Accurate Protein Dynamic Conformational Ensembles: Combining AlphaFold, MD, and Amide 15N(1H) NMR Relaxation
Abstract
1. Introduction
2. Results
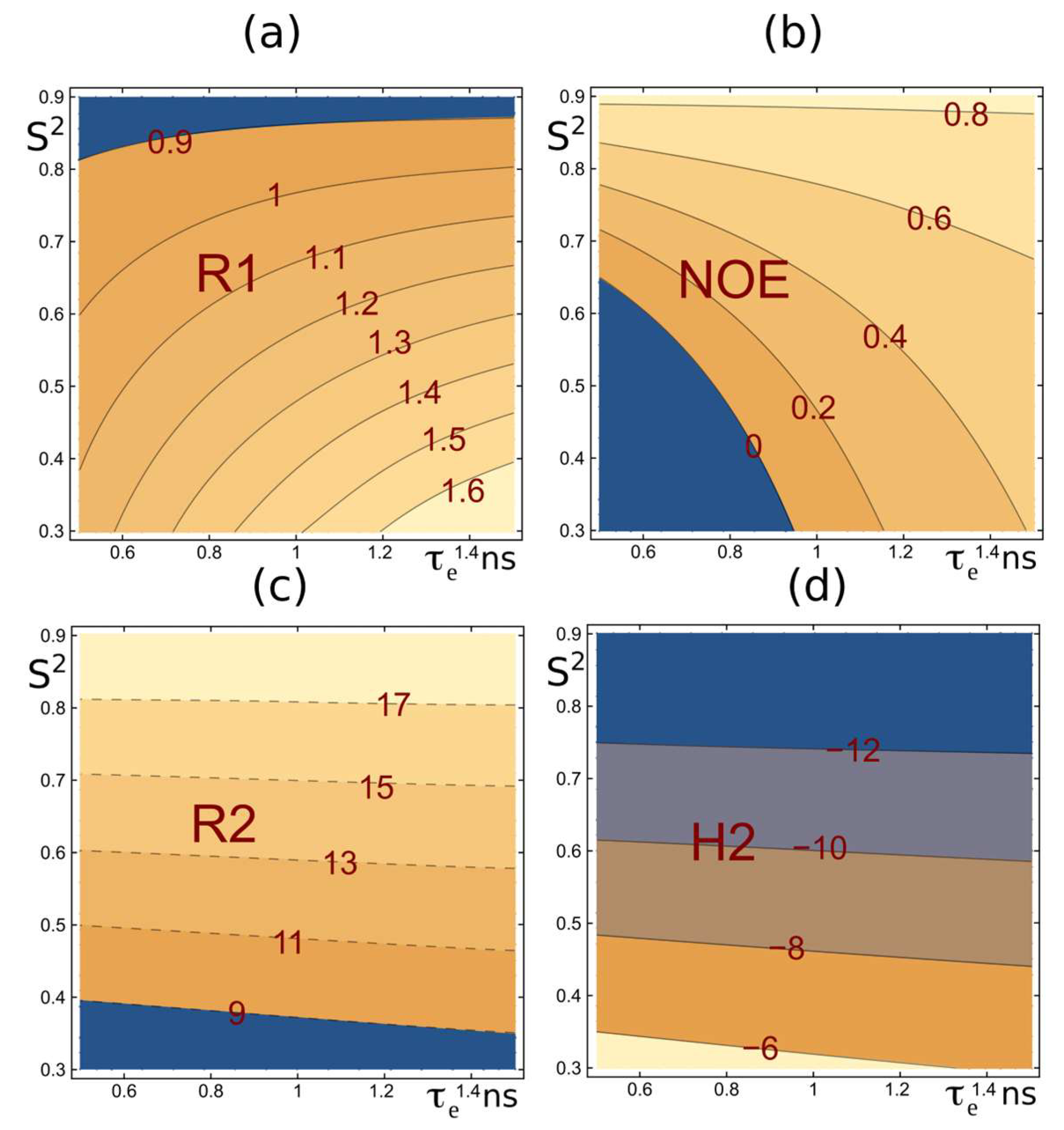
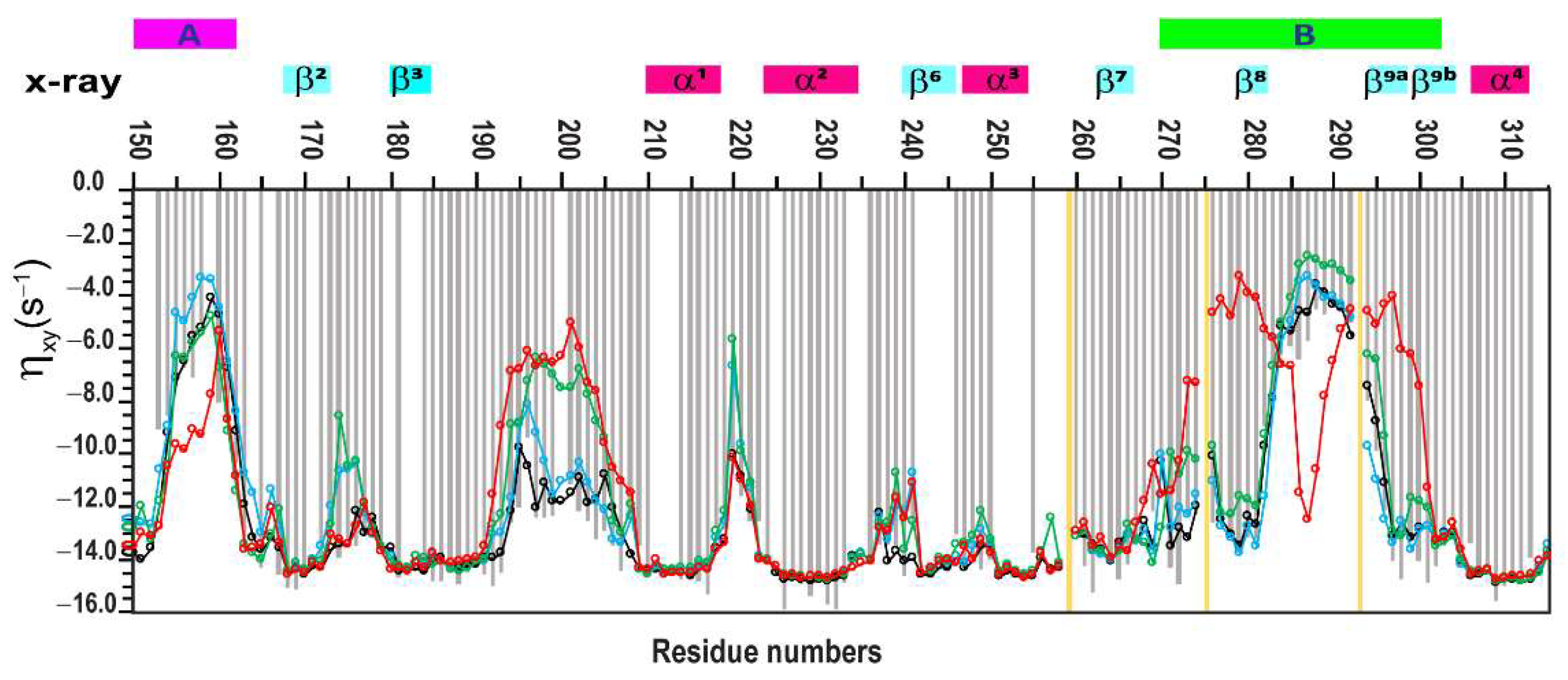
2.1. Selection of Relaxation Parameters for MD Trajectory Verification: Comparison of R2 and ηxy Relaxation Data
2.2. Determination of the Isotropic Rotational Tumbling Time of Protein
2.3. Identification and Validation of Structural Ensembles of the PsrSp Protein Based on Backbone Relaxation Dynamic
2.4. Validation of Structural Ensembles of the PsrSp Protein by Alternative Methods
3. Discussion
