The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure
Abstract
1. Introduction
1.1. Gut Microbiota as a Reservoir for Colistin Resistance Genes
1.2. Impact of Colistin on Gut Microbial Diversity and Composition
1.3. Horizontal Gene Transfer of Colistin Resistance Genes in the Gut
1.4. Early-Life Colonization and Neonatal Implications
- Routine screening of mothers with recognized risk factors for antimicrobial resistance—such as prior antibiotic exposure, recent hospitalization, or documented carriage of resistant organisms;
- Enhanced hygiene protocols in delivery and NICU environments;
- Minimization of unnecessary antibiotic use in both mothers and neonates; Awareness and education initiatives for clinicians, healthcare workers, and family units regarding AMR and colistin resistance, especially in neonatal intensive care settings.
1.5. Clinical and Public Health Risks of Gut Carriage of Colistin-Resistant Enterobacterales
1.5.1. Clinical Risks
1.5.2. Public Health Risks
1.6. Alternative Strategies to Colistin for Managing Multidrug-Resistant Enterobacterales
- Cefiderocol, a siderophore cephalosporin with potent activity against Gram-negative pathogens, including mcr-positive strains [99].
- Ceftazidime-avibactam and meropenem-vaborbactam, which are active against KPC-producing organisms, although their efficacy may be limited in mcr-co-harboring isolates [92].
- Tigecycline and eravacycline (new-generation tetracyclines) often retain activity against colistin-resistant isolates and are used for intra-abdominal and complicated soft tissue infections [100].
- Fosfomycin has also shown synergistic potential in combination regimens, especially for urinary tract infections [101].
1.7. Decolonization Strategies for mcr-Positive Gut Colonization
1.7.1. Antibiotic-Based Approaches
1.7.2. Fecal Microbiota Transplantation (FMT)
1.7.3. Bacteriophage Therapy
1.7.4. CRISPR-Cas-Based Systems
1.7.5. Probiotics and Microbiota Modulators
1.7.6. Targeted Antimicrobial Peptides and Nanoparticles
1.8. Artificial Intelligence–Driven Prediction of mcr Emergence
1.9. Antimicrobial Stewardship and Infection Control Implications
2. Conclusions
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Stobberingh, E.E.; Savelkoul, P.H.M.; Wolffs, P.F.G. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 2013, 4, 87. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Liu, J.-H.; Liu, Y.-Y.; Shen, Y.-B.; Yang, J.; Walsh, T.R.; Wang, Y.; Shen, J. Plasmid-Mediated Colistin-Resistance Genes: mcr. Trends Microbiol. 2024, 32, 365–378. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa-Carrillo, D.; Suárez-Cuartín, G.; Golpe, R.; Máiz Carro, L.; Martinez-Garcia, M.A. Inhaled colistimethate sodium in the management of patients with bronchiectasis infected by Pseudomonas aeruginosa: A narrative review of current evidence. Infect. Drug Resist. 2022, 15, 7271–7292. [Google Scholar] [CrossRef]
- Falagas, M.E.; Kasiakou, S.K. Colistin: The revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI). Pathogen Detection Isolates Browser. mcr-11.1 AMR Genotype. Available online: https://www.ncbi.nlm.nih.gov/pathogens/isolates/#AMR_genotypes:mcr-11.1 (accessed on 29 August 2025).
- Sun, J.; Zhang, H.; Liu, Y.H.; Feng, Y. Towards understanding mcr-like colistin resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, H.; Xu, J.; Wang, Y.; Zhang, Q.; Walsh, T.R.; Shao, B.; Wu, C.; Hu, Y.; Gao, H.; et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat. Microbiol. 2018, 3, 1054–1062. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Tawfick, M.M.; Elshamy, A.A.; Mohamed, K.T.; El Menofy, N.G. Gut Commensal Escherichia coli, a High-Risk Reservoir of Transferable Plasmid-Mediated Antimicrobial Resistance Traits. Infect. Drug Resist. 2022, 15, 1077–1091. [Google Scholar] [CrossRef]
- Smelikova, E.; Tkadlec, J.; Krutova, M. How to: Screening for mcr-Mediated Resistance to Colistin. Clin. Microbiol. Infect. 2022, 28, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Turlej-Rogacka, A.; Xavier, B.B.; Janssens, L.; Lammens, C.; Zarkotou, O.; Pournaras, S.; Goossens, H.; Malhotra-Kumar, S. Evaluation of Colistin Stability in Agar and Comparison of Four Methods for MIC Testing of Colistin. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Witherell, K.S.; Price, J.; Bandaranayake, A.D.; Olson, J.; Call, D.R. Circumventing Colistin Resistance by Combining Colistin and Antimicrobial Peptides to Kill Colistin-Resistant and Multidrug-Resistant Gram-Negative Bacteria. J. Glob. Antimicrob. Resist. 2020, 22, 706–712. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Feng, Y.; Zhang, X.; McNally, A.; Zong, Z. New Variant of mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying mcr-1 and blaNDM-5. Antimicrob. Agents Chemother. 2017, 61, e01757-17. [Google Scholar] [CrossRef]
