Proteomic Analysis Uncovers Enhanced Inflammatory Phenotype and Distinct Metabolic Changes in IDH1 Mutant Glioma Cells
Abstract
1. Introduction
2. Results and Discussion
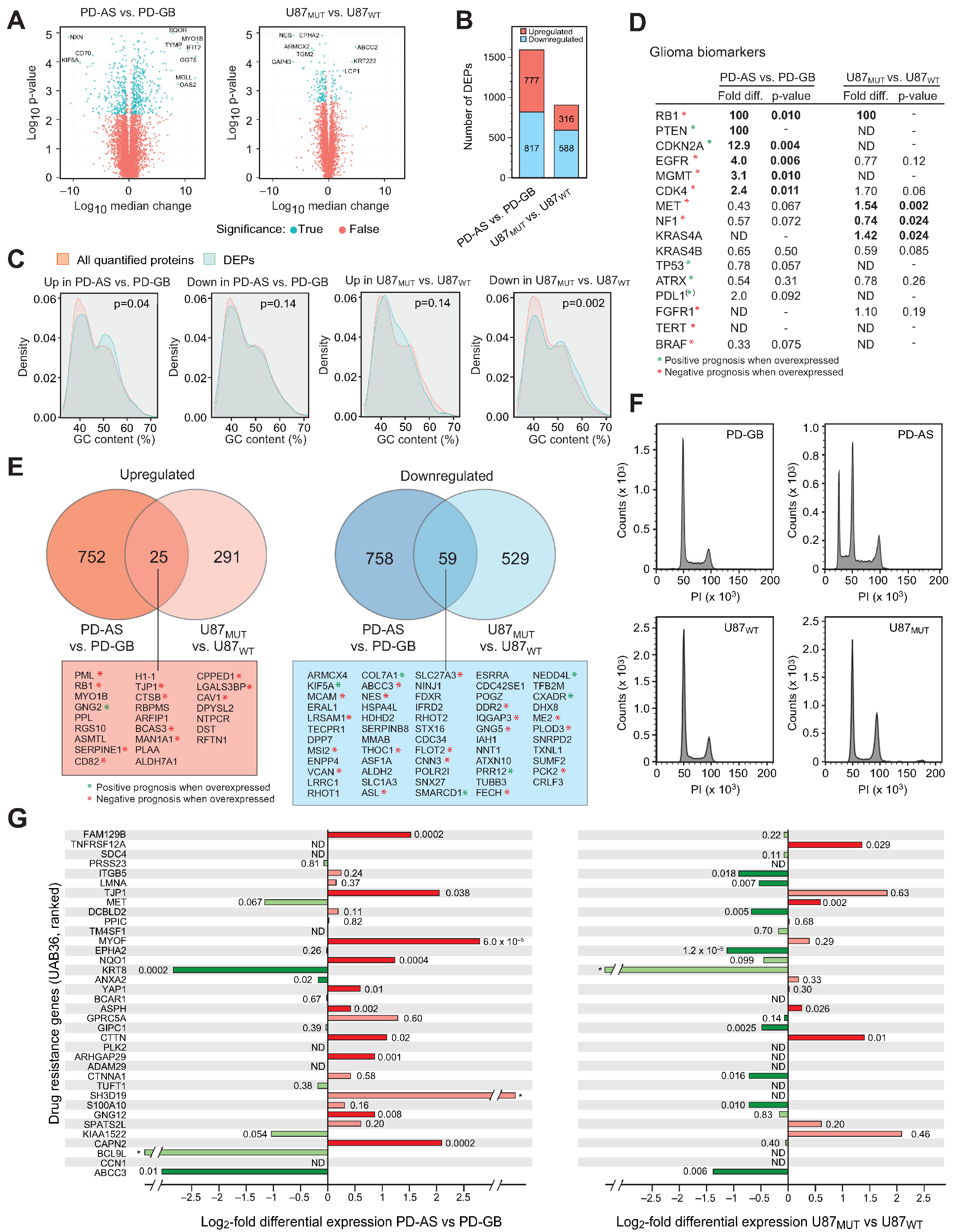
2.1. Common DEPs in IDH1 Mutant Versus IDH1 Wild-Type Glioma Cells Are Associated with Tumor Aggressiveness and Response to Treatment
2.2. IDH Mutant Glioma Cells Display Elevated MHC Antigen Presentation and Interferon Signaling
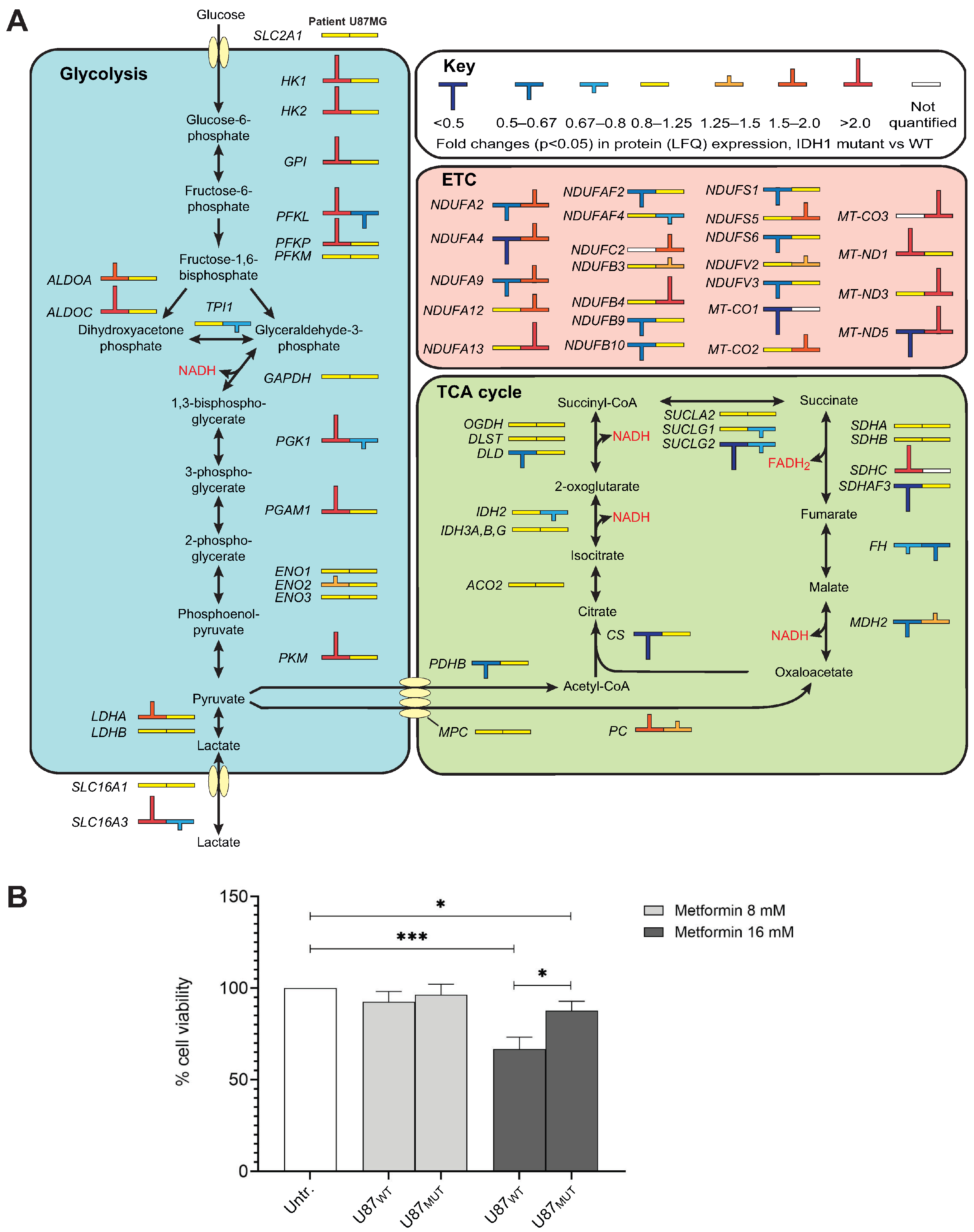
2.3. IDH1 Mutations Are Associated with Distinct Metabolic Shifts Depending on Genetic Background
2.4. DNA Repair Processes Are Differentially Affected in IDH-Mutant Glioma Cells
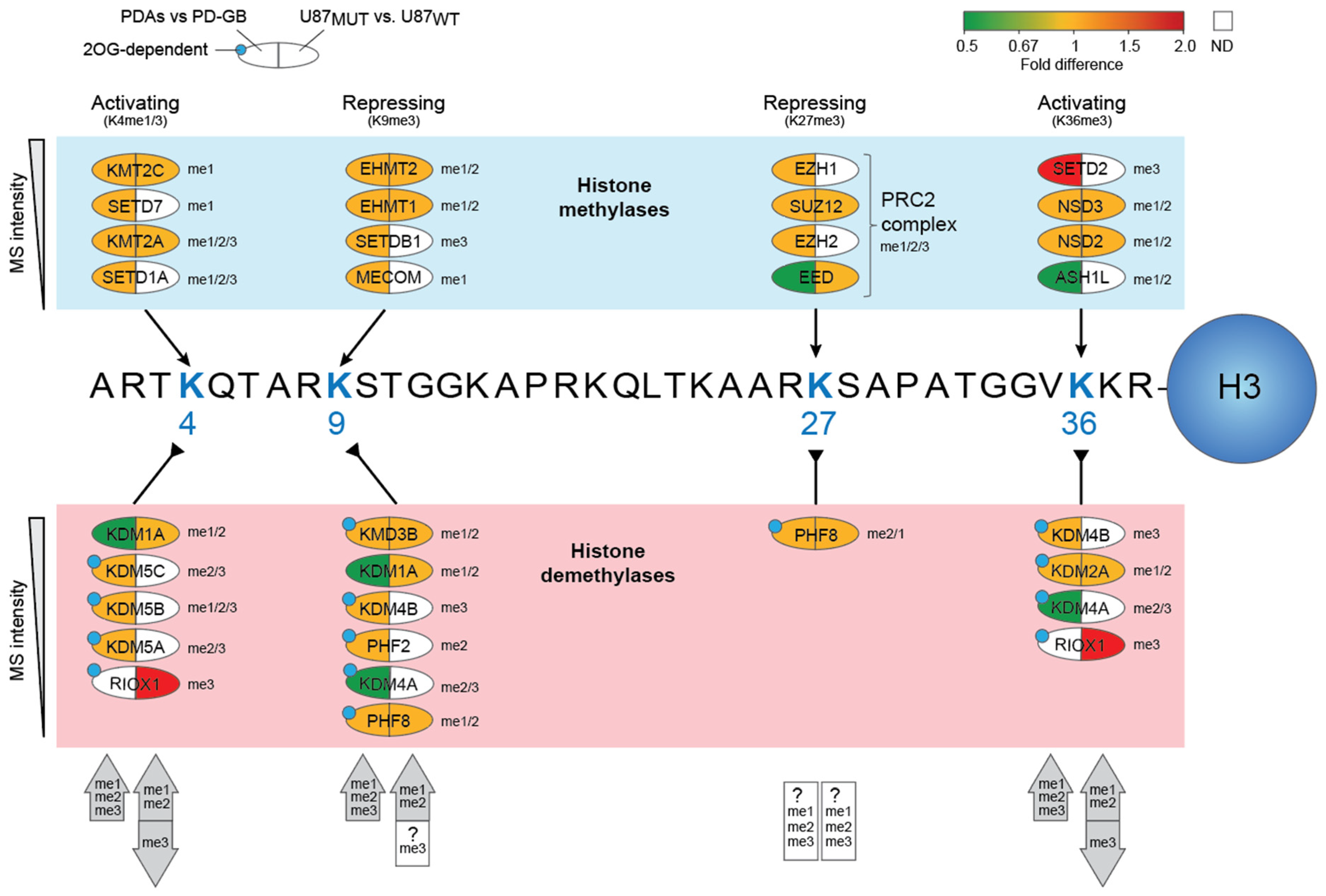
2.5. Potential Contribution of Altered Histone Lysine Methylation to DNA Damage Responses in the IDH-Mutant Cell Models
3. Materials and Methods
3.1. Cell Culture
3.2. Cell Cycle Analyses
3.3. Viability Assay
3.4. LC-MS/MS Analyses
3.5. Bioinformatic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 2OG | α-ketoglutarate, 2-oxoglutarate |
| 2OHG | D-2-hydroxyglutarate |
| DEP | Differentially expressed protein |
| ETC | Electron transport chain |
| GO | Gene ontology |
| HLA | Human leukocyte antigen |
| IDH | Isocitrate dehydrogenase |
| KMT | Lysine methyltransferase |
| KDM | Lysine demethylase |
| LC-MS/MS | Liquid chromatography with tandem mass spectrometry |
| LFQ | Label-free quantitative |
| MHC | Major histocompatibility complex |
| PD-AS | Patient-derived astrocytoma, IDH1 mutant |
| PD-GB | Patient-derived glioblastoma, IDH wild-type |
| TMZ | Temozolomide |
References
- Messaoudi, K.; Clavreul, A.; Lagarce, F. Toward an effective strategy in glioblastoma treatment. Part I: Resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov. Today 2015, 20, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.R.; Wen, P.Y.; Lang-Orsini, M.; Chukwueke, U.N. World Health Organization 2021 Classification of Central Nervous System Tumors and Implications for Therapy for Adult-Type Gliomas: A Review. JAMA Oncol. 2022, 8, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Mohile, N.A.; Messersmith, H.; Gatson, N.T.; Hottinger, A.F.; Lassman, A.; Morton, J.; Ney, D.; Nghiemphu, P.L.; Olar, A.; Olson, J.; et al. Therapy for Diffuse Astrocytic and Oligodendroglial Tumors in Adults: ASCO-SNO Guideline. J. Clin. Oncol. 2022, 40, 403–426. [Google Scholar] [CrossRef]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Tesileanu, C.M.S.; Vallentgoed, W.R.; Sanson, M.; Taal, W.; Clement, P.M.; Wick, W.; Brandes, A.A.; Baurain, J.F.; Chinot, O.L.; Wheeler, H.; et al. Non-IDH1-R132H IDH1/2 mutations are associated with increased DNA methylation and improved survival in astrocytomas, compared to IDH1-R132H mutations. Acta Neuropathol. 2021, 141, 945–957. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Curry, R.N.; Glasgow, S.M. The Role of Neurodevelopmental Pathways in Brain Tumors. Front. Cell Dev. Biol. 2021, 9, 659055. [Google Scholar] [CrossRef]
- Kolodziejczak-Guglas, I.; Simoes, R.L.S.; de Souza Santos, E.; Demicco, E.G.; Lazcano Segura, R.N.; Ma, W.; Wang, P.; Geffen, Y.; Storrs, E.; Petralia, F.; et al. Proteomic-based stemness score measures oncogenic dedifferentiation and enables the identification of druggable targets. Cell Genom. 2025, 5, 100851. [Google Scholar] [CrossRef]
- Statoulla, E.; Chalkiadaki, K.; Karozis, D.; Gkogkas, C.G. Regulation of mRNA translation in stem cells; links to brain disorders. Cell Signal 2021, 88, 110166. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593, Erratum in Science 2019, 363, eaax1384. https://doi.org/10.1126/science.aax1384. [Google Scholar] [CrossRef]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Lv, W.; Yang, F.; Ge, Z.; Xin, L.; Zhang, L.; Zhai, Y.; Liu, X.; Guo, Q.; Mao, X.; Luo, P.; et al. Aberrant overexpression of myosin 1b in glioblastoma promotes angiogenesis via VEGF-myc-myosin 1b-Piezo1 axis. J. Biol. Chem. 2024, 300, 107807. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, R.; Lyu, Y.; Liu, C.; Liu, Y.; Feng, Y.; Fu, M.; Wong, P.J.C.; Du, Z.; Qiu, T.; et al. Proteomic profiling of gliomas unveils immune and metabolism-driven subtypes with implications for anti-nucleotide metabolism therapy. Nat. Commun. 2024, 15, 10005. [Google Scholar] [CrossRef] [PubMed]