3.1. Experimental Validation of Conformational Ensembles
3.2. Relaxation-Based Validation of Conformational Ensembles
3.3. Identification of Conformational Ensembles
3.4. Experimental Validation and Functional Implications
3.5. Deposition of Data and Structures
4. Methods
4.1. Sample Preparation
4.2. NMR Experiments
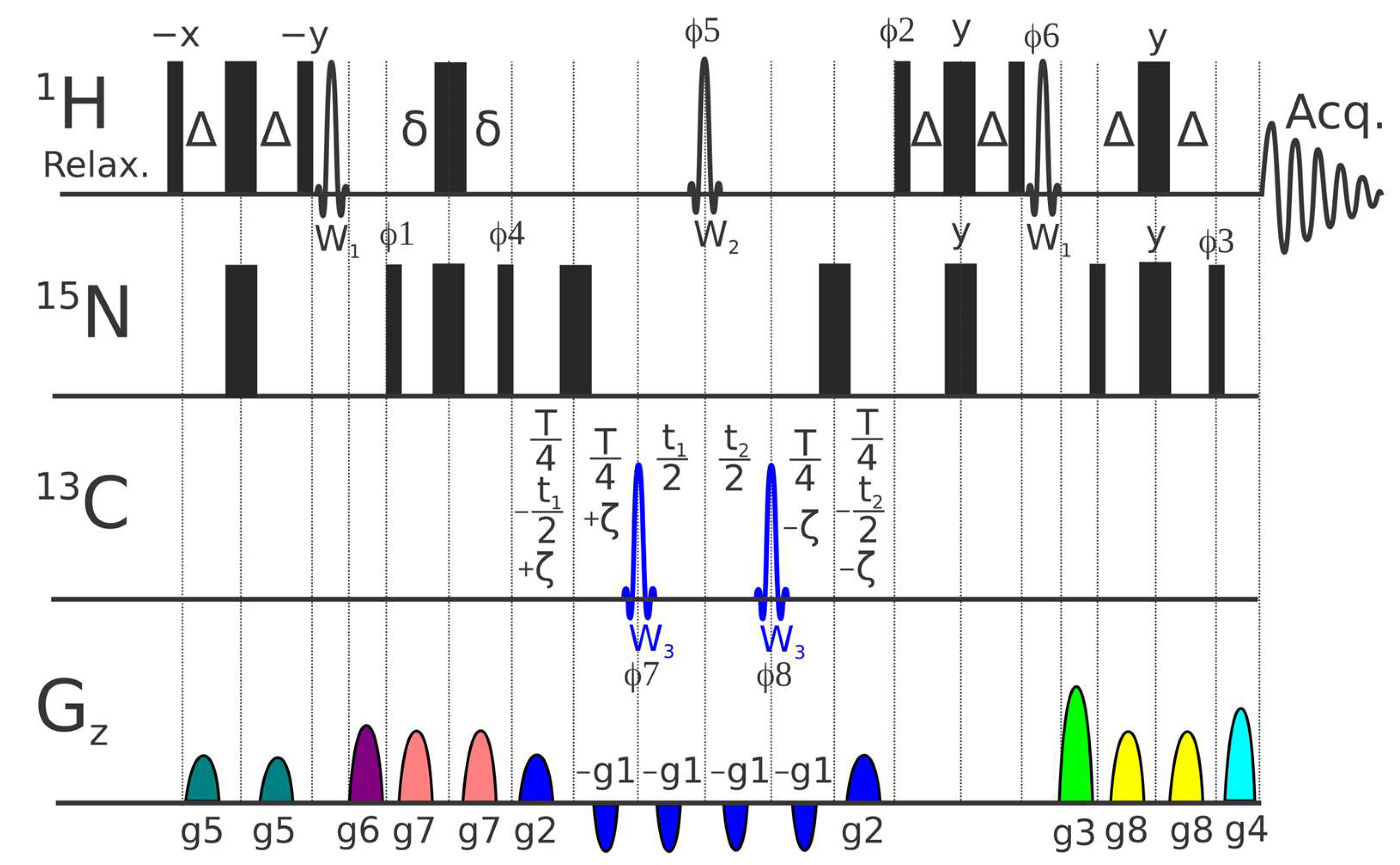
4.3. Pulse Sequence for 1H-15N CSA/DD Cross-Correlation
- (a)
- We found that application of 15N coding echo–anti-echo gradients (g1 and g2) across all six intervals, where CSA/DD effects and sampling occur, minimizes the possible systematic errors of shaped and hard pulses with defocusing of residual undesired coherences.