- Yin, W.; Ling, Z.; Dong, Y.; Qiao, L.; Shen, Y.; Liu, Z.; Wu, Y.; Li, W.; Zhang, R.; Walsh, T.R.; et al. Mobile Colistin Resistance Enzyme MCR-3 Facilitates Bacterial Evasion of Host Phagocytosis. Adv. Sci. 2021, 8, 2101336. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel Plasmid-Mediated Colistin Resistance mcr-4 Gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a Novel Transposon-Associated Phosphoethanolamine Transferase Gene, mcr-5, Conferring Colistin Resistance in d-Tartrate Fermenting Salmonella enterica subsp. enterica Serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef]
- AbuOun, M.; Stubberfield, E.J.; Duggett, N.A.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, L.P.; Lemma, F.; Crook, D.W.; Teale, C.; et al. mcr-1 and mcr-2 (mcr-6.1) Variant Genes Identified in Moraxella Species Isolated from Pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017, 72, 2745–2749, Erratum in J. Antimicrob. Chemother. 2018, 73, 2904. [Google Scholar] [CrossRef]
- Phetburom, N.; Boueroy, P.; Chopjitt, P.; Hatrongjit, R.; Akeda, Y.; Hamada, S.; Nuanualsuwan, S.; Kerdsin, A. Klebsiella pneumoniae Complex Harboring mcr-1, mcr-7, and mcr-8 Isolates from Slaughtered Pigs in Thailand. Microorganisms 2021, 9, 2436. [Google Scholar] [CrossRef] [PubMed]
- Eltai, N.O.; Kelly, B.; Al-Mana, H.A.; Ibrahim, E.B.; Yassine, H.M.; Al Thani, A.; Al Maslmani, M.; Lammens, C.; Xavier, B.B.; Malhotra-Kumar, S. Identification of mcr-8 in Clinical Isolates from Qatar and Evaluation of Their Antimicrobial Profiles. Front. Microbiol. 2020, 11, 1954. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Wongsurawat, T.; Jenjaroenpun, P.; Chopjitt, P.; Boueroy, P.; Akeda, Y.; Okada, K.; Iida, T.; Hamada, S.; Kerdsin, A. Genomic Analysis of Carbapenem- and Colistin-Resistant Klebsiella pneumoniae Complex Harbouring mcr-8 and mcr-9 from Individuals in Thailand. Sci. Rep. 2024, 14, 16836. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef]
- Macesic, N.; Blakeway, L.V.; Stewart, J.D.; Hawkey, J.; Wyres, K.L.; Judd, L.M.; Wick, R.R.; Jenney, A.W.; Holt, K.E.; Peleg, A.Y. Silent Spread of Mobile Colistin Resistance Gene mcr-9.1 on IncHI2 ‘Superplasmids’ in Clinical Carbapenem-Resistant Enterobacterales. Clin. Microbiol. Infect. 2021, 27, 1856.e7–1856.e13. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of Novel Mobile Colistin Resistance Gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Cui, Y.; Quan, J.; Zhao, D.; Han, X.; Shi, Q.; Wang, Q.; Jiang, Y.; Du, X.; Li, X.; et al. High Prevalence of Colistin Resistance and mcr-9/10 Genes in Enterobacter spp. in a Tertiary Hospital over a Decade. Int. J. Antimicrob. Agents 2022, 59, 106573. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Campos-Madueno, E.I.; Moradi, M.; Eddoubaji, Y.; Shahi, F.; Moradi, S.; Bernasconi, O.J.; Moser, A.I.; Endimiani, A. Intestinal colonization with multidrug-resistant Enterobacterales: Screening, epidemiology, clinical impact, and strategies to decolonize carriers. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 229–254. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Mirceta, M.; Wick, R.R.; Edwards, D.J.; Thomson, N.R.; Strugnell, R.A.; Pratt, N.; Garlick, J.S.; Watson, K.M.; Pilcher, D.V.; et al. Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 2017, 65, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Portal, E.A.R.; Sands, K.; Farley, C.; Boostrom, I.; Jones, E.; Barrell, M.; Carvalho, M.J.; Milton, R.; Iregbu, K.; Modibbo, F.; et al. Characterisation of colistin resistance in Gram-negative microbiota of pregnant women and neonates in Nigeria. Nat. Commun. 2024, 15, 2302. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in pig production: Chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef]
- EMA & ECDC. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2018. EMA/24309/2020. Available online: https://share.google/5MgbK8Un3eCsnM8YH (accessed on 30 August 2025).