- Dogan, E.; Yildirim, Z.; Akalin, T.; Ozgiray, E.; Akinturk, N.; Aktan, C.; Solmaz, A.E.; Biceroglu, H.; Caliskan, K.E.; Ertan, Y.; et al. Investigating the effects of PTEN mutations on cGAS-STING pathway in glioblastoma tumours. J. Neuro-Oncol. 2024, 166, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Ge, H.; Long, Y.; Yang, C.; Chang, Y.E.; Mu, L.; Sayour, E.J.; De Leon, G.; Wang, Q.J.; Yang, J.C.; et al. CD70, a novel target of CAR T-cell therapy for gliomas. Neuro-Oncology 2018, 20, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, X.; Tang, X.; Yuan, Y.; Wang, W. Lactylation-related gene signatures identify glioma molecular subtypes with prognostic, immunological, and therapeutic implications. Front. Oncol. 2025, 15, 1613423. [Google Scholar] [CrossRef]
- Biserova, K.; Jakovlevs, A.; Uljanovs, R.; Strumfa, I. Cancer Stem Cells: Significance in Origin, Pathogenesis and Treatment of Glioblastoma. Cells 2021, 10, 621. [Google Scholar] [CrossRef]
- Selvaraj, S.; Srinivas, B.H.; Verma, S.K.; Ms, G. Significance of Nestin and CD133 as cancer stem cell markers in diffuse glioma and association with p53 expression and IDH status. Int. J. Clin. Exp. Pathol. 2024, 17, 208–218. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Liu, P.; Yan, X.; Ma, C.; Gu, J.; Tian, F.; Qu, J. Prognostic value of m6A regulators and the nomogram construction in glioma patients. Medicine 2022, 101, e30643. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhong, X.; Xia, M.; Zhong, J. The roles and mechanisms of the m6A reader protein YTHDF1 in tumor biology and human diseases. Mol. Ther. Nucleic Acids 2021, 26, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Elcheva, I.A.; Gowda, C.P.; Bogush, D.; Gornostaeva, S.; Fakhardo, A.; Sheth, N.; Kokolus, K.M.; Sharma, A.; Dovat, S.; Uzun, Y.; et al. IGF2BP family of RNA-binding proteins regulate innate and adaptive immune responses in cancer cells and tumor microenvironment. Front. Immunol. 2023, 14, 1224516. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Yu, J.; Lai, M.; Zhou, Z.; Zhou, J.; Hu, Q.; Li, J.; Li, H.; Chen, L.; Wen, L.; Zhou, M.; et al. The PTEN-associated immune prognostic signature reveals the landscape of the tumor microenvironment in glioblastoma. J. Neuroimmunol. 2023, 376, 578034. [Google Scholar] [CrossRef] [PubMed]
- Lu, V.M.; O’Connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neuro-Oncol. 2020, 148, 221–229. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT Status as a Clinical Biomarker in Glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Haase, S.; Garcia-Fabiani, M.B.; Carney, S.; Altshuler, D.; Nunez, F.J.; Mendez, F.M.; Nunez, F.; Lowenstein, P.R.; Castro, M.G. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert. Opin. Ther. Targets 2018, 22, 599–613. [Google Scholar] [CrossRef]
- Leeb, M.; Wutz, A. Haploid genomes illustrate epigenetic constraints and gene dosage effects in mammals. Epigenet. Chromatin 2013, 6, 41. [Google Scholar] [CrossRef][Green Version]
- Jentus, M.M.; Bakker, L.; Verstegen, M.; Pelsma, I.; van Wezel, T.; Ruano, D.; Kapiteijn, E.; Crobach, S.; Biermasz, N.; Morreau, H. Chromosomal alteration patterns in PitNETs: Massive losses in aggressive tumors. Endocr. Relat. Cancer 2025, 32, e240070. [Google Scholar] [CrossRef]
- Ghongane, P.; Kapanidou, M.; Asghar, A.; Elowe, S.; Bolanos-Garcia, V.M. The dynamic protein Knl1—A kinetochore rendezvous. J. Cell Sci. 2014, 127, 3415–3423. [Google Scholar] [CrossRef] [PubMed]
- Polley, S.; Raisch, T.; Ghetti, S.; Korner, M.; Terbeck, M.; Grater, F.; Raunser, S.; Aponte-Santamaria, C.; Vetter, I.R.; Musacchio, A. Structure of the human KMN complex and implications for regulation of its assembly. Nat. Struct. Mol. Biol. 2024, 31, 861–873. [Google Scholar] [CrossRef]
- Fellows, B.J.; Tolezano, G.C.; Pires, S.F.; Ruegg, M.S.G.; Knapp, K.M.; Krepischi, A.C.V.; Bicknell, L.S. A novel KNL1 intronic splicing variant likely destabilizes the KMN complex, causing primary microcephaly. Am. J. Med. Genet. A 2024, 194, e63468. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, K.; Zhao, S.; Li, Y.; Qiu, W.; Zhu, C.; Wang, Y.; Dong, C.; Liu, J.; Lu, Y.; et al. Role of Kinetochore Scaffold 1 (KNL1) in Tumorigenesis and Tumor Immune Microenvironment in Pan-Cancer: Bioinformatics Analyses and Validation of Expression. Int. J. Gen. Med. 2023, 16, 4883–4906. [Google Scholar] [CrossRef]
- Sahin, U.; Koslowski, M.; Tureci, O.; Eberle, T.; Zwick, C.; Romeike, B.; Moringlane, J.R.; Schwechheimer, K.; Feiden, W.; Pfreundschuh, M. Expression of cancer testis genes in human brain tumors. Clin. Cancer Res. 2000, 6, 3916–3922. [Google Scholar] [PubMed]
