, 288, 6095–6111. [Google Scholar] [CrossRef]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; Degraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef]
- He, J.; Jiang, B.H. Interplay between Reactive oxygen Species and MicroRNAs in Cancer. Curr. Pharmacol. Rep. 2016, 2, 82–90. [Google Scholar] [CrossRef]
- Shin, B.; Feser, R.; Nault, B.; Hunter, S.; Maiti, S.; Ugwuagbo, K.C.; Majumder, M. miR526b and miR655 Induce Oxidative Stress in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4039. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Dunn, L.; Liu, L.; Hasan, A.; Vincent, K.; Brackstone, M.; Hess, D.; Lala, P.K. COX-2 induces oncogenic micro RNA miR655 in human breast cancer. Sci. Rep. 2018, 8, 327. [Google Scholar] [CrossRef]
- Majumder, M.; Landman, E.; Liu, L.; Hess, D.; Lala, P.K. COX-2 Elevates Oncogenic miR-526b in Breast Cancer by EP4 Activation. Mol. Cancer Res. 2015, 13, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.M.; Nieto, N. CYP2E1 and oxidant stress in alcoholic and non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 395–398. [Google Scholar] [CrossRef]
- Formisano, L.; Lu, Y.; Servetto, A.; Hanker, A.B.; Jansen, V.M.; Bauer, J.A.; Sudhan, D.R.; Guerrero-Zotano, A.L.; Croessmann, S.; Guo, Y.; et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat. Commun. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Li, C.; Liu, J.; He, D.; Mao, F.; Rao, X.; Zhao, Y.; Lanman, N.A.; Kazemian, M.; Farah, E.; Liu, J.; et al. GSTM2 is a key molecular determinant of resistance to SG-ARIs. Oncogene 2022, 41, 4498–4511. [Google Scholar] [CrossRef]
- Ho, W.J.; Law, A.M.K.; Masle-Farquhar, E.; Castillo, L.E.; Mawson, A.; O’Bryan, M.K.; Goodnow, C.C.; Gallego-Ortega, D.; Oakes, S.R.; Ormandy, C.J. Activation of the viral sensor oligoadenylate synthetase 2 (Oas2) prevents pregnancy-driven mammary cancer metastases. Breast Cancer Res. 2022, 24, 31. [Google Scholar] [CrossRef]
- Lu, J.; Yang, L.; Yang, X.; Chen, B.; Liu, Z. Investigating the clinical significance of OAS family genes in breast cancer: An in vitro and in silico study. Hereditas 2024, 161, 50. [Google Scholar] [CrossRef]
- Majumder, M.; Xin, X.; Liu, L.; Tutunea-Fatan, E.; Rodriguez-Torres, M.; Vincent, K.; Postovit, L.M.; Hess, D.; Lala, P.K. COX-2 Induces Breast Cancer Stem Cells via EP4/PI3K/AKT/NOTCH/WNT Axis. Stem Cells 2016, 34, 2290–2305. [Google Scholar] [CrossRef]
- Yusupov, M.; Privat-Maldonado, A.; Cordeiro, R.M.; Verswyvel, H.; Shaw, P.; Razzokov, J.; Smits, E.; Bogaerts, A. Oxidative damage to hyaluronan-CD44 interactions as an underlying mechanism of action of oxidative stress-inducing cancer therapy. Redox Biol. 2021, 43, 101968. [Google Scholar] [CrossRef]
- Ando, T.; Yamasaki, J.; Saya, H.; Nagano, O. CD44: A key regulator of iron metabolism, redox balance, and therapeutic resistance in cancer stem cells. Stem Cells 2025, 43, sxaf024. [Google Scholar] [CrossRef] [PubMed]
- Khan, E.M.; Heidinger, J.M.; Levy, M.; Lisanti, M.P.; Ravid, T.; Goldkorn, T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J. Biol. Chem. 2006, 281, 14486–14493. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pennock, S.; Chen, X.; Wang, Z. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell Biol. 2002, 22, 7279–7290. [Google Scholar] [CrossRef]
- Wu, Q.; Zhen, Y.; Shi, L.; Vu, P.; Greninger, P.; Adil, R.; Merritt, J.; Egan, R.; Wu, M.J.; Yin, X.; et al. EGFR Inhibition Potentiates FGFR Inhibitor Therapy and Overcomes Resistance in FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2022, 12, 1378–1395. [Google Scholar] [CrossRef]