- (b)
- (c)
- A 1H Reburp inversion pulse (W2), selective on amide protons, preserves water magnetization along the +Z-axis and ensures uniform water (saturation) state for all ζ delays.
- (d)
- W2 additionally ensures uniformity with respect to the 2JN-Hα scalar coupling evolution across all ζ delays. Note that a non-negligible fraction of Hα protons are typically present even in deuterated protein samples.
- (e)
- All water flip-back pulses (W1) are placed outside the periods of the magnetization transfer over the 1JNH coupling and are followed by gradients.
4.4. Determination of 1H-15N CSA/DD Cross-Correlation (ηxy), Relaxation Rate R1 and 1H-15N Nuclear Overhauser Effect (NOE)
4.5. Theoretical Simulations: AlphaFold3 as a Starting Point for Full Atomic Molecular Dynamic
4.6. Individual MD Trajectory Analyses with Back-Calculation of Theoretical 15N Relaxation Parameters
4.7. Calculation of Chemical Shift Procedure
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Friedland, G.D.; Lakomek, N.A.; Griesinger, C.; Meiler, J.; Kortemme, T. A Correspondence Between Solution-State Dynamics of an Individual Protein and the Sequence and Conformational Diversity of its Family. PLoS Comput. Biol. 2009, 5, e1000393. [Google Scholar] [CrossRef]
- Boehr, D.D.; Nussinov, R.; Wright, P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009, 5, 789–796, Erratum in Nat. Chem. Biol. 2009, 5, 954. [Google Scholar] [CrossRef]
- Nussinov, R. Introduction to Protein Ensembles and Allostery. Chem. Rev. 2016, 116, 6263–6266. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Liu, Y.L.; Zhang, W.A.; Jang, H. Protein conformational ensembles in function: Roles and mechanisms. RSC Chem. Biol. 2023, 4, 850–864. [Google Scholar] [CrossRef]
- Brunger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S.; et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1998, 54, 905–921. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Guntert, P. Automated NMR Structure Calculation with CYANA; Humana Press: Totowa, NJ, USA, 2004. [Google Scholar]
- Klukowski, P.; Damberger, F.F.; Allain, F.H.T.; Iwai, H.; Kadavath, H.; Ramelot, T.A.; Montelione, G.T.; Riek, R.; Güntert, P. The 100-protein NMR spectra dataset: A resource for biomolecular NMR data analysis. Sci. Data 2024, 11, 30. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Homology modeling of larger proteins guided by chemical shifts. Nat. Methods 2015, 12, U747–U777. [Google Scholar] [CrossRef]
- Shapiro, Y.E. NMR spectroscopy on domain dynamics in biomacromolecules. Prog. Biophys. Mol. Biol. 2013, 112, 58–117, Erratum in Prog. Biophys. Mol. Biol. 2014, 114, 13. [Google Scholar] [CrossRef]
- Torchia, D.A. NMR studies of dynamic biomolecular conformational ensembles. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 84, 14–32, Erratum in Prog. Biophys. Mol. Biol. 2014, 114, 13. [Google Scholar] [CrossRef] [PubMed]
- van den Bedem, H.; Fraser, J.S. Integrative, dynamic structural biology at atomic resolution-it’s about time. Nat. Methods 2015, 12, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.H.; Xi, W.H.; Nussinov, R.; Ma, B.Y. Protein Ensembles: How Does Nature Harness Thermodynamic Fluctuations for Life? The Diverse Functional Roles of Conformational Ensembles in the Cell. Chem. Rev. 2016, 116, 6516–6551. [Google Scholar] [CrossRef]
- Ravera, E.; Sgheri, L.; Parigi, G.; Luchinat, C. A critical assessment of methods to recover information from averaged data. Phys. Chem. Chem. Phys. 2016, 18, 5686–5701. [Google Scholar] [CrossRef]
- Costa, R.G.L.; Fushman, D. Reweighting methods for elucidation of conformation ensembles of proteins. Curr. Opin. Struct. Biol. 2022, 77, 102470. [Google Scholar] [CrossRef]
- Ramelot, T.A.; Tejero, R.; Montelione, G.T. Representing structures of the multiple conformational states of proteins Theresa A. Ramelot, Roberto Tejero and. Curr. Opin. Struct. Biol. 2023, 83, 102703. [Google Scholar] [CrossRef]
- Schwalbe, H.; Audergon, P.; Haley, N.; Amaro, C.A.; Agirre, J.; Baldus, M.; Banci, L.; Baumeister, W.; Blackledge, M.; Carazo, J.M.; et al. The future of integrated structural biology. Structure 2024, 32, 1563–1580. [Google Scholar] [CrossRef]
- Dokholyan, N.V. Experimentally-driven protein structure modeling. J. Proteom. 2020, 220, 103777. [Google Scholar] [CrossRef]
- Jensen, M.R.; Salmon, L.; Nodet, G.; Blackledge, M. Defining Conformational Ensembles of Intrinsically Disordered and Partially Folded Proteins Directly from Chemical Shifts. J. Am. Chem. Soc. 2010, 132, 1270–1272. [Google Scholar] [CrossRef]