- Nobili, G.; La Bella, G.; Basanisi, M.G.; Damato, A.M.; Coppola, R.; Migliorelli, R.; Rondinone, V.; Leekitcharoenphon, P.; Bortolaia, V.; La Salandra, G. Occurrence and Characterisation of Colistin-Resistant Escherichia coli in Raw Meat in Southern Italy in 2018–2020. Microorganisms 2022, 10, 1805. [Google Scholar] [CrossRef]
- Ahmed, S.; Das, T.; Islam, M.Z.; Herrero-Fresno, A.; Biswas, P.K.; Olsen, J.E. High prevalence of mcr-1-encoded colistin resistance in commensal Escherichia coli from broiler chicken in Bangladesh. Sci. Rep. 2020, 10, 18637. [Google Scholar] [CrossRef]
- Shi, J.; Zhu, H.; Liu, C.; Xie, H.; Li, C.; Cao, X.; Shen, H. Epidemiological and genomic characteristics of global mcr-positive Escherichia coli isolates. Front. Microbiol. 2023, 13, 1105401. [Google Scholar] [CrossRef]
- Azzam, A.; Salem, H.; Nazih, M.; Lotfy, E.M.; Hassan, F.E.; Khaled, H. Prevalence, trends, and molecular insights into colistin resistance among gram-negative bacteria in Egypt: A systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2025, 24, 32. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: A review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef]
- Zhao, J.; Duan, G.; Chang, J.; Wang, H.; Zhu, D.; Li, J.; Zhu, Y. Co-Exposure to Cyazofamid and Polymyxin E: Variations in Microbial Community and Antibiotic Resistance in the Soil–Animal–Plant System. Environ. Res. 2025, 242, 121160. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; Available online: https://www.who.int/publications/i/item/9789241509763 (accessed on 29 August 2025).
- Skov, R.L.; Monnet, D.L. Plasmid-mediated colistin resistance (mcr-1 gene): Three months later, the story unfolds. Eurosurveillance 2016, 21, 30155. [Google Scholar] [CrossRef]
- Andrade, B.G.N.; Goris, T.; Afli, H.; Coutinho, F.H.; Dávila, A.M.R.; Cuadrat, R.R.C. Putative mobilized colistin resistance genes in the human gut microbiome. BMC Microbiol. 2021, 21, 220. [Google Scholar] [CrossRef]
- Dalmasso, G.; Beyrouthy, R.; Brugiroux, S.; Ruppé, E.; Guillouard, L.; Bonnin, V.; Saint-Sardos, P.; Ghozlane, A.; Gaumet, V.; Barnich, N.; et al. Genes mcr improve the intestinal fitness of pathogenic Escherichia coli and balance their lifestyle to commensalism. Microbiome 2023, 11, 12. [Google Scholar] [CrossRef]
- Avellán-Llaguno, R.D.; Xie, A.; Obeten, A.U.; Pan, Z.; Zhang, Y.; Ye, G.; Sun, X.; Huang, Q. Composition, Antibiotic Resistance, and Functionality of the Gut Microbiome in Urban Cats. ACS Environ. Sci. Technol. 2025, 59, 12345–12356. [Google Scholar] [CrossRef]
- Hassan, J.; Osman, M.; Xu, T.; Naas, T.; Schiff, S.J.; Mann, D.; Esseili, M.A.; Deng, X.; Kassem, I.I. Monitoring Sewage and Effluent Water Is an Effective Approach for the Detection of the Mobile Colistin Resistance Genes (mcr) and Associated Bacterial Hosts in the Human Population and Environment in the USA. Environ. Pollut. 2024, 366, 125515. [Google Scholar] [CrossRef]
- Dwiyanto, J.; Huët, M.A.L.; Hussain, M.H.; Chong, C.W.; Rahman, S.; Choo, S.W.; Chin, V.K.; Rahman, R.A.; Chua, E.G.; Foo, S.C.; et al. Social Demographics Determinants for Resistome and Microbiome Variation of a Multiethnic Community in Southern Malaysia. npj Biofilms Microbiomes 2023, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, J.; Li, S.C. Understanding Horizontal Gene Transfer Network in Human Gut Microbiota. Gut Pathog. 2020, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global Monitoring of Antimicrobial Resistance Based on Metagenomics Analyses of Urban Sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.N.; Gaston, J.M.; Dai, C.L.; Zhao, S.; Poyet, M.; Groussin, M.; Yin, X.; Li, L.-G.; van Loosdrecht, M.C.M.; Topp, E.; et al. An Omics-Based Framework for Assessing the Health Risk of Antimicrobial Resistance Genes. Nat. Commun. 2021, 12, 4765. [Google Scholar] [CrossRef]
- Lee, D.-H.; Cha, J.-H.; Kim, D.-W.; Lee, K.; Kim, Y.-S.; Oh, H.-Y.; Cho, Y.-H.; Cha, C.-J. Colistin-degrading proteases confer collective resistance to microbial communities during polymicrobial infections. Microbiome 2022, 10, 129. [Google Scholar] [CrossRef]