- Sahu, D.; Shi, J.; Segura Rueda, I.A.; Chatrath, A.; Dutta, A. Development of a polygenic score predicting drug resistance and patient outcome in breast cancer. npj Precis. Oncol. 2024, 8, 219. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Hou, W.; Wang, F.; Cheng, W.; Qiu, F.; Wu, P.; Zhang, G. Identification of Immune-Related lncRNA Prognostic Signature and Molecular Subtypes for Glioblastoma. Front. Immunol. 2021, 12, 706936. [Google Scholar] [CrossRef]
- Shen, L.; Shen, H.; Wang, T.; Chen, G.; Yu, Z.; Liu, F. Analysis of ABCC3 in glioma progression: Implications for prognosis, immunotherapy, and drug resistance. Discov. Oncol. 2025, 16, 179. [Google Scholar] [CrossRef]
- Zagzag, D.; Salnikow, K.; Chiriboga, L.; Yee, H.; Lan, L.; Ali, M.A.; Garcia, R.; Demaria, S.; Newcomb, E.W. Downregulation of major histocompatibility complex antigens in invading glioma cells: Stealth invasion of the brain. Lab. Investig. 2005, 85, 328–341. [Google Scholar] [CrossRef]
- Zhao, B.; Meng, L.Q.; Huang, H.N.; Pan, Y.; Xu, Q.Q. A novel functional polymorphism, 16974 A/C, in the interleukin-12-3′ untranslated region is associated with risk of glioma. DNA Cell Biol. 2009, 28, 335–341. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Chen, K.; Shan, D.; Cui, G.; Yuan, W.; Lin, Q.; Gimple, R.C.; Dixit, D.; Lu, C.; et al. IFI35 regulates non-canonical NF-kappaB signaling to maintain glioblastoma stem cells and recruit tumor-associated macrophages. Cell Death Differ. 2024, 31, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guan, Q.; Nan, Y.; Ma, Q.; Zhong, Y. Overexpression of human MX2 gene suppresses cell proliferation, migration, and invasion via ERK/P38/NF-kappaB pathway in glioblastoma cells. J. Cell. Biochem. 2019, 120, 18762–18770. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Salas, S.X.; Macias-Silva, M.; Tecalco-Cruz, A.C. Upregulation of the canonical signaling pathway of interferon-gamma is associated with glioblastoma progression. Mol. Biol. Rep. 2024, 51, 64. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Mahalingam, R.; Sen, S.; Martinez-Ledesma, E.; Khan, A.; Gandy, K.; Lang, F.F.; Sulman, E.P.; Alfaro-Munoz, K.D.; Majd, N.K.; et al. Intrinsic Interferon Signaling Regulates the Cell Death and Mesenchymal Phenotype of Glioblastoma Stem Cells. Cancers 2021, 13, 5284. [Google Scholar] [CrossRef]
- Miao, B.; Ge, L.; He, C.; Wang, X.; Wu, J.; Li, X.; Chen, K.; Wan, J.; Xing, S.; Ren, L.; et al. SMYD5 is a ribosomal methyltransferase that catalyzes RPL40 lysine methylation to enhance translation output and promote hepatocellular carcinoma. Cell Res. 2024, 34, 648–660. [Google Scholar] [CrossRef]
- Park, J.; Wu, J.; Szkop, K.J.; Jeong, J.; Jovanovic, P.; Husmann, D.; Flores, N.M.; Francis, J.W.; Chen, Y.C.; Benitez, A.M.; et al. SMYD5 methylation of rpL40 links ribosomal output to gastric cancer. Nature 2024, 632, 656–663. [Google Scholar] [CrossRef]
- Tasab, M.; Batten, M.R.; Bulleid, N.J. Hsp47: A molecular chaperone that interacts with and stabilizes correctly-folded procollagen. EMBO J. 2000, 19, 2204–2211. [Google Scholar] [CrossRef]
- Hansen, L.J.; Sun, R.; Yang, R.; Singh, S.X.; Chen, L.H.; Pirozzi, C.J.; Moure, C.J.; Hemphill, C.; Carpenter, A.B.; Healy, P.; et al. MTAP Loss Promotes Stemness in Glioblastoma and Confers Unique Susceptibility to Purine Starvation. Cancer Res. 2019, 79, 3383–3394. [Google Scholar] [CrossRef]
- Umemura, Y.; Clarke, N.; Al-Holou, W.; Elaimy, A.; Scott, A.; Leung, D.; Kim, M.; Ferris, S.; Thomas, J.; Heth, J.; et al. AB036. Targeting glioblastoma de-novo purine metabolism to overcome chemoradiation resistance: An interim result of phase 0/1 clinical trial in newly diagnosed and recurrent glioblastoma. Chin. Clin. Oncol. 2024, 13, AB036. [Google Scholar] [CrossRef]
- Cortes Ballen, A.I.; Amosu, M.; Ravinder, S.; Chan, J.; Derin, E.; Slika, H.; Tyler, B. Metabolic Reprogramming in Glioblastoma Multiforme: A Review of Pathways and Therapeutic Targets. Cells 2024, 13, 1574. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chin, R.M.; Vergnes, L.; Hwang, H.; Deng, G.; Xing, Y.; Pai, M.Y.; Li, S.; Ta, L.; Fazlollahi, F.; et al. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metab. 2015, 22, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol. Cell 2008, 30, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Yokota, A.; Harada, H.; Huang, G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1alpha in cancer. Cancer Sci. 2019, 110, 1510–1517. [Google Scholar] [CrossRef]
- Hirsila, M.; Koivunen, P.; Gunzler, V.; Kivirikko, K.I.; Myllyharju, J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003, 278, 30772–30780. [Google Scholar] [CrossRef]