- Robustelli, P.; Trbovic, N.; Friesner, R.A.; Palmer, A.G. Conformational Dynamics of the Partially Disordered Yeast Transcription Factor GCN4. J. Chem. Theory Comput. 2013, 9, 5190–5200. [Google Scholar] [CrossRef]
- Palmer, A.G. A dynamic look backward and forward. J. Magn. Reson. 2016, 266, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Clore, G.M.; Schwieters, C.D. Amplitudes of protein backbone dynamics and correlated motions in a small α/β protein: Correspondence of dipolar coupling and heteronuclear relaxation measurements. Biochemistry 2004, 43, 10678–10691. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Best, R.B.; DePristo, M.A.; Dobson, C.M.; Vendruscolo, M. Simultaneous determination of protein structure and dynamics. Nature 2005, 433, 128–132. [Google Scholar] [CrossRef]
- Markwick, P.R.L.; Bouvignies, G.; Blackledge, M. Exploring multiple timescale motions in protein GB3 using accelerated molecular dynamics and NMR spectroscopy. J. Am. Chem. Soc. 2007, 129, 4724–4730. [Google Scholar] [CrossRef]
- Showalter, S.A.; Bruschweiler, R. Validation of molecular dynamics simulations of biomolecules using NMR spin relaxation as benchmarks: Application to the AMBER99SB force field. J. Chem. Theory Comput. 2007, 3, 961–975. [Google Scholar] [CrossRef]
- Lange, O.F.; Lakomek, N.A.; Farès, C.; Schröder, G.F.; Walter, K.F.A.; Becker, S.; Meiler, J.; Grubmüller, H.; Griesinger, C.; de Groot, B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 2008, 320, 1471–1475. [Google Scholar] [CrossRef]
- Nodet, G.; Salmon, L.; Ozenne, V.; Meier, S.; Jensen, M.R.; Blackledge, M. Quantitative Description of Backbone Conformational Sampling of Unfolded Proteins at Amino Acid Resolution from NMR Residual Dipolar Couplings. J. Am. Chem. Soc. 2009, 131, 17908–17918. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Synergism between x-ray crystallography and NMR residual dipolar couplings in characterizing protein dynamics. Struct. Dyn. 2023, 10, 040901. [Google Scholar] [CrossRef]
- Allison, J.R.; Varnai, P.; Dobson, C.M.; Vendruscolo, M. Determination of the Free Energy Landscape of α-Synuclein Using Spin Label Nuclear Magnetic Resonance Measurements. J. Am. Chem. Soc. 2009, 131, 18314–18326. [Google Scholar] [CrossRef]
- Bertini, I.; Luchinat, C.; Parigi, G.; Ravera, E.; Bertini, I.; Luchinat, C.; Parigi, G.; Ravera, E. Chapter 12—Hints on Experimental Techniques; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Palmer, A.G. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004, 104, 3623–3640. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, J.; Fairbrother, W.; Palmer, A., III; Rance, M.; Skelton, N. Principles and Practice: Protein NMR Spectroscopy; Elsevier: Amsterdam, The Netherlands; Academic Press: Cambridge, MA, USA, 2007. [Google Scholar]
- Trbovic, N.; Kim, B.; Friesner, R.A.; Palmer, A.G. Structural analysis of protein dynamics by MD simulations and NMR spin-relaxation. Proteins 2008, 71, 684–694. [Google Scholar] [CrossRef]
- Palmer, A.G. Enzyme Dynamics from NMR Spectroscopy. Acc. Chem. Res. 2015, 48, 457–465. [Google Scholar] [CrossRef]
- Kay, L.E.; Torchia, D.A.; Bax, A. Backbone dynamics of proteins as studied by nitrogen-15 inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry 1989, 28, 8972–8979. [Google Scholar] [CrossRef] [PubMed]
- Korzhnev, D.M.; Billeter, M.; Arseniev, A.S.; Orekhov, V.Y. NMR studies of Brownian tumbling and internal motions in proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2001, 38, 197–266. [Google Scholar] [CrossRef]
- Zumpfe, K.; Smith, A.A. Model-Free or Not? Front. Mol. Biosci. 2021, 8, 727553. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, C.; Zawadzka-Kazimierczuk, A.; Kontaxis, G.; Konrat, R. Using Cross-Correlated Spin Relaxation to Characterize Backbone Dihedral Angle Distributions of Flexible Protein Segments. Chemphyschem 2021, 22, 18–28. [Google Scholar] [CrossRef]
- Stenstroem, O.; Champion, C.; Lehner, M.; Bouvignies, G.; Riniker, S.; Ferrage, F. How does it really move? Recent progress in the investigation of protein nanosecond dynamics by NMR and simulation. Curr. Opin. Struct. Biol. 2022, 77, 102459. [Google Scholar] [CrossRef]
- Dauber-Osguthorpe, P.; Hagler, A.T. Biomolecular force fields: Where have we been, where are we now, where do we need to go and how do we get there? J. Comput.-Aided Mol. Des. 2019, 33, 133–203. [Google Scholar] [CrossRef]