- Barlaam, A.; Parisi, A.; Spinelli, E.; Caruso, M.; Di Taranto, P.; Normanno, G. Global Emergence of Colistin-Resistant Escherichia coli in Food Chains and Associated Food Safety Implications: A Review. J. Food Prot. 2019, 82, 1440–1448. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, D.; Fu, S.; Zhang, J.; Zhang, X.; He, J.; Peng, C.; Zhang, Y.; Qiu, Y.; Ye, C.; et al. Metagenomic Sequencing Analysis of the Effects of Colistin Sulfate on the Pig Gut Microbiome. Front. Vet. Sci. 2021, 8, 663820. [Google Scholar] [CrossRef]
- Nation, R.L.; Li, J.; Cars, O.; Couet, W.; Dudley, M.N.; Kaye, K.S.; Mouton, J.W.; Paterson, D.L.; Tam, V.H.; Theuretzbacher, U.; et al. Framework for optimisation of the clinical use of colistin and polymyxin B: The Prato polymyxin consensus. Lancet Infect. Dis. 2015, 15, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Q.; Gao, Y.; Liu, L.; Duan, Y.; Mao, D.; Luo, Y. Colistin and amoxicillin combinatorial exposure alters the human intestinal microbiota and antibiotic resistome in the simulated human intestinal microbiota. Sci. Total Environ. 2021, 750, 141415. [Google Scholar] [CrossRef] [PubMed]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen 2022, 11, e1260. [Google Scholar] [CrossRef]
- Lathakumari, R.H.; Vajravelu, L.K.; Satheesan, A.; Ravi, S.; Thulukanam, J. Antibiotics and the gut microbiome: Understanding the impact on human health. Med. Microecol. 2024, 20, 100106. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2021, 46, fuab049. [Google Scholar] [CrossRef]
- Shayista, H.; Prasad, M.N.N.; Raj, S.N.; Prasad, A.; Lakshmi, S.; Ranjini, H.K.; Manju, K.; Ravikumara; Chouhan, R.S.; Khohlova, O.Y.; et al. Complexity of Antibiotic Resistance and Its Impact on Gut Microbiota Dynamics. Eng. Microbiol. 2025, 5, 100187. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Zhang, J.; Yang, Z.; Liu, L.; Liu, X.; Zheng, Z.; Chan, E.W.; Chen, S. Genetic Characterization of mcr-1-Bearing Plasmids to Depict Molecular Mechanisms Underlying Dissemination. J. Antimicrob. Chemother. 2017, 72, 393–401. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, G.-B.; Zhang, R.; Shen, Y.; Tyrrell, J.M.; Huang, X.; Zhou, H.; Lei, L.; Li, H.; Doi, Y.; et al. Prevalence, Risk Factors, Outcomes, and Molecular Epidemiology of mcr-1-Positive Enterobacteriaceae in Patients and Healthy Adults from China: An Epidemiological and Clinical Study. Lancet Infect. Dis. 2017, 17, 390–399, Erratum in Lancet Infect. Dis. 2017, 17, 897. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. IS26 and the IS6 family: Versatile resistance gene movers and genome reorganizers. Microbiol. Mol. Biol. Rev. 2024, 88, e00119-22. [Google Scholar] [CrossRef]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17, Erratum in mBio 2017, 8, e01166-17. [Google Scholar] [CrossRef] [PubMed]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C.; Mgbeahuruike, A.C.; Ikpendu, C.N.; Okafor, N.A.; Oguttu, J.W. Epidemiology and Traits of Mobile Colistin Resistance (mcr) Gene-Bearing Organisms from Horses. Microorganisms 2022, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Beaber, J.W.; Hochhut, B.; Waldor, M.K. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004, 427, 72–74. [Google Scholar] [CrossRef]
- Kosterlitz, O.; Muñiz Tirado, A.; Wate, C.; Elg, C.; Bozic, I.; Top, E.M.; Kerr, B. Estimating the Transfer Rates of Bacterial Plasmids with an Adapted Luria–Delbrück Fluctuation Analysis. PLoS Biol. 2022, 20, e3001732. [Google Scholar] [CrossRef]
- Ridenhour, B.J.; Metzger, G.A.; France, M.; Gliniewicz, K.; Millstein, J.; Forney, L.J.; Top, E.M. Persistence of Antibiotic Resistance Plasmids in Bacterial Biofilms. Evol. Appl. 2017, 10, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Press, M.O.; Sullivan, S.; Liachko, I.; Top, E.M. Linking the Resistome and Plasmidome to the Microbiome. ISME J. 2019, 13, 2437–2446. [Google Scholar] [CrossRef]
- Yaffe, E.; Relman, D.A. Tracking Microbial Evolution in the Human Gut Using Hi-C Reveals Extensive Horizontal Gene Transfer, Persistence and Adaptation. Nat. Microbiol. 2020, 5, 343–353. [Google Scholar] [CrossRef]
- Tamburini, F.B.; Andermann, T.M.; Bhatt, A.S. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef]
- Catassi, G.; Garcia Mateo, S.; Occhionero, A.S.; Esposito, C.; Giorgio, V.; Aloi, M.; Gasbarrini, A.; Cammarota, G.; Ianiro, G. The importance of gut microbiome in the perinatal period. Eur. J. Pediatr. 2024, 183, 5085–5101. [Google Scholar] [CrossRef]