- De Rosa, V.; Iommelli, F.; Monti, M.; Fonti, R.; Votta, G.; Stoppelli, M.P.; Del Vecchio, S. Reversal of Warburg Effect and Reactivation of Oxidative Phosphorylation by Differential Inhibition of EGFR Signaling Pathways in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 5110–5120. [Google Scholar] [CrossRef]
- Shen, T.; Wang, H.; Tang, B.; Zhu, G.; Wang, X. The impact of RNA binding proteins and the associated long non-coding RNAs in the TCA cycle on cancer pathogenesis. RNA Biol. 2023, 20, 223–234. [Google Scholar] [CrossRef]
- Sang, L.; Ju, H.Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 2021, 3, 90–106. [Google Scholar] [CrossRef]
- Liao, Y.; Du, L.; Qiu, E.; Zeng, Y. Characterization of growth arrest-specific transcript 5 and growth arrest-specific transcript 5-related m6A gene signature in glioma: An observational study. Medicine 2024, 103, e39414. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, L.; Peng, R. Hypoxia-Inducible Factor 1 and Mitochondria: An Intimate Connection. Biomolecules 2022, 13, 50. [Google Scholar] [CrossRef]
- Wang, Y.; Yen, F.S.; Zhu, X.G.; Timson, R.C.; Weber, R.; Xing, C.; Liu, Y.; Allwein, B.; Luo, H.; Yeh, H.W.; et al. SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature 2021, 599, 136–140. [Google Scholar] [CrossRef]
- Sperry, J.; Condro, M.C.; Guo, L.; Braas, D.; Vanderveer-Harris, N.; Kim, K.K.O.; Pope, W.B.; Divakaruni, A.S.; Lai, A.; Christofk, H.; et al. Glioblastoma Utilizes Fatty Acids and Ketone Bodies for Growth Allowing Progression during Ketogenic Diet Therapy. iScience 2020, 23, 101453. [Google Scholar] [CrossRef] [PubMed]
- Foretz, M.; Guigas, B.; Viollet, B. Metformin: Update on mechanisms of action and repurposing potential. Nat. Rev. Endocrinol. 2023, 19, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhang, Y.; Akter, K.A.; Nozohouri, S.; Archie, S.R.; Patel, D.; Villalba, H.; Abbruscato, T. Permeability of Metformin across an In Vitro Blood-Brain Barrier Model during Normoxia and Oxygen-Glucose Deprivation Conditions: Role of Organic Cation Transporters (Octs). Pharmaceutics 2023, 15, 1357. [Google Scholar] [CrossRef] [PubMed]
- Porper, K.; Shpatz, Y.; Plotkin, L.; Pechthold, R.G.; Talianski, A.; Champ, C.E.; Furman, O.; Shimoni-Sebag, A.; Symon, Z.; Amit, U.; et al. A Phase I clinical trial of dose-escalated metabolic therapy combined with concomitant radiation therapy in high-grade glioma. J. Neuro-Oncol. 2021, 153, 487–496. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Kobayashi, A.; Cahill, D.P.; Wakimoto, H.; Tanaka, S. Molecular biology and novel therapeutics for IDH mutant gliomas: The new era of IDH inhibitors. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 189102. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Y.; Yang, R.; Sun, C.; Gao, X.; Gu, X.; Yuan, Y.; Nie, Y.; Xu, S.; Han, R.; et al. IDH1 mutation inhibits differentiation of astrocytes and glioma cells with low oxoglutarate dehydrogenase expression by disturbing alpha-ketoglutarate-related metabolism and epigenetic modification. Life Metab. 2024, 3, loae002. [Google Scholar] [CrossRef]
- Avellaneda Matteo, D.; Grunseth, A.J.; Gonzalez, E.R.; Anselmo, S.L.; Kennedy, M.A.; Moman, P.; Scott, D.A.; Hoang, A.; Sohl, C.D. Molecular mechanisms of isocitrate dehydrogenase 1 (IDH1) mutations identified in tumors: The role of size and hydrophobicity at residue 132 on catalytic efficiency. J. Biol. Chem. 2017, 292, 7971–7983. [Google Scholar] [CrossRef]
- Erasimus, H.; Gobin, M.; Niclou, S.; Van Dyck, E. DNA repair mechanisms and their clinical impact in glioblastoma. Mutat. Res. Rev. Mutat. Res. 2016, 769, 19–35. [Google Scholar] [CrossRef]
- Duncan, B.K.; Miller, J.H. Mutagenic deamination of cytosine residues in DNA. Nature 1980, 287, 560–561. [Google Scholar] [CrossRef]
- Nunez, F.J.; Mendez, F.M.; Kadiyala, P.; Alghamri, M.S.; Savelieff, M.G.; Garcia-Fabiani, M.B.; Haase, S.; Koschmann, C.; Calinescu, A.A.; Kamran, N.; et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci. Transl. Med. 2019, 11, eaaq1427. [Google Scholar] [CrossRef] [PubMed]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drablos, F.; Saetrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.; Lowery, M. Human DNA Repair Genes. Update of the Table Cited in Wood RD, Mitchell M, & Lindahl T Mutation Research, 2005, in Science, 2001, in the Reference Book DNA Repair and Mutagenesis, 2nd Edition, 2006, and in Nature Reviews Cancer, 2011. Available online: https://www.mdanderson.org/documents/Labs/Wood-Laboratory/human-dna-repair-genes.html#chr (accessed on 14 February 2023).