- Schlick, T.; Portillo-Ledesma, S. Biomolecular modeling thrives in the age of technology. Nat. Comput. Sci. 2021, 1, 321–331, Erratum in Nat. Comput. Sci. 2021, 1, 767. [Google Scholar] [CrossRef]
- Schlick, T.; Portillo-Ledesma, S.; Myers, C.G.; Beljak, L.; Chen, J.; Dakhel, S.; Darling, D.; Ghosh, S.; Hall, J.; Jan, M.; et al. Biomolecular Modeling and Simulation: A Prospering Multidisciplinary Field. Annu. Rev. Biophys. 2021, 50, 267–301. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.Q.; Wu, P.; Wu, S.; Lee, T.Y.; Bai, C. Application of Computational Biology and Artificial Intelligence in Drug Design. Int. J. Mol. Sci. 2022, 23, 13568. [Google Scholar] [CrossRef]
- Kang, W.; Jiang, F.; Wu, Y.D. How to strike a conformational balance in protein force fields for molecular dynamics simulations? Wires Comput. Mol. Sci. 2022, 12, 1578. [Google Scholar] [CrossRef]
- Orioli, S.; Larsen, A.H.; Bottaro, S.; Lindorff-Larsen, K. How to learn from inconsistencies: Integrating molecular simulations with experimental data. Prog. Mol. Biol. Transl. 2020, 170, 123–176. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Fushman, D.; Cowburn, D. Simulated and NMR-derived backbone dynamics of a protein with significant flexibility:: A comparison of spectral densities for the βARK PH domain. J. Am. Chem. Soc. 2001, 123, 3021–3036. [Google Scholar] [CrossRef] [PubMed]
- Nederveen, A.J.; Bonvin, A.M.J.J. NMR relaxation and internal dynamics of ubiquitin from a 0.2 μs MD simulation. J. Chem. Theory Comput. 2005, 1, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Salvi, N.; Abyzov, A.; Blackledge, M. Analytical Description of NMR Relaxation Highlights Correlated Dynamics in Intrinsically Disordered Proteins. Angew. Chem. Int. Ed. 2017, 56, 14020–14024. [Google Scholar] [CrossRef] [PubMed]
- Kämpf, K.; Izmailov, S.A.; Rabdano, S.O.; Groves, A.T.; Podkorytov, I.S.; Skrynnikov, N.R. What Drives 15N Spin Relaxation in Disordered Proteins? Combined NMR/MD Study of the H4 Histone Tail. Biophys. J. 2018, 115, 2348–2367. [Google Scholar] [CrossRef]
- Salvi, N.; Zapletal, V.; Jasenáková, Z.; Zachrdla, M.; Padrta, P.; Narasimhan, S.; Marquardsen, T.; Tyburn, J.M.; Zídek, L.; Blackledge, M.; et al. Convergent views on disordered protein dynamics from NMR and computational approaches. Biophys. J. 2022, 121, 3785–3794. [Google Scholar] [CrossRef]
- Agback, T.; Lesovoy, D.; Han, X.; Lomzov, A.; Sun, R.H.; Sandalova, T.; Orekhov, V.Y.; Achour, A.; Agback, P. Combined NMR and molecular dynamics conformational filter identifies unambiguously dynamic ensembles of Dengue protease NS2B/NS3pro. Commun. Biol. 2023, 6, 1193. [Google Scholar] [CrossRef]
- Koller, A.N.; Schwalbe, H.; Gohlke, H. Starting structure dependence of NMR order parameters derived from MD simulations: Implications for judging force-field quality. Biophys. J. 2008, 95, L4–L6. [Google Scholar] [CrossRef]
- Kummerer, F.; Orioli, S.; Lindorff-Larsen, K. Fitting Force Field Parameters to NMR Relaxation Data. J. Chem. Theory Comput. 2023, 19, 3741–3751. [Google Scholar] [CrossRef]
- Kummerer, F.; Orioli, S.; Harding-Larsen, D.; Hoffmann, F.; Gavrilov, Y.; Teilum, K.; Lindorff-Larsen, K. Fitting Side-Chain NMR Relaxation Data Using Molecular Simulations. J. Chem. Theory Comput. 2021, 17, 5262–5275. [Google Scholar] [CrossRef] [PubMed]
- Bonomi, M.; Heller, G.T.; Camilloni, C.; Vendruscolo, M. Principles of protein structural ensemble determination. Curr. Opin. Struct. Biol. 2017, 42, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Ge, L.Y.; Chen, X.; Lv, Z.X.; Wang, S.H.; Zhou, X.G.; Zhang, G.J. Beyond static structures: Protein dynamic conformations modeling in the post-AlphaFold era. Brief. Bioinform. 2025, 26, bbaf340. [Google Scholar] [CrossRef] [PubMed]
- Banayan, N.E.; Hsu, A.; Hunt, J.F.; Palmer, A.I.I.I.; Friesner, R.A. Parsing Dynamics of Protein Backbone NH and Side-Chain Methyl Groups using Molecular Dynamics Simulations. J. Chem. Theory Comput. 2024, 20, 6316–6327. [Google Scholar] [CrossRef]
- Kaczmarski, J.A.; Mahawaththa, M.C.; Feintuch, A.; Clifton, B.E.; Adams, L.A.; Goldfarb, D.; Otting, G.; Jackson, C.J. Altered conformational sampling along an evolutionary trajectory changes the catalytic activity of an enzyme. Nat. Commun. 2020, 11, 5945. [Google Scholar] [CrossRef]
- Nussinov, R.; Liu, Y.L.; Zhang, W.A.; Jang, H. Cell phenotypes can be predicted from propensities of protein conformations. Curr. Opin. Struct. Biol. 2023, 83, 102722. [Google Scholar] [CrossRef]