- Li, W.; Tapiainen, T.; Brinkac, L.; Lorenzi, H.A.; Moncera, K.; Tejesvi, M.V.; Salo, J.; Nelson, K.E. Vertical transmission of gut microbiome and antimicrobial resistance genes in infants exposed to antibiotics at birth. J. Infect. Dis. 2020, 224, 1236–1246. [Google Scholar] [CrossRef]
- Rallis, D.; Giapros, V.; Serbis, A.; Baltogianni, M. Fighting Antimicrobial Resistance in Neonatal Intensive Care Units: Rational Use of Antibiotics in Neonatal Sepsis. Antibiotics 2023, 12, 508. [Google Scholar] [CrossRef] [PubMed]
- Nakwan, N.; Chokephaibulkit, K.; Imberti, R. The Use of Colistin for the Treatment of Multidrug-Resistant Gram-Negative Infections in Neonates and Infants: A Review of the Literature. Pediatr. Infect. Dis. J. 2019, 38, 1107–1112. [Google Scholar] [CrossRef]
- Fouhy, F.; Watkins, C.; Hill, C.J.; O’Shea, C.A.; Nagle, B.; Dempsey, E.M.; O’Toole, P.W.; Stanton, C.; Ross, R.P.; Ryan, C.A. Perinatal factors affect the gut microbiota up to four years after birth. Nat. Commun. 2019, 10, 1517. [Google Scholar] [CrossRef]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Jin, L.; Cai, M.; Wang, Q.; Wu, X.; Wang, S.; Zhang, Y.; Li, H.; Chen, Z.; Liu, P.; et al. Decoding the Origins, Spread, and Global Risks of mcr-9 Gene. eBioMedicine 2024, 108, 105326. [Google Scholar] [CrossRef] [PubMed]
- Farooq, S.; Talat, A.; Khan, A.U. Letter to the Editor: Identification of mcr-9.1 and mcr-10.1 Colistin Resistance Genes in Neonates from Publicly Available Gut Metagenomic Data. Microb. Drug Resist. 2024, 30, 314–316. [Google Scholar] [CrossRef]
- Lee, Y.Q.; Kamar, A.A.; Velayuthana, R.D.; Chong, C.W.; Teh, C.S.J. Clonal relatedness in the acquisition of intestinal carriage and transmission of multidrug resistant (MDR) Klebsiella pneumoniae and Escherichia coli and its risk factors among preterm infants admitted to the neonatal intensive care unit (NICU). Pediatr. Neonatol. 2021, 62, 129–137. [Google Scholar] [CrossRef]
- Bamford, A.; Masini, T.; Williams, P.; Sharland, M.; Gigante, V.; Dixit, D.; Sati, H.; Huttner, B.; Bin Nisar, Y.; Cappello, B.; et al. Tackling the Threat of Antimicrobial Resistance in Neonates and Children: Outcomes from the First WHO-Convened Paediatric Drug Optimisation Exercise for Antibiotics. Lancet Child Adolesc. Health 2024, 8, 456–466. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; Watson, R.L.J.; Chu, M.L.J.N.; Arp, K.; de Waal, W.J.; Schiering, I.; Plötz, F.B.; Willems, R.J.L.; van Schaik, W.; et al. Effect of early-life antibiotics for suspected neonatal sepsis on gut microbiome trajectory and resistance gene profiles: A prospective randomized study. Nat. Commun. 2022, 13, 893. [Google Scholar] [CrossRef]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P.; et al. Stunted Microbiota and Opportunistic Pathogen Colonization in Caesarean-Section Birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef]
- Kiu, R.; Darby, E.M.; Alcon-Giner, C.; Acuna-Gonzalez, A.; Camargo, A.; Lamberte, L.E.; Phillips, S.; Sim, K.; Shaw, A.G.; Clarke, P.; et al. Impact of Early Life Antibiotic and Probiotic Treatment on Gut Microbiome and Resistome of Very-Low-Birth-Weight Preterm Infants. Nat. Commun. 2025, 16, 7569. [Google Scholar] [CrossRef]
- Korpela, K.; Helve, O.; Kolho, K.L.; Saisto, T.; Skogberg, K.; Dikareva, E.; Stefanovic, V.; Salonen, A.; Andersson, S.; de Vos, W.M. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: A proof-of-concept study. Cell 2020, 183, 324–334.e5. [Google Scholar] [CrossRef]
- Dutra, S.V.O.; Sarkar, A.; Yoo, J.Y.; Shaffer-Hudkins, E.; Groer, M. Premature Infant Gut Microbiome Relationships with Childhood Behavioral Scales: Preliminary Insights. Front. Nutr. 2024, 10, 1294549. [Google Scholar] [CrossRef]
- Sarkar, A.; Yoo, J.Y.; Dutra, S.V.O.; Morgan, K.H.; Groer, M. The Association between Early-Life Gut Microbiota and Long-Term Health and Diseases. J. Clin. Med. 2021, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of AmpC β-Lactamase–Producing Enterobacterales. Clin. Infect. Dis. 2021, 72, e169–e183. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Piffanelli, M.; Mazzacane, S. Spread of mcr-1-Driven Colistin Resistance on Hospital Surfaces, Italy. Emerg. Infect. Dis. 2018, 24, 1141–1144. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 2014, 27, 543–574. [Google Scholar] [CrossRef]