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef] [PubMed]
- Thangavel, C.; Boopathi, E.; Ciment, S.; Liu, Y.; O’Neill, R.; Sharma, A.; McMahon, S.B.; Mellert, H.; Addya, S.; Ertel, A.; et al. The retinoblastoma tumor suppressor modulates DNA repair and radioresponsiveness. Clin. Cancer Res. 2014, 20, 5468–5482, Erratum in Clin. Cancer Res. 2024, 30, 5695. https://doi.org/10.1158/1078-0432.CCR-24-3735. [Google Scholar] [CrossRef]
- Nakamura, M.; Konishi, N.; Hiasa, Y.; Tsunoda, S.; Fukushima, Y.; Tsuzuki, T.; Takemura, K.; Aoki, H.; Kobitsu, K.; Sakaki, T. Immunohistochemical detection of CDKN2, retinoblastoma and p53 gene products in primary astrocytic tumors. Int. J. Oncol. 1996, 8, 889–893. [Google Scholar] [CrossRef]
- Xu, H.J.; Quinlan, D.C.; Davidson, A.G.; Hu, S.X.; Summers, C.L.; Li, J.; Benedict, W.F. Altered retinoblastoma protein expression and prognosis in early-stage non-small-cell lung carcinoma. J. Natl. Cancer Inst. 1994, 86, 695–699. [Google Scholar] [CrossRef]
- Kornblau, S.M.; Andreeff, M.; Hu, S.X.; Xu, H.J.; Patel, S.; Theriault, A.; Koller, C.; Kantarjian, H.; Estey, E.; Deisseroth, A.B.; et al. Low and maximally phosphorylated levels of the retinoblastoma protein confer poor prognosis in newly diagnosed acute myelogenous leukemia: A prospective study. Clin. Cancer Res. 1998, 4, 1955–1963. [Google Scholar]
- Tchigvintsev, A.; Tchigvintsev, D.; Flick, R.; Popovic, A.; Dong, A.; Xu, X.; Brown, G.; Lu, W.; Wu, H.; Cui, H.; et al. Biochemical and structural studies of conserved Maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides. Chem. Biol. 2013, 20, 1386–1398. [Google Scholar] [CrossRef][Green Version]
- Dai, D.P.; Gan, W.; Hayakawa, H.; Zhu, J.L.; Zhang, X.Q.; Hu, G.X.; Xu, T.; Jiang, Z.L.; Zhang, L.Q.; Hu, X.D.; et al. Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells. Proc. Natl. Acad. Sci. USA 2018, 115, 4218–4222. [Google Scholar] [CrossRef]
- Hayakawa, H.; Hofer, A.; Thelander, L.; Kitajima, S.; Cai, Y.; Oshiro, S.; Yakushiji, H.; Nakabeppu, Y.; Kuwano, M.; Sekiguchi, M. Metabolic fate of oxidized guanine ribonucleotides in mammalian cells. Biochemistry 1999, 38, 3610–3614. [Google Scholar] [CrossRef]
- Resende, F.F.B.; Titze-de-Almeida, S.S.; Titze-de-Almeida, R. Function of neuronal nitric oxide synthase enzyme in temozolomide-induced damage of astrocytic tumor cells. Oncol. Lett. 2018, 15, 4891–4899. [Google Scholar] [CrossRef] [PubMed]
- Narendrula, R.; Mispel-Beyer, K.; Guo, B.; Parissenti, A.M.; Pritzker, L.B.; Pritzker, K.; Masilamani, T.; Wang, X.; Lanner, C. RNA disruption is associated with response to multiple classes of chemotherapy drugs in tumor cell lines. BMC Cancer 2016, 16, 146. [Google Scholar] [CrossRef]
- Pettersen, H.S.; Visnes, T.; Vagbo, C.B.; Svaasand, E.K.; Doseth, B.; Slupphaug, G.; Kavli, B.; Krokan, H.E. UNG-initiated base excision repair is the major repair route for 5-fluorouracil in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA incorporation. Nucleic Acids Res. 2011, 39, 8430–8444. [Google Scholar] [CrossRef] [PubMed]
- Vagbo, C.B.; Slupphaug, G. RNA in DNA repair. DNA Repair. 2020, 95, 102927. [Google Scholar] [CrossRef] [PubMed]
- Simms, N.; Knight, J.R.P. RNA damage: The forgotten target of clinical compounds. Front. RNA Res. 2023, 1, 1248236. [Google Scholar] [CrossRef]
- Huang, T.H.; Fowler, F.; Chen, C.C.; Shen, Z.J.; Sleckman, B.; Tyler, J.K. The Histone Chaperones ASF1 and CAF-1 Promote MMS22L-TONSL-Mediated Rad51 Loading onto ssDNA during Homologous Recombination in Human Cells. Mol. Cell 2018, 69, 879–892.e5. [Google Scholar] [CrossRef]
- McKenzie, L.D.; LeClair, J.W.; Miller, K.N.; Strong, A.D.; Chan, H.L.; Oates, E.L.; Ligon, K.L.; Brennan, C.W.; Chheda, M.G. CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci. Rep. 2019, 9, 4444. [Google Scholar] [CrossRef]
- Zhao, J.; Tian, S.; Guo, Q.; Bao, K.; Yu, G.; Wang, X.; Shen, X.; Zhang, J.; Chen, J.; Yang, Y.; et al. A PARylation-phosphorylation cascade promotes TOPBP1 loading and RPA-RAD51 exchange in homologous recombination. Mol. Cell 2022, 82, 2571–2587. [Google Scholar] [CrossRef]
- Heath, J.; Cheyou, E.S.; Findlay, S.; Luo, V.M.; Carpio, E.P.; Lee, J.; Djerir, B.; Chen, X.; Morin, T.; Lebeau, B.; et al. POGZ promotes homology-directed DNA repair in an HP1-dependent manner. EMBO Rep. 2022, 23, e51041. [Google Scholar] [CrossRef]
- Vancevska, A.; Ahmed, W.; Pfeiffer, V.; Feretzaki, M.; Boulton, S.J.; Lingner, J. SMCHD1 promotes ATM-dependent DNA damage signaling and repair of uncapped telomeres. EMBO J. 2020, 39, e102668. [Google Scholar] [CrossRef]
- Mu, Y.; Lou, J.; Srivastava, M.; Zhao, B.; Feng, X.H.; Liu, T.; Chen, J.; Huang, J. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016, 17, 94–109. [Google Scholar] [CrossRef]
- Zoppoli, G.; Regairaz, M.; Leo, E.; Reinhold, W.C.; Varma, S.; Ballestrero, A.; Doroshow, J.H.; Pommier, Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. USA 2012, 109, 15030–15035. [Google Scholar] [CrossRef]
- Wei, S.; Li, C.; Yin, Z.; Wen, J.; Meng, H.; Xue, L.; Wang, J. Histone methylation in DNA repair and clinical practice: New findings during the past 5-years. J. Cancer 2018, 9, 2072–2081. [Google Scholar] [CrossRef] [PubMed]
- McCornack, C.; Woodiwiss, T.; Hardi, A.; Yano, H.; Kim, A.H. The function of histone methylation and acetylation regulators in GBM pathophysiology. Front. Oncol. 2023, 13, 1144184. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Kheradpour, P.; Mikkelsen, T.S.; Shoresh, N.; Ward, L.D.; Epstein, C.B.; Zhang, X.; Wang, L.; Issner, R.; Coyne, M.; et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011, 473, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef]
- Fang, H.; Zhu, X.; Yang, H.; Oh, J.; Barbour, J.A.; Wong, J.W.H. Deficiency of replication-independent DNA mismatch repair drives a 5-methylcytosine deamination mutational signature in cancer. Sci. Adv. 2021, 7, eabg4398. [Google Scholar] [CrossRef]
- Zhang, J.; Stevens, M.F.; Bradshaw, T.D. Temozolomide: Mechanisms of action, repair and resistance. Curr. Mol. Pharmacol. 2012, 5, 102–114. [Google Scholar] [CrossRef]
- Smythies, J.A.; Sun, M.; Masson, N.; Salama, R.; Simpson, P.D.; Murray, E.; Neumann, V.; Cockman, M.E.; Choudhry, H.; Ratcliffe, P.J.; et al. Inherent DNA-binding specificities of the HIF-1alpha and HIF-2alpha transcription factors in chromatin. EMBO Rep. 2019, 20, e46401. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Geiszler, D.J.; Kong, A.T.; Avtonomov, D.M.; Yu, F.; Leprevost, F.D.V.; Nesvizhskii, A.I. PTM-Shepherd: Analysis and Summarization of Post-Translational and Chemical Modifications From Open Search Results. Mol. Cell. Proteom. 2021, 20, 100018. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Aldaz, P.; Martin-Martin, N.; Saenz-Antonanzas, A.; Carrasco-Garcia, E.; Alvarez-Satta, M.; Elua-Pinin, A.; Pollard, S.M.; Lawrie, C.H.; Moreno-Valladares, M.; Sampron, N.; et al. High SOX9 Maintains Glioma Stem Cell Activity through a Regulatory Loop Involving STAT3 and PML. Int. J. Mol. Sci. 2022, 23, 4511. [Google Scholar] [CrossRef]