- Wallerstein, J.; Han, X.; Levkovets, M.; Lesovoy, D.; Malmodin, D.; Mirabello, C.; Wallner, B.; Sun, R.H.; Sandalova, T.; Agback, P.; et al. Insights into mechanisms of MALT1 allostery from NMR and AlphaFold dynamic analyses. Commun. Biol. 2024, 7, 868. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Ojoawo, A.; Otten, R.; Apitz, J.M.; Pitsawong, W.; Hömberger, M.; Ovchinnikov, S.; Colwell, L.; Kern, D. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature 2024, 625, 832–839. [Google Scholar] [CrossRef]
- Bryant, P.; Noé, F. Structure prediction of alternative protein conformations. Nat. Commun. 2024, 15, 7328. [Google Scholar] [CrossRef]
- Heo, L.; Feig, M. High-accuracy protein structures by combining machine-learning with physics-based refinement. Proteins 2020, 88, 637–642. [Google Scholar] [CrossRef]
- Ma, P.Y.; Li, D.W.; Bruschweiler, R. Predicting protein flexibility with AlphaFold. Proteins 2023, 91, 847–855. [Google Scholar] [CrossRef]
- Vani, B.P.; Aranganathan, A.; Wang, D.; Tiwary, P. AlphaFold2-RAVE: From Sequence to Boltzmann Ranking. J. Chem. Theory Comput. 2023, 19, 4351–4354. [Google Scholar] [CrossRef]
- Lewis, S.; Hempel, T.; Jimenez-Luna, J.; Gastegger, M.; Xie, Y.; Foong, A.Y.K.; Satorras, V.G.; Abdin, O.; Veeling, B.S.; Zaporozhets, I.; et al. Scalable emulation of protein equilibrium ensembles with generative deep learning. Science 2025, 389, eadv9817. [Google Scholar] [CrossRef]
- He, J.; Lin, P.; Chen, J.; Cao, H.; Huang, S.Y. Model building of protein complexes from intermediate-resolution cryo-EM maps with deep learning-guided automatic assembly. Nat. Commun. 2022, 13, 4066. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, H.; Terashi, G.; Taluja, M.; Kihara, D. DiffModeler: Large macromolecular structure modeling for cryo-EM maps using a diffusion model. Nat. Methods 2024, 21, 2307–2317. [Google Scholar] [CrossRef]
- Zhou, X.G.; Li, Y.; Zhang, C.X.; Zheng, W.; Zhang, G.J.; Zhang, Y. Progressive assembly of multi-domain protein structures from cryo-EM density maps. Nat. Comput. Sci. 2022, 2, 265–275. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Cai, Y.X.; Zhang, B.; Zheng, W.; Freddolino, L.; Zhang, G.J.; Zhou, X.G. DEMO-EM2: Assembling protein complex structures from cryo-EM maps through intertwined chain and domain fitting. Brief. Bioinform. 2024, 25, bbae113. [Google Scholar] [CrossRef]
- Zhao, K.L.; Zhao, P.X.; Wang, S.H.; Xia, Y.H.; Zhang, G.J. FoldPAthreader: Predicting protein folding pathway using a novel folding force field model derived from known protein universe. Genome Biol. 2024, 25, 152. [Google Scholar] [CrossRef]
- Stefanovi’c, C.; Hager, F.F.; Schäffer, C. LytR-CpsA-Psr Glycopolymer Transferases: Essential Bricks in Gram-Positive Bacterial Cell Wall Assembly. Int. J. Mol. Sci. 2021, 22, 908. [Google Scholar] [CrossRef]
- Rajaei, A.; Rowe, H.M.; Neely, M.N. The LCP Family Protein, Psr, Is Required for Cell Wall Integrity and Virulence in Streptococcus agalactiae. Microorganisms 2022, 10, 217. [Google Scholar] [CrossRef]
- Sandalova, T.; Sala, B.M.; Moche, M.; Ljunggren, H.G.; Alici, E.; Henriques-Normark, B.; Agback, T.; Lesovoy, D.; Agback, P.; Achour, A. Crystallographic and NMR Study of Streptococcus pneumonia LCP Protein Psrsp Indicate the Importance of Dynamics in Four Long Loops for Ligand Specificity. Crystals 2024, 14, 1094. [Google Scholar] [CrossRef]
- Kovermann, M.; Rogne, P.; Wolf-Watz, M. Protein dynamics and function from solution state NMR spectroscopy. Q Rev. Biophys. 2016, 49, e6. [Google Scholar] [CrossRef]
- Kumari, P.; Frey, L.; Sobol, A.; Lakomek, N.A.; Riek, R. 15N transverse relaxation measurements for the characterization of μs-ms dynamics are deteriorated by the deuterium isotope effect on 15N resulting from solvent exchange. J. Biomol. NMR 2018, 72, 125–137. [Google Scholar] [CrossRef]
- Kichik, N.; Tarrago, T.; Claasen, B.; Gairi, M.; Millet, O.; Giralt, E. 15N relaxation NMR studies of prolyl oligopeptidase, an 80 kDa enzyme, reveal a pre-existing equilibrium between different conformational states. Chembiochem 2011, 12, 2737–2739. [Google Scholar] [CrossRef]
- Korzhnev, D.M.; Tischenko, E.V.; Arseniev, A.S. Off-resonance effects in 15N T2 CPMG measurements. J. Biomol. NMR 2000, 17, 231–237. [Google Scholar] [CrossRef]
- Lipari, G.; Szabo, A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 2. Analysis of Experimental Results. J. Am. Chem. Soc. 1982, 104, 4559–4570. [Google Scholar] [CrossRef]