- D’Souza, A.W.; Boolchandani, M.; Patel, S.; Owens, S.M.; Siegel, M.; Mathur, S.; Engevik, K.; Atwood, C.; Rood, J.I.; Dantas, G.; et al. Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers. Genome Med. 2021, 13, 79. [Google Scholar] [CrossRef]
- Giani, T.; Sennati, S.; Antonelli, A.; Di Pilato, V.; di Maggio, T.; Mantella, A.; Niccolai, C.; Spinicci, M.; Monasterio, J.; Castellanos, P. High Prevalence of Carriage of mcr-1-Positive Enteric Bacteria among Healthy Children from Rural Communities in the Chaco Region, Bolivia, September to October 2016. Eurosurveillance 2018, 23, 1800115. [Google Scholar] [CrossRef]
- Bich, V.T.N.; Thanh, L.V.; Thai, P.D.; Hoa, N.T.; Tuan, P.Q.; Hien, V.T.; Lan, N.T.; Anh, P.H.; Hanh, T.T.; Dung, T.T.; et al. An exploration of the gut and environmental resistome in a community in northern Vietnam in relation to antibiotic use. Antimicrob. Resist. Infect. Control 2019, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, H.; Hao, L.; Kayani, M.U.R. Identification of Antibiotic Resistance Genes from Whole Genome and Metagenome Sequencing Datasets. One Health Adv. 2025, 3, 14. [Google Scholar] [CrossRef]
- Salleh, M.Z. Addressing antimicrobial resistance: Structural insights into cefiderocol’s mode of action and emerging resistance mechanisms. J. Infect. Public Health 2025, 18, 102871. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Cheung, D.; Adam, H.; Zelenitsky, S.; Golden, A.; Schweizer, F.; Gorityala, B.; Lagacé-Wiens, P.R.S.; Walkty, A.; Gin, A.S.; et al. Review of eravacycline, a novel fluorocycline antibacterial agent. Drugs 2016, 76, 567–588. [Google Scholar] [CrossRef]
- Antonello, R.M.; Principe, L.; Maraolo, A.E.; Viaggi, V.; Pol, R.; Fabbiani, M.; Montagnani, F.; Lovecchio, A.; Luzzati, R.; Di Bella, S. Fosfomycin as Partner Drug for Systemic Infection Management. A Systematic Review of Its Synergistic Properties from In Vitro and In Vivo Studies. Antibiotics 2020, 9, 500. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Rousaki, M.; Kritsotakis, E.I. Cefiderocol: Systematic Review of Mechanisms of Resistance, Heteroresistance and In Vivo Emergence of Resistance. Antibiotics 2022, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Boutin, S.; Kocer, K.; Fiedler, M.O.; Störzinger, D.; Weigand, M.A.; Tan, B.; Richter, D.; Rupp, C.; Mieth, M.; et al. Rapid Development of Cefiderocol Resistance in Carbapenem-Resistant Enterobacter cloacae during Therapy Is Associated with cirA Mutations. Clin. Infect. Dis. 2022, 74, 905–908. [Google Scholar] [CrossRef]
- Wunderink, R.G.; Matsunaga, Y.; Ariyasu, M.; Clevenbergh, P.; Echols, R.; Kaye, K.S.; Kollef, M.; Menon, A.; Pogue, J.M.; Shorr, A.F.; et al. Cefiderocol versus High-Dose, Extended-Infusion Meropenem for the Treatment of Gram-Negative Nosocomial Pneumonia (APEKS-NP): A Randomised, Double-Blind, Phase 3, Non-Inferiority Trial. Lancet Infect. Dis. 2020, 21, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Takemura, M.; Yamano, Y.; Matsunaga, Y.; Ariyasu, M.; Echols, R.; Nagata, T.D. Characterization of Shifts in Minimum Inhibitory Concentrations during Treatment with Cefiderocol or Comparators in the Phase 3 CREDIBLE-CR and APEKS-NP Studies. Open Forum Infect. Dis. 2020, 7 (Suppl. S1), S649–S650. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Lima, P.G.; Oliveira, J.T.A.; Amaral, J.L.; Freitas, C.D.T.; Souza, P.F.N. Synthetic antimicrobial peptides: Characteristics, design, and potential as alternative molecules to overcome microbial resistance. Life Sci. 2021, 278, 119647. [Google Scholar] [CrossRef] [PubMed]
- Rieg, S.; Küpper, M.F.; de With, K.; Schwab, F.; Pfeifer, Y.; Heudorf, U.; Kola, A.; Seifert, H.; Klupp, E.M.; Bohnert, J.A.; et al. Intestinal decolonization of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBL): A retrospective observational study in patients at risk for infection and a brief review of the literature. BMC Infect. Dis. 2015, 15, 475. [Google Scholar] [CrossRef]
- Xenofontos, E.; Renieris, G.; Kalogridi, M.; Papageorgiou, E.; Christodoulou, S.; Spanakis, N.; Rapti, V.; Triantafyllou, I.; Tsagris, V.; Giamarellou, H.; et al. An animal model of limitation of gut colonization by carbapenemase-producing Klebsiella pneumoniae using rifaximin. Sci. Rep. 2022, 12, 3789. [Google Scholar] [CrossRef]