- Nisole, S.; Maroui, M.A.; Mascle, X.H.; Aubry, M.; Chelbi-Alix, M.K. Differential Roles of PML Isoforms. Front. Oncol. 2013, 3, 125. [Google Scholar] [CrossRef] [PubMed]
- Dono, A.; Ramesh, A.V.; Wang, E.; Shah, M.; Tandon, N.; Ballester, L.Y.; Esquenazi, Y. The role of RB1 alteration and 4q12 amplification in IDH-WT glioblastoma. Neurooncol Adv. 2021, 3, vdab050. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, R.; Li, X.; Yu, L.; Hua, D.; Sun, C.; Shi, C.; Luo, W.; Rao, C.; Jiang, Z.; et al. Splicing factor SRSF1 promotes gliomagenesis via oncogenic splice-switching of MYO1B. J. Clin. Invest. 2019, 129, 676–693. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.; Mallick, B. LncRNA-associated competing endogenous RNA network analysis uncovered key lncRNAs involved in temozolomide resistance and tumor recurrence of glioblastoma. J. Mol. Recognit. 2023, 36, e3060. [Google Scholar] [CrossRef]
- Edmonds, N.L.; Flores, S.E.; Mahmutovic, A.; Young, S.J.; Mauldin, I.S.; Slingluff, C.L., Jr. CD103 and periplakin are potential biomarkers for response of metastatic melanoma to pembrolizumab. Melanoma Res. 2022, 32, 440–450. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, Y.; Qiu, P.; Ma, T.; Bai, Y.; Bu, J.; Hu, Y.; Jin, M.; Zhu, T.; Gu, X. RGS10 deficiency facilitates distant metastasis by inducing epithelial-mesenchymal transition in breast cancer. Elife 2024, 13. [Google Scholar] [CrossRef]
- Connolly, N.P.; Galisteo, R.; Xu, S.; Bar, E.E.; Peng, S.; Tran, N.L.; Ames, H.M.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A. Elevated fibroblast growth factor-inducible 14 expression transforms proneural-like gliomas into more aggressive and lethal brain cancer. Glia 2021, 69, 2199–2214. [Google Scholar] [CrossRef]
- Paradowski, M.; Bilinska, M.; Bar, J. Characteristics of the expression of KAI1/CD82 and PDGFRbeta and their impact on glioma progression. Folia Neuropathol. 2016, 54, 241–248. [Google Scholar] [CrossRef]
- Leng, X.; Ma, J.; Liu, Y.; Shen, S.; Yu, H.; Zheng, J.; Liu, X.; Liu, L.; Chen, J.; Zhao, L.; et al. Mechanism of piR-DQ590027/MIR17HG regulating the permeability of glioma conditioned normal BBB. J. Exp. Clin. Cancer Res. 2018, 37, 246. [Google Scholar] [CrossRef]
- Zhang, R.; Tremblay, T.L.; McDermid, A.; Thibault, P.; Stanimirovic, D. Identification of differentially expressed proteins in human glioblastoma cell lines and tumors. Glia 2003, 42, 194–208. [Google Scholar] [CrossRef]
- Gyorffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Sun, Q.; Yuan, F.; Xu, Y.; Tong, S.; Li, Y.; Yi, S.; Yan, T.; Chen, Q.; et al. BCAS3 accelerates glioblastoma tumorigenesis by restraining the P53/GADD45alpha signaling pathway. Exp. Cell Res. 2022, 417, 113231. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, Y.; Zhang, N.; Lin, X.; Zhu, D.; Shen, C.; Wang, X.; Li, H.; Xue, J.; Yu, Q.; et al. The Antipsychotic Drug Penfluridol Inhibits N-Linked Glycoprotein Processing and Enhances T-cell-Mediated Tumor Immunity. Mol. Cancer Ther. 2024, 23, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Suarez, G.; Sha, J.; Sierra, J.C.; Peterson, J.W.; Chopra, A.K. Phospholipase A2-activating protein (PLAA) enhances cisplatin-induced apoptosis in HeLa cells. Cell Signal 2009, 21, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Gu, L.; Liu, Y.; Wang, L.; Zhu, J.; Tang, S.; Wei, X.; Wang, J.; Zhang, S.; Wang, X.; et al. PLAA suppresses ovarian cancer metastasis via METTL3-mediated m(6)A modification of TRPC3 mRNA. Oncogene 2022, 41, 4145–4158. [Google Scholar] [CrossRef]
- Sauers, M.E.; Abraham, R.T.; Alvin, J.D.; Zemaitis, M.A. Factors influencing dimethylphenobarbital N-demethylation by isolated hepatocytes from untreated and phenobarbital-treated rats. Drug Metab. Dispos. 1980, 8, 208–211. [Google Scholar] [CrossRef]
- Rosi, A.; Ricci-Vitiani, L.; Biffoni, M.; Grande, S.; Luciani, A.M.; Palma, A.; Runci, D.; Cappellari, M.; De Maria, R.; Guidoni, L.; et al. (1) H NMR spectroscopy of glioblastoma stem-like cells identifies alpha-aminoadipate as a marker of tumor aggressiveness. NMR Biomed. 2015, 28, 317–326. [Google Scholar] [CrossRef]
- Sun, G.; Liu, W. The neutrophil extracellular traps-related gene signature predicts the prognosis of glioblastoma multiforme. Folia Neuropathol. 2024, 62, 59–75. [Google Scholar] [CrossRef]
- Li, J.; Cao, H.; Yang, J.; Wang, B. CircCDK1 blocking IGF2BP2-mediated m6A modification of CPPED1 promotes laryngeal squamous cell carcinoma metastasis via the PI3K/AKT signal pathway. Gene 2023, 884, 147686. [Google Scholar] [CrossRef]
- Hudson, A.L.; Cho, A.; Colvin, E.K.; Hayes, S.A.; Wheeler, H.R.; Howell, V.M. CA9, CYFIP2 and LGALS3BP-A Novel Biomarker Panel to Aid Prognostication in Glioma. Cancers 2024, 16, 1069. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, G.; Yuan, D.; Wu, P.; Guo, J.; Lu, Y.; Wang, Z. Caveolin-1 promotes glioma proliferation and metastasis by enhancing EMT via mediating PAI-1 activation and its correlation with immune infiltrates. Heliyon 2024, 10, e24464. [Google Scholar] [CrossRef]
- Se, Y.B.; Kim, S.H.; Kim, J.Y.; Kim, J.E.; Dho, Y.S.; Kim, J.W.; Kim, Y.H.; Woo, H.G.; Kim, S.H.; Kang, S.H.; et al. Underexpression of HOXA11 Is Associated with Treatment Resistance and Poor Prognosis in Glioblastoma. Cancer Res. Treat. 2017, 49, 387–398. [Google Scholar] [CrossRef]
- Yu, J.; Deng, X.; Lin, X.; Xie, L.; Guo, S.; Lin, X.; Lin, D. DST regulates cisplatin resistance in colorectal cancer via PI3K/Akt pathway. J. Pharm. Pharmacol. 2024, 77, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhang, L.; Ma, X.; Cai, S.; Jia, Y.; Zhao, L. RFTN1 facilitates gastric cancer progression by modulating AKT/p38 signaling pathways. Pathol. Res. Pract. 2022, 234, 153902. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Yu, X.J.; Wei, K.; Wang, S.X.; Liu, Q.K.; Wang, Y.G.; Li, H.; Huang, C. Mesenchymal stem cells shuttling miR-503 via extracellular vesicles enhance glioma immune escape. Oncoimmunology 2022, 11, 1965317. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Voshart, D.; Paridaen, J.; Oosterhof, N.; Liang, D.; Thiruvalluvan, A.; Zuhorn, I.S.; den Dunnen, W.F.A.; Zhang, G.; Lin, H.; et al. CD146 increases stemness and aggressiveness in glioblastoma and activates YAP signaling. Cell Mol. Life Sci. 2022, 79, 398. [Google Scholar] [CrossRef]
- Mansuer, M.; Zhou, L.; Wang, C.; Gao, L.; Jiang, Y. Erianin induces ferroptosis in GSCs via REST/LRSAM1 mediated SLC40A1 ubiquitination to overcome TMZ resistance. Cell Death Dis. 2024, 15, 522. [Google Scholar] [CrossRef]