- Lipari, G.; Szabo, A. Model-Free Approach to the Interpretation of Nuclear Magnetic-Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. J. Am. Chem. Soc. 1982, 104, 4546–4559. [Google Scholar] [CrossRef]
- Chevelkov, V.; Zhuravleva, A.V.; Xue, Y.; Reif, B.; Skrynnikov, N.R. Combined analysis of 15N relaxation data from solid- and solution-state NMR Spectroscopy. J. Am. Chem. Soc. 2007, 129, 12594–12595. [Google Scholar] [CrossRef]
- del Alamo, D.; DeSousa, L.; Nair, R.M.; Rahman, S.; Meiler, J.; Mchaourab, H.S. Integrated AlphaFold2 and DEER investigation of the conformational dynamics of a pH-dependent APC antiporter. Proc. Natl. Acad. Sci. USA 2022, 119, e2206129119. [Google Scholar] [CrossRef]
- del Alamo, D.; Sala, D.; Mchaourab, H.S.; Meiler, J. Sampling alternative conformational states of transporters and receptors with AlphaFold2. eLife 2022, 11, e80140. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mann, H.B.; Whitney, D.R. On a Test of Whether One of 2 Random Variables Is Stochastically Larger Than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Han, B.; Liu, Y.F.; Ginzinger, S.W.; Wishart, D.S. SHIFTX2: Significantly improved protein chemical shift prediction. J. Biomol. NMR 2011, 50, 43–57. [Google Scholar] [CrossRef]
- Gampp, O.; Kadavath, H.; Riek, R. NMR tools to detect protein allostery. Curr. Opin. Struct. Biol. 2024, 86, 102792. [Google Scholar] [CrossRef]
- Zerbe, B.S.; Hall, D.R.; Vajda, S.; Whitty, A.; Kozakov, D. Relationship between Hot Spot Residues and Ligand Binding Hot Spots in Protein-Protein Interfaces. J. Chem. Inf. Model 2012, 52, 2236–2244. [Google Scholar] [CrossRef]
- Rosell, M.; Fernandez-Recio, J. Hot-spot analysis for drug discovery targeting protein-protein interactions. Expert Opin. Drug Discov. 2018, 13, 327–338. [Google Scholar] [CrossRef]
- Yu, H.R.; Ma, S.; Li, Y.W.; Dalby, P.A. Hot spots-making directed evolution easier. Biotechnol. Adv. 2022, 56, 107926. [Google Scholar] [CrossRef]
- Paquete-Ferreira, J.; Freire, F.; Fernandes, H.S.; Muthukumaran, J.; Ramos, J.; Bryton, J.; Panjkovich, A.; Svergun, D.; Santos, M.F.A.; Correia, M.A.S.; et al. Structural insights of an LCP protein–LytR–from Streptococcus dysgalactiae subs. dysgalactiae through biophysical and in silico methods. Front. Chem. 2024, 12, 1379914. [Google Scholar] [CrossRef]
- Nussinov, R.; Zhang, M.Z.; Liu, Y.L.; Jang, H. AlphaFold, allosteric, and orthosteric drug discovery: Ways forward. Drug Discov. Today 2023, 28, 103551. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; del Sol, A.; Nussinov, R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol. 2008, 378, 1–11. [Google Scholar] [CrossRef]
- Lexa, K.W.; Carlson, H.A. Full Protein Flexibility Is Essential for Proper Hot-Spot Mapping. J. Am. Chem. Soc. 2011, 133, 200–202. [Google Scholar] [CrossRef]
- Robertson, A.J.; Courtney, J.M.; Shen, Y.; Ying, J.F.; Bax, A. Concordance of X-ray and AlphaFold2 Models of SARS-CoV-2 Main Protease with Residual Dipolar Couplings Measured in Solution. J. Am. Chem. Soc. 2021, 143, 19306–19310. [Google Scholar] [CrossRef]
- Lomize, A.; VIu, O.; Arsen’ev, A. Refinement of the spatial structure of the gramicidin A ion channel. Bioorganicheskaia Khimiia 1992, 18, 182–200. [Google Scholar]
- Vögeli, B.; Kazemi, S.; Güntert, P.; Riek, R. Spatial elucidation of motion in proteins by ensemble-based structure calculation using exact NOEs. Nat. Struct. Mol. Biol. 2012, 19, 1053–1057. [Google Scholar] [CrossRef]
- Vögeli, B.; Olsson, S.; Güntert, P.; Riek, R. The Exact NOE as an Alternative in Ensemble Structure Determination. Biophys. J. 2016, 110, 113–126. [Google Scholar] [CrossRef]
- Wenchel, L.; Gampp, O.; Riek, R. Super-resolution NMR spectroscopy. J. Magn. Reson. 2024, 366, 107746. [Google Scholar] [CrossRef]
- Schaefer, K.; Owens, T.W.; Kahne, D.; Walker, S. Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus. J. Am. Chem. Soc. 2018, 140, 2442–2445. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, A.; Hoyland, C.N.; Vollmer, D.; Bisle, S.; Cleverley, R.M.; Johnsborg, O.; Håvarstein, L.S.; Lewis, R.J.; Vollmer, W. Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae. Microb. Drug Resist. 2012, 18, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Li, F.K.K.; Rosell, F.I.; Gale, R.T.; Simorre, J.P.; Brown, E.D.; Strynadka, N.C.J. Crystallographic analysis of Staphylococcus aureus LcpA, the primary wall teichoic acid ligase. J. Biol. Chem. 2020, 295, 2629–2639. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Prestegard, J.H. Direct measurement of dipole-dipole/CSA cross-correlated relaxation by a constant-time experiment. J. Magn. Reson. 2008, 193, 23–31. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ferrage, F.; Dorai, K. Cross-correlation in Biomolecules. In Cross-Relaxation and Cross-Correlation Parameters in NMR: Molecular Approaches; Canet, D., Ed.; Royal Society of Chemistry: London, UK, 2018. [Google Scholar]