- Alagna, L.; Palomba, E.; Mangioni, D.; Bozzi, G.; Lombardi, A.; Ungaro, R.; Castelli, V.; Prati, D.; Vecchi, M.; Muscatello, A.; et al. Multidrug-Resistant Gram-Negative Bacteria Decolonization in Immunocompromised Patients: A Focus on Fecal Microbiota Transplantation. Int. J. Mol. Sci. 2020, 21, 5619. [Google Scholar] [CrossRef]
- Shin, J.; Lee, J.-H.; Park, S.-H.; Cha, B.; Kwon, K.S.; Kim, H.; Shin, Y.W. Efficacy and Safety of Fecal Microbiota Transplantation for Clearance of Multidrug-Resistant Organisms under Multiple Comorbidities: A Prospective Comparative Trial. Biomedicines 2022, 10, 2404. [Google Scholar] [CrossRef]
- Nooij, S.; Vendrik, K.E.W.; Zwittink, R.D.; Ducarmon, Q.R.; Keller, J.J.; Kuijper, E.J.; Terveer, E.M.; Netherlands Donor Feces Bank Study Group. Long-Term Beneficial Effect of Faecal Microbiota Transplantation on Colonisation of Multidrug-Resistant Bacteria and Resistome Abundance in Patients with Recurrent Clostridioides difficile Infection. Genome Med. 2024, 16, 37. [Google Scholar] [CrossRef]
- Rasmussen, T.S.; Mao, X.; Forster, S.; Larsen, S.B.; Von Münchow, A.; Tranæs, K.D.; Brunse, A.; Larsen, F.; Castro Mejia, J.L.; Adamberg, S.; et al. Overcoming Donor Variability and Risks Associated with Fecal Microbiota Transplants through Bacteriophage-Mediated Treatments. Microbiome 2024, 12, 119. [Google Scholar] [CrossRef]
- Bénard, M.V.; de Bruijn, C.M.A.; Fenneman, A.C.; Wortelboer, K.; Zeevenhoven, J.; Rethans, B.; Herrema, H.J.; van Gool, T.; Nieuwdorp, M.; Benninga, M.A.; et al. Challenges and Costs of Donor Screening for Fecal Microbiota Transplantations. PLoS ONE 2022, 17, e0276323. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Babakhani, S.; Moradi, L.; Karami, S.; Shahbandeh, M.; Mirshekar, M.; Mohebi, S.; Moghadam, M.T. Bacteriophage as a Novel Therapeutic Weapon for Killing Colistin-Resistant MDR and XDR Gram-Negative Bacteria: A Review. Curr. Microbiol. 2021, 78, 4023–4036. [Google Scholar] [CrossRef]
- Geng, X.; Zhang, Z.-D.; Li, Y.-X.; Hao, R.-C.; Yang, Y.-J.; Liu, X.-W.; Li, J.-Y. Fingolimod synergizes and reverses Klebsiella pneumoniae resistance to colistin. Front. Microbiol. 2024, 15, 1396663. [Google Scholar] [CrossRef]
- Wang, X.; Loh, B.; Gordillo Altamirano, F.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 2021, 10, 2205–2219. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, stabilisation and encapsulation of bacteriophage for phage therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef] [PubMed]
- Nale, J.Y.; Spencer, J.; Hargreaves, K.R.; Buckley, A.M.; Trzepinski, P.; Douce, G.R.; Clokie, M.R.J. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob. Agents Chemother. 2016, 60, 968–981. [Google Scholar] [CrossRef]
- Zhao, M.; Li, H.; Gan, D.; Wang, M.; Deng, H.; Yang, Q.E. Antibacterial effect of phage cocktails and phage-antibiotic synergy against pathogenic Klebsiella pneumoniae. mSystems 2024, 9, e00607-24. [Google Scholar] [CrossRef]
- Khambhati, K.; Bhattacharjee, G.; Gohil, N.; Dhanoa, G.K.; Sagona, A.P.; Mani, I.; Bui, N.L.; Chu, D.-T.; Karapurkar, J.K.; Jang, S.H.; et al. Phage engineering and phage-assisted CRISPR-Cas delivery to combat multidrug-resistant pathogens. Bioeng. Transl. Med. 2022, 8, e10381. [Google Scholar] [CrossRef]
- Kim, M.K.; Suh, G.A.; Cullen, G.D.; Perez Rodriguez, S.; Dharmaraj, T.; Chang, T.H.W.; Li, Z.; Chen, Q.; Green, S.I.; Lavigne, R.; et al. Bacteriophage Therapy for Multidrug-Resistant Infections: Current Technologies and Therapeutic Approaches. J. Clin. Investig. 2025, 135, e187996. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Yang, Y.; Zheng, R. Challenges and Opportunities of Phage Therapy for Klebsiella pneumoniae Infections. Appl. Environ. Microbiol. 2024, 90, e01353-24. [Google Scholar] [CrossRef] [PubMed]
- Van Nieuwenhuyse, B.; Merabishvili, M.; Goeders, N.; Vanneste, K.; Bogaerts, B.; de Jode, M.; Ravau, J.; Wagemans, J.; Belkhir, L.; Van der Linden, D. Phage-Mediated Digestive Decolonization in a Gut-On-A-Chip Model: A Tale of Gut-Specific Bacterial Prosperity. Viruses 2024, 16, 1047. [Google Scholar] [CrossRef]