- Holdman, X.B.; Welte, T.; Rajapakshe, K.; Pond, A.; Coarfa, C.; Mo, Q.; Huang, S.; Hilsenbeck, S.G.; Edwards, D.P.; Zhang, X.; et al. Upregulation of EGFR signaling is correlated with tumor stroma remodeling and tumor recurrence in FGFR1-driven breast cancer. Breast Cancer Res. 2015, 17, 141. [Google Scholar] [CrossRef]
- Chibaya, L.; Karim, B.; Zhang, H.; Jones, S.N. Mdm2 phosphorylation by Akt regulates the p53 response to oxidative stress to promote cell proliferation and tumorigenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2003193118. [Google Scholar] [CrossRef]
- Beucher, L.; Gabillard-Lefort, C.; Baris, O.R.; Mialet-Perez, J. Monoamine oxidases: A missing link between mitochondria and inflammation in chronic diseases ? Redox Biol. 2024, 77, 103393. [Google Scholar] [CrossRef]
- Delierneux, C.; Kouba, S.; Shanmughapriya, S.; Potier-Cartereau, M.; Trebak, M.; Hempel, N. Mitochondrial Calcium Regulation of Redox Signaling in Cancer. Cells 2020, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Boveris, A.; Cadenas, E. Mitochondrial energy metabolism and redox signaling in brain aging and neurodegeneration. Antioxid. Redox Signal 2014, 20, 353–371. [Google Scholar] [CrossRef] [PubMed]
- Alsaad, I.; Abdel Rahman, D.M.A.; Al-Tamimi, O.; Alhaj, S.A.; Sabbah, D.A.; Hajjo, R.; Bardaweel, S.K. Targeting MAO-B with Small-Molecule Inhibitors: A Decade of Advances in Anticancer Research (2012–2024). Molecules 2024, 30, 126. [Google Scholar] [CrossRef]
- Nault, B.D.; Majumder, M. miR-526b enhances glucose metabolism in breast cancer cells, an effect reversed by targeting the COX-2/EP4 pathway. Mol. Biol. Rep. 2025, 52, 351. [Google Scholar] [CrossRef] [PubMed]
- Gervin, E.; Shin, B.; Opperman, R.; Cullen, M.; Feser, R.; Maiti, S.; Majumder, M. Chemically Induced Hypoxia Enhances miRNA Functions in Breast Cancer. Cancers 2020, 12, 2008. [Google Scholar] [CrossRef]
- Zhang, Z.; Lei, A.; Xu, L.; Chen, L.; Chen, Y.; Zhang, X.; Gao, Y.; Yang, X.; Zhang, M.; Cao, Y. Similarity in gene-regulatory networks suggests that cancer cells share characteristics of embryonic neural cells. J. Biol. Chem. 2017, 292, 12842–12859. [Google Scholar] [CrossRef]
- Ma, N.; Wang, Y.; Li, X.; Xu, M.; Tan, D. Reactive oxygen species in cancer: Mechanistic insights and therapeutic innovations. Cell Stress. Chaperones 2025, 30, 100108. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.V. OpenComet: An automated tool for comet assay image analysis. Redox Biol. 2014, 2, 457–465, Erratum in Redox Biol. 2021, 40, 101876. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 July 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis, 2nd edition. Meas-Interdiscip. Res. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Gyorffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Opperman, R.M.; Maiti, S.; Majumder, M. Distinct Oxidative Stress Adaptations Driven by the Overexpression of miR-526b, miR-655, and COX-2 in Breast Cancer. Int. J. Mol. Sci. 2025, 26, 9103. https://doi.org/10.3390/ijms26189103
Opperman RM, Maiti S, Majumder M. Distinct Oxidative Stress Adaptations Driven by the Overexpression of miR-526b, miR-655, and COX-2 in Breast Cancer. International Journal of Molecular Sciences. 2025; 26(18):9103. https://doi.org/10.3390/ijms26189103
Chicago/Turabian StyleOpperman, Reid M., Sujit Maiti, and Mousumi Majumder. 2025. "Distinct Oxidative Stress Adaptations Driven by the Overexpression of miR-526b, miR-655, and COX-2 in Breast Cancer" International Journal of Molecular Sciences 26, no. 18: 9103. https://doi.org/10.3390/ijms26189103
APA StyleOpperman, R. M., Maiti, S., & Majumder, M. (2025). Distinct Oxidative Stress Adaptations Driven by the Overexpression of miR-526b, miR-655, and COX-2 in Breast Cancer. International Journal of Molecular Sciences, 26(18), 9103. https://doi.org/10.3390/ijms26189103