- Pervushin, K.V.; Wider, G.; Wüthrich, K. Single transition-to-single transition polarization transfer (ST2-PT) in [15N,1H]-TROSY. J. Biomol. NMR 1998, 12, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Brutscher, B. PRESERVE: Adding variable flip-angle excitation to transverse relaxation-optimized NMR spectroscopy. Magn. Reson. 2024, 5, 131–142. [Google Scholar] [CrossRef]
- Lakomek, N.A.; Ying, J.F.; Bax, A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J. Biomol. NMR 2012, 53, 209–221. [Google Scholar] [CrossRef]
- Zhu, G.; Xia, Y.; Nicholson, L.K.; Sze, K.H. Protein dynamics measurements by TROSY-based NMR experiments. J. Magn. Reson. 2000, 143, 423–426. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493, Erratum in Nature 2024, 636, E4. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Schwerdtfeger, P.; Wales, D.J. 100 Years of the Lennard-Jones Potential. J. Chem. Theory Comput. 2024, 20, 3379–3405. [Google Scholar] [CrossRef]
- Qiu, Y.J.; Shan, W.J.; Zhang, H.Y. Force Field Benchmark of Amino Acids. 3. Hydration with Scaled Lennard-Jones Interactions. J. Chem. Inf. Model. 2021, 61, 3571–3582. [Google Scholar] [CrossRef]
- Yoo, J.; Aksimentiev, A. New tricks for old dogs: Improving the accuracy of biomolecular force fields by pair-specific corrections to non-bonded interactions. Phys. Chem. Chem. Phys. 2018, 20, 8432–8449. [Google Scholar] [CrossRef]
- Robson, S.A.; Dag, Ç.; Wu, H.W.; Ziarek, J.J. TRACT revisited: An algebraic solution for determining overall rotational correlation times from cross-correlated relaxation rates. J. Biomol. NMR 2021, 75, 293–302. [Google Scholar] [CrossRef]
- Yao, L.S.; Vögeli, B.; Ying, J.F.; Bax, A. NMR Determination of Amide N-H Equilibrium Bond Length from Concerted Dipolar Coupling Measurements. J. Am. Chem. Soc. 2008, 130, 16518–16520. [Google Scholar] [CrossRef]
- Chill, J.H.; Louis, J.M.; Baber, J.L.; Bax, A. Measurement of 15N relaxation in the detergent-solubilized tetrameric KcsA potassium channel. J. Biomol. NMR 2006, 36, 123–136. [Google Scholar] [CrossRef]
- Andrae, R.; Schulze-Hartung, T.; Melchior, P. Dos and don’ts of reduced chi-squared. arXiv 2010, arXiv:1012.3754. [Google Scholar] [CrossRef]


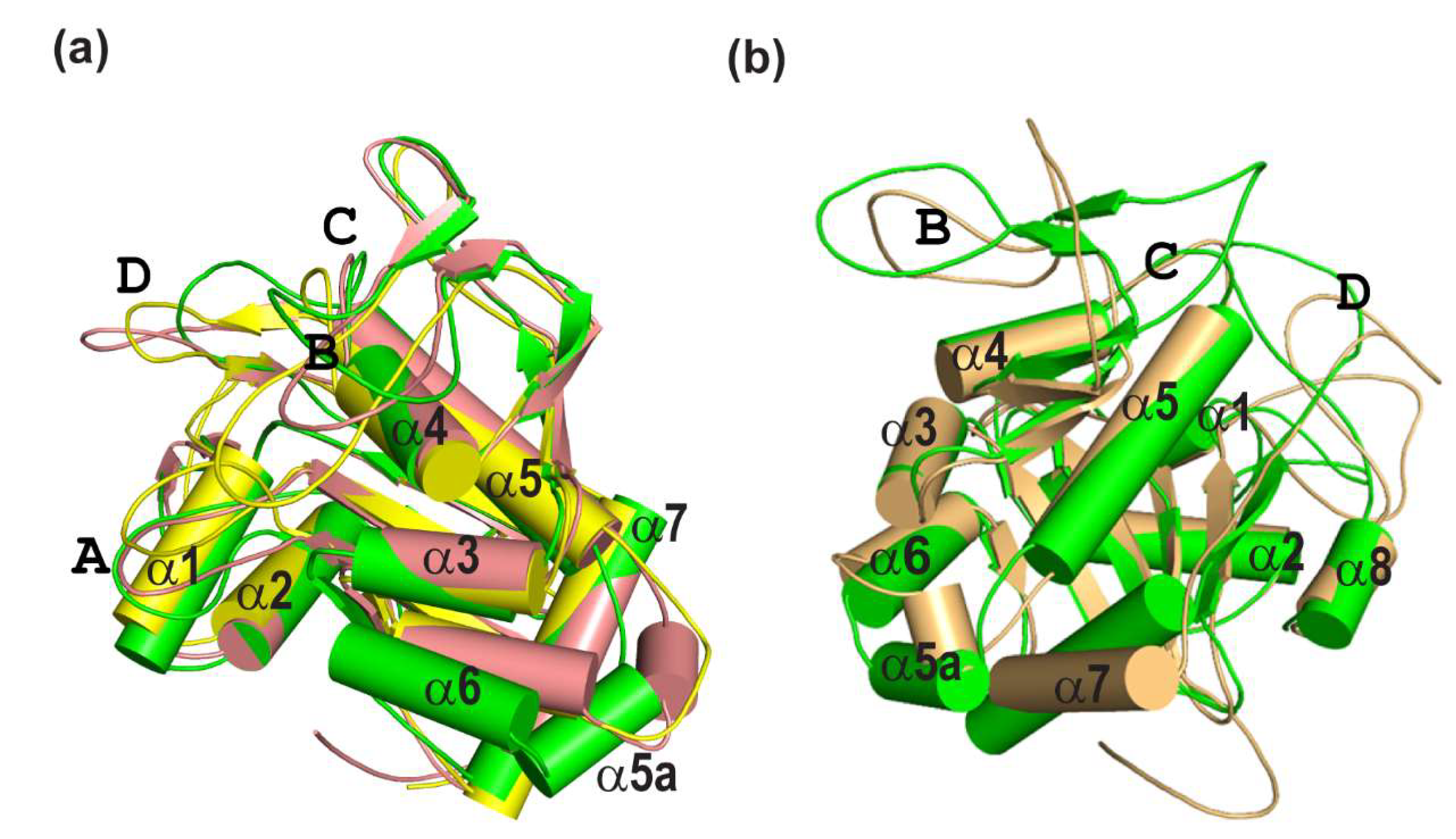
| X-Ray Subunit | (I) 700–1250 ns | (II) 1750–2250 ns | (III) 2500–3000 ns | (IV) 4650–5150 ns |
|---|---|---|---|---|
| A | 1.14 Å (239 Cα) | 1.74 Å (222 Cα) | 1.68 Å (217 Cα) | 1.815 Å (196 Cα) |
| B | 1.02 Å (237 Cα) | 1.55 Å (217 Cα) | 1.645 Å (210 Cα) | 1.778 Å (199 Cα) |
| C | 1.13 Å (241 Cα) | 1.52 Å (205 Cα) | 1.65 Å (201 Cα) | 2.015 Å (201 Cα) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lesovoy, D.; Roshchin, K.; Sala, B.M.; Sandalova, T.; Achour, A.; Agback, T.; Agback, P.; Orekhov, V. Accurate Protein Dynamic Conformational Ensembles: Combining AlphaFold, MD, and Amide 15N(1H) NMR Relaxation. Int. J. Mol. Sci. 2025, 26, 8917. https://doi.org/10.3390/ijms26188917
Lesovoy D, Roshchin K, Sala BM, Sandalova T, Achour A, Agback T, Agback P, Orekhov V. Accurate Protein Dynamic Conformational Ensembles: Combining AlphaFold, MD, and Amide 15N(1H) NMR Relaxation. International Journal of Molecular Sciences. 2025; 26(18):8917. https://doi.org/10.3390/ijms26188917
Chicago/Turabian StyleLesovoy, Dmitry, Konstantin Roshchin, Benedetta Maria Sala, Tatyana Sandalova, Adnane Achour, Tatiana Agback, Peter Agback, and Vladislav Orekhov. 2025. "Accurate Protein Dynamic Conformational Ensembles: Combining AlphaFold, MD, and Amide 15N(1H) NMR Relaxation" International Journal of Molecular Sciences 26, no. 18: 8917. https://doi.org/10.3390/ijms26188917
APA StyleLesovoy, D., Roshchin, K., Sala, B. M., Sandalova, T., Achour, A., Agback, T., Agback, P., & Orekhov, V. (2025). Accurate Protein Dynamic Conformational Ensembles: Combining AlphaFold, MD, and Amide 15N(1H) NMR Relaxation. International Journal of Molecular Sciences, 26(18), 8917. https://doi.org/10.3390/ijms26188917