- Shang, Z.; Lai, Y.; Cheng, H. DPP2/7 is a Potential Predictor of Prognosis and Target in Immunotherapy in Colorectal Cancer: An Integrative Multi-omics Analysis. Comb. Chem. High. Throughput Screen. 2024, 27, 1642–1660. [Google Scholar] [CrossRef]
- Jiang, X.; Tan, J.; Wen, Y.; Liu, W.; Wu, S.; Wang, L.; Wangou, S.; Liu, D.; Du, C.; Zhu, B.; et al. MSI2-TGF-beta/TGF-beta R1/SMAD3 positive feedback regulation in glioblastoma. Cancer Chemother. Pharmacol. 2019, 84, 415–425. [Google Scholar] [CrossRef]
- Mohd Amin, A.; Panneerselvan, N.; Md Noor, S.; Mohtaruddin, N.; Sathar, J.; Norbaya, W.S.; Osman, R.; Kee, L.H.; Mohd Yaakub, W.H.; Cheong, S.K.; et al. ENPP4 and HOXA3 as potential leukaemia stem cell markers in acute myeloid leukaemia. Malays. J. Pathol. 2023, 45, 65–76. [Google Scholar]
- Kong, F.; Yang, S.; Shi, R.; Peng, Y. The Up-Regulated Expression of Mitochondrial Membrane Molecule RHOT1 in Gastric Cancer Predicts the Prognosis of Patients and Promotes the Malignant Biological Behavior of Cells. Mol. Biotechnol. 2024, 67, 1095–1108. [Google Scholar] [CrossRef]
- Motaln, H.; Gruden, K.; Hren, M.; Schichor, C.; Primon, M.; Rotter, A.; Lah, T.T. Human mesenchymal stem cells exploit the immune response mediating chemokines to impact the phenotype of glioblastoma. Cell Transplant. 2012, 21, 1529–1545. [Google Scholar] [CrossRef]
- Ruiz-Lopez, E.; Jovcevska, I.; Gonzalez-Gomez, R.; Tejero, H.; Al-Shahrour, F.; Muyldermans, S.; Schuhmacher, A.J. Nanobodies targeting ABCC3 for immunotargeted applications in glioblastoma. Sci. Rep. 2022, 12, 22581. [Google Scholar] [CrossRef]
- An, S.; Song, I.H.; Woo, C.G. Diagnostic Value of Nestin Expression in Adult Gliomas. Int. J. Surg. Pathol. 2023, 31, 1014–1020. [Google Scholar] [CrossRef] [PubMed]
- Budhiraja, S.; Baisiwala, S.; Cho, S.; Chojak, R.; Kazi, H.A.; Stepniak, A.; Perrault, E.N.; Chen, L.; Park, C.H.; Dmello, C.; et al. THOC1 complexes with SIN3A to regulate R-loops and promote glioblastoma progression. bioRxiv 2024. [Google Scholar] [CrossRef]
- Tang, M.; Chen, Z.; Wang, C.; Feng, X.; Lee, N.; Huang, M.; Zhang, H.; Li, S.; Xiong, Y.; Chen, J. Histone chaperone ASF1 acts with RIF1 to promote DNA end joining in BRCA1-deficient cells. J. Biol. Chem. 2022, 298, 101979. [Google Scholar] [CrossRef] [PubMed]
- Rieckher, M.; Gallrein, C.; Alquezar-Artieda, N.; Bourached-Silva, N.; Vaddavalli, P.L.; Mares, D.; Backhaus, M.; Blindauer, T.; Greger, K.; Wiesner, E.; et al. Distinct DNA repair mechanisms prevent formaldehyde toxicity during development, reproduction and aging. Nucleic Acids Res. 2024, 52, 8271–8285. [Google Scholar] [CrossRef] [PubMed]
- Restall, I.J.; Cseh, O.; Richards, L.M.; Pugh, T.J.; Luchman, H.A.; Weiss, S. Brain Tumor Stem Cell Dependence on Glutaminase Reveals a Metabolic Vulnerability through the Amino Acid Deprivation Response Pathway. Cancer Res. 2020, 80, 5478–5490. [Google Scholar] [CrossRef]
- Shi, Z.; Ge, X.; Li, M.; Yin, J.; Wang, X.; Zhang, J.; Chen, D.; Li, X.; Wang, X.; Ji, J.; et al. Argininosuccinate lyase drives activation of mutant TERT promoter in glioblastomas. Mol. Cell 2022, 82, 3919–3931.e3917. [Google Scholar] [CrossRef]
- Kolar, E.A.; Shi, X.; Clay, E.M.; Liu, Y.; Xia, S.; Zhang, C.; Le, A.; Watkins, P.A. Depleting glioblastoma cells of very long-chain acyl-CoA synthetase 3 (ACSVL3) produces metabolic alterations in non-lipid pathways. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chen, S.Y.; Wu, J.; Chen, Y.; Wang, Y.E.; Setayeshpour, Y.; Federico, C.; Mestre, A.A.; Lin, C.C.; Chi, J.T. NINJ1 regulates ferroptosis via xCT antiporter interaction and CoA modulation. Cell Death Dis. 2024, 15, 755. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, Y.; Yang, C.; Zhang, X.; Xu, C.; Qiao, X.; Xu, J.; Tian, S.; Fang, C.; Kang, C. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol. Med. 2019, 16, 784–796. [Google Scholar] [CrossRef]
- Zhao, X.C.; Wang, G.Z.; Wen, Z.S.; Zhou, Y.C.; Hu, Q.; Zhang, B.; Qu, L.W.; Gao, S.H.; Liu, J.; Ma, L.; et al. Systematic identification of CDC34 that functions to stabilize EGFR and promote lung carcinogenesis. EBioMedicine 2020, 53, 102689. [Google Scholar] [CrossRef]
- Song, T.; Hu, Z.; Liu, J.; Huang, W. FLOT2 upregulation promotes growth and invasion by interacting and stabilizing EphA2 in gliomas. Biochem. Biophys. Res. Commun. 2021, 548, 67–73. [Google Scholar] [CrossRef]
- Xie, Y.; Ding, W.; Xiang, Y.; Wang, X.; Yang, J. Calponin 3 Acts as a Potential Diagnostic and Prognostic Marker and Promotes Glioma Cell Proliferation, Migration, and Invasion. World Neurosurg. 2022, 165, e721–e731. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, H.; Fei, M.; Tang, T.; Niu, W.; Zhang, L. Smarcd1 Inhibits the Malignant Phenotypes of Human Glioblastoma Cells via Crosstalk with Notch1. Mol. Neurobiol. 2021, 58, 1438–1452. [Google Scholar] [CrossRef] [PubMed]
- Dings, M.P.G.; van der Zalm, A.P.; Bootsma, S.; van Maanen, T.F.J.; Waasdorp, C.; van den Ende, T.; Liu, D.; Bailey, P.; Koster, J.; Zwijnenburg, D.A.; et al. Estrogen-related receptor alpha drives mitochondrial biogenesis and resistance to neoadjuvant chemoradiation in esophageal cancer. Cell Rep. Med. 2022, 3, 100802. [Google Scholar] [CrossRef] [PubMed]
- Kalailingam, P.; Tan, H.B.; Pan, J.Y.; Tan, S.H.; Thanabalu, T. Overexpression of CDC42SE1 in A431 Cells Reduced Cell Proliferation by Inhibiting the Akt Pathway. Cells 2019, 8, 117. [Google Scholar] [CrossRef]
- Heath, J.; Mirabelli, C.; Annis, M.G.; Sabourin, V.; Hebert, S.; Findlay, S.; Kim, H.; Witcher, M.; Kleinman, C.L.; Siegel, P.M.; et al. The Neurodevelopmental Protein POGZ Suppresses Metastasis in Triple Negative Breast Cancer by Attenuating TGFbeta Signaling. Cancer Res. 2024, 84, 3743–3760. [Google Scholar] [CrossRef]
- El Husseini, K.; Marguet, F.; Lamy, A.; Magne, N.; Fontanilles, M. Major response to temozolomide as first-line treatment for newly-diagnosed DDR2-mutated glioblastoma: A case report. Rev. Neurol. 2020, 176, 402–404. [Google Scholar] [CrossRef]
- Gao, X.; Ge, J.; Gao, X.; Mei, N.A.; Su, Y.; Shan, S.; Qian, W.; Guan, J.; Zhang, Z.; Wang, L. IQGAP3 promotes the progression of glioma as an immune and prognostic marker. Oncol. Res. 2024, 32, 659–678. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Liu, B.; Jiang, M.; Yan, S.; Han, X.; Shen, H.; Na, M.; Wang, Y.; Ren, Z.; et al. GNG5 is a novel oncogene associated with cell migration, proliferation, and poor prognosis in glioma. Cancer Cell Int. 2021, 21, 297. [Google Scholar] [CrossRef]
- Liang, S.; Zhu, L.; Yang, F.; Dong, H. Transcription factor YY1-activated GNG5 facilitates glioblastoma cell growth, invasion, stemness and glycolysis through Wnt/beta-catenin pathway. Sci. Rep. 2024, 14, 25234. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, Y.Y.; Pan, Y.Q.; Zheng, X.J.; Liao, K.; Mo, H.Y.; Sheng, H.; Wu, Q.N.; Liu, Z.X.; Zeng, Z.L.; et al. IL-1beta-associated NNT acetylation orchestrates iron-sulfur cluster maintenance and cancer immunotherapy resistance. Mol. Cell 2023, 83, 1887–1902.e8. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Shen, G.; Su, Z.; Du, J.; Xu, F.; Yu, Y. RAD21 inhibited transcription of tumor suppressor MIR4697HG and led to glioma tumorigenesis. Biomed. Pharmacother. 2020, 123, 109759. [Google Scholar] [CrossRef]
- Teng, L.; Nakada, M.; Zhao, S.G.; Endo, Y.; Furuyama, N.; Nambu, E.; Pyko, I.V.; Hayashi, Y.; Hamada, J.I. Silencing of ferrochelatase enhances 5-aminolevulinic acid-based fluorescence and photodynamic therapy efficacy. Br. J. Cancer 2011, 104, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.H.; Qiu, S.; Xing, Y.; Xu, J.; Lu, B.; Zhao, S.F.; Li, Y.T.; Su, Z.Z. Paeoniflorin Regulates NEDD4L/STAT3 Pathway to Induce Ferroptosis in Human Glioma Cells. J. Oncol. 2022, 2022, 6093216. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, B.; Bian, E.; Zong, G.; He, J.; Wang, Y.; Ma, C.; Wan, J. Ubiquitination Destabilizes Protein Sphingosine Kinase 2 to Regulate Glioma Malignancy. Front. Cell Neurosci. 2021, 15, 660354. [Google Scholar] [CrossRef]
- Geng, X.; Geng, Z.; Li, H.; Zhang, Y.; Li, J.; Chang, H. Over-expression of TFB2M facilitates cell growth and metastasis via activating ROS-Akt-NF-kappaB signalling in hepatocellular carcinoma. Liver Int. 2020, 40, 1756–1769. [Google Scholar] [CrossRef]
- Fok, P.T.; Huang, K.C.; Holland, P.C.; Nalbantoglu, J. The Coxsackie and adenovirus receptor binds microtubules and plays a role in cell migration. J. Biol. Chem. 2007, 282, 7512–7521. [Google Scholar] [CrossRef]
- Yang, M.; Chen, X.; Zhang, J.; Xiong, E.; Wang, Q.; Fang, W.; Li, L.; Fei, F.; Gong, A. ME2 Promotes Proneural-Mesenchymal Transition and Lipogenesis in Glioblastoma. Front. Oncol. 2021, 11, 715593. [Google Scholar] [CrossRef]
- Tsai, C.K.; Huang, L.C.; Tsai, W.C.; Huang, S.M.; Lee, J.T.; Hueng, D.Y. Overexpression of PLOD3 promotes tumor progression and poor prognosis in gliomas. Oncotarget 2018, 9, 15705–15720. [Google Scholar] [CrossRef]
- Hu, Z.; Li, M.; Chen, Y.; Chen, L.; Han, Y.; Chen, C.; Lu, X.; You, N.; Lou, Y.; Huang, Y.; et al. Disruption of PABPN1 phase separation by SNRPD2 drives colorectal cancer cell proliferation and migration through promoting alternative polyadenylation of CTNNBIP1. Sci. China Life Sci. 2024, 67, 1212–1225. [Google Scholar] [CrossRef]
- Ni, P.; Xu, W.; Zhang, Y.; Chen, Q.; Li, A.; Wang, S.; Xu, S.; Zhou, J. TXNL1 induces apoptosis in cisplatin resistant human gastric cancer cell lines. Curr. Cancer Drug Targets 2015, 14, 850–859. [Google Scholar] [CrossRef]
- Xue, S.; Cai, Y.; Liu, J.; Ji, K.; Yi, P.; Long, H.; Zhang, X.; Li, P.; Song, Y. Dysregulation of phosphoenolpyruvate carboxykinase in cancers: A comprehensive analysis. Cell Signal 2024, 120, 111198. [Google Scholar] [CrossRef]
- Fnu, S.; Williamson, E.A.; De Haro, L.P.; Brenneman, M.; Wray, J.; Shaheen, M.; Radhakrishnan, K.; Lee, S.H.; Nickoloff, J.A.; Hromas, R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2011, 108, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, X.; Wang, J.; Shang, W.; Wang, X.; Zhang, L.; Li, J. Mechanism of NURP1 in temozolomide resistance in hypoxia-treated glioma cells via the KDM3A/TFEB axis. Oncol. Res. 2023, 31, 345–359. [Google Scholar] [CrossRef]
- Yoo, J.; Kim, G.W.; Jeon, Y.H.; Lee, S.W.; Kwon, S.H. Epigenetic roles of KDM3B and KDM3C in tumorigenesis and their therapeutic implications. Cell Death Dis. 2024, 15, 451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, H.; Li, Z.; Sun, W.; Zhao, E.; Cui, H. Histone demethylase KDM4B accelerates the progression of glioblastoma via the epigenetic regulation of MYC stability. Clin. Epigenet. 2023, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Mallm, J.P.; Windisch, P.; Biran, A.; Gal, Z.; Schumacher, S.; Glass, R.; Herold-Mende, C.; Meshorer, E.; Barbus, M.; Rippe, K. Glioblastoma initiating cells are sensitive to histone demethylase inhibition due to epigenetic deregulation. Int. J. Cancer 2020, 146, 1281–1292. [Google Scholar] [CrossRef]
- Long, C.; Song, Y.; Pan, Y.; Wu, C. Identification of molecular subtypes and a risk model based on inflammation-related genes in patients with low grade glioma. Heliyon 2023, 9, e22429. [Google Scholar] [CrossRef] [PubMed]
- Laukka, T.; Myllykoski, M.; Looper, R.E.; Koivunen, P. Cancer-associated 2-oxoglutarate analogues modify histone methylation by inhibiting histone lysine demethylases. J. Mol. Biol. 2018, 430, 3081–3092. [Google Scholar] [CrossRef]
- Shao, P.; Liu, Q.; Qi, H.H. KDM7 Demethylases: Regulation, Function and Therapeutic Targeting. Adv. Exp. Med. Biol. 2023, 1433, 167–184. [Google Scholar] [CrossRef]
- Xiao, Y.; Li, J.; Liao, X.; He, Y.; He, T.; Yang, C.; Jiang, L.; Jeon, S.M.; Lee, J.H.; Chen, Y.; et al. RIOX1-demethylated cGAS regulates ionizing radiation-elicited DNA repair. Bone Res. 2022, 10, 19. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravn Berg, S.; Brambilla, A.; Hagen, L.; Sharma, A.; Vågbø, C.B.; Liabakk, N.B.; Kissova, M.; Arano Barenys, M.; Bjørås, M.; Torp, S.H.; et al. Proteomic Analysis Uncovers Enhanced Inflammatory Phenotype and Distinct Metabolic Changes in IDH1 Mutant Glioma Cells. Int. J. Mol. Sci. 2025, 26, 9075. https://doi.org/10.3390/ijms26189075
Ravn Berg S, Brambilla A, Hagen L, Sharma A, Vågbø CB, Liabakk NB, Kissova M, Arano Barenys M, Bjørås M, Torp SH, et al. Proteomic Analysis Uncovers Enhanced Inflammatory Phenotype and Distinct Metabolic Changes in IDH1 Mutant Glioma Cells. International Journal of Molecular Sciences. 2025; 26(18):9075. https://doi.org/10.3390/ijms26189075
Chicago/Turabian StyleRavn Berg, Sigrid, Alessandro Brambilla, Lars Hagen, Animesh Sharma, Cathrine Broberg Vågbø, Nina Beate Liabakk, Miroslava Kissova, Miquel Arano Barenys, Magnar Bjørås, Sverre Helge Torp, and et al. 2025. "Proteomic Analysis Uncovers Enhanced Inflammatory Phenotype and Distinct Metabolic Changes in IDH1 Mutant Glioma Cells" International Journal of Molecular Sciences 26, no. 18: 9075. https://doi.org/10.3390/ijms26189075
APA StyleRavn Berg, S., Brambilla, A., Hagen, L., Sharma, A., Vågbø, C. B., Liabakk, N. B., Kissova, M., Arano Barenys, M., Bjørås, M., Torp, S. H., & Slupphaug, G. (2025). Proteomic Analysis Uncovers Enhanced Inflammatory Phenotype and Distinct Metabolic Changes in IDH1 Mutant Glioma Cells. International Journal of Molecular Sciences, 26(18), 9075. https://doi.org/10.3390/ijms26189075