- Ahmed, M.M.; Kayode, H.H.; Okesanya, O.J.; Ukoaka, B.M.; Eshun, G.; Mourid, M.R.; Adigun, O.A.; Ogaya, J.B.; Mohamed, Z.O.; Lucero-Prisno III, D.E.; et al. CRISPR-Cas systems in the fight against antimicrobial resistance: Current status, potentials, and future directions. Infect. Drug Resist. 2024, 17, 5229–5245. [Google Scholar] [CrossRef]
- Kadkhoda, H.; Gholizadeh, P.; Samadi Kafil, H.; Ghotaslou, R.; Pirzadeh, T.; Ahangarzadeh Rezaee, M.; Nabizadeh, E.; Feizi, H.; Aghazadeh, M. Role of CRISPR-Cas systems and anti-CRISPR proteins in bacterial antibiotic resistance. Heliyon 2024, 10, e34692. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Roy, P.; Grenier, F.; Menendez, A.; Burrus, V.; Rodrigue, S. High-Efficiency Delivery of CRISPR-Cas9 by Engineered Probiotics Enables Precise Microbiome Editing. Mol. Syst. Biol. 2021, 17, e10335. [Google Scholar] [CrossRef]
- Duan, C.; Cao, J.; Gu, J.; Zhang, Y.; Wang, Q.; Liang, J.; Xu, L.; Zhang, T. Harnessing the CRISPR-Cas Systems to Combat Antimicrobial Resistance. Front. Microbiol. 2021, 12, 716064. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.N.; Spanogiannopoulos, P.; Soto-Perez, P.; Alexander, M.; Nalley, M.J.; Bisanz, J.E.; Nayak, R.R.; Weakley, A.M.; Yu, F.B.; Turnbaugh, P.J. Phage-Delivered CRISPR-Cas9 for Strain-Specific Depletion and Genomic Deletions in the Gut Microbiome. Cell Rep. 2021, 37, 109930. [Google Scholar] [CrossRef] [PubMed]
- Pursey, E.; Sünderhauf, D.; Gaze, W.H.; Westra, E.R.; van Houte, S. CRISPR-Cas Antimicrobials: Challenges and Future Prospects. PLoS Pathog. 2018, 14, e1006990. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente Tagarro, C.; Martín-González, D.; de Lucas, A.; Bordel, S.; Santos-Beneit, F. Current Knowledge on CRISPR Strategies against Antimicrobial-Resistant Bacteria. Antibiotics 2024, 13, 1141. [Google Scholar] [CrossRef]
- Sachdeva, A.; Tomar, T.; Malik, T.; Bains, A.; Karnwal, A. Exploring probiotics as a sustainable alternative to antimicrobial growth promoters: Mechanisms and benefits in animal health. Front. Sustain. Food Syst. 2025, 8, 1523678. [Google Scholar] [CrossRef]
- Paranga, T.G.; Mitu, I.; Pavel-Tanasa, M.; Rosu, M.F.; Miftode, I.-L.; Constantinescu, D.; Obreja, M.; Plesca, C.E.; Miftode, E. Cytokine Storm in COVID-19: Exploring IL-6 Signaling and Cytokine-Microbiome Interactions as Emerging Therapeutic Approaches. Int. J. Mol. Sci. 2024, 25, 11411. [Google Scholar] [CrossRef]
- Anton-Păduraru, D.-T.; Trofin, F.; Nastase, E.V.; Miftode, R.S.; Miftode, I.-L.; Trandafirescu, M.F.; Cojocaru, E.; Țarcă, E.; Mindru, D.E.; Dorneanu, O.S. The Role of the Gut Microbiota in Anorexia Nervosa in Children and Adults—Systematic Review. Int. J. Mol. Sci. 2024, 25, 41. [Google Scholar] [CrossRef] [PubMed]
- Zollner-Schwetz, I.; Scarpatetti, M.; Pichler, G.; Pux, C.; Klymiuk, I.; Trajanoski, S.; Krause, R. Effect of a multispecies probiotic on intestinal and skin colonization by multidrug-resistant gram-negative bacteria in patients in a long-term care facility: A pilot study. Nutrients 2020, 12, 1586. [Google Scholar] [CrossRef]
- Karbalaei, M.; Keikha, M. Probiotics and intestinal decolonization of antibiotic-resistant microorganisms; a reality or fantasy? Ann. Med. Surg. 2022, 80, 104269. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk Between Akkermansia muciniphila and the Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef]
- Zheng, M.; Han, R.; Yuan, Y.; Xing, Y.; Zhang, W.; Sun, Z.; Liu, Y.; Li, J.; Mao, T. The role of Akkermansia muciniphila in inflammatory bowel disease: Current knowledge and perspectives. Front. Immunol. 2023, 13, 1089600. [Google Scholar] [CrossRef]
- Ljungquist, O.; Kampmann, C.; Resman, F.; Riesbeck, K.; Tham, J. Probiotics for Intestinal Decolonization of ESBL-Producing Enterobacteriaceae: A Randomized, Placebo-Controlled Clinical Trial. Clin. Microbiol. Infect. 2020, 26, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Rauseo, A.M.; Hink, T.; Reske, K.A.; Seiler, S.M.; Bommarito, K.M.; Fraser, V.J.; Burnham, C.A.D.; Dubberke, E.R. A Randomized Controlled Trial of Lactobacillus rhamnosus GG on Antimicrobial-Resistant Organism Colonization. Infect. Control Hosp. Epidemiol. 2022, 43, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.-P.; Lee, C.-C.; Lee, J.-C.; Tsai, P.-J.; Hsueh, P.-R.; Ko, W.-C. The Potential of Probiotics to Eradicate Gut Carriage of Pathogenic or Antimicrobial-Resistant Enterobacterales. Antibiotics 2021, 10, 1086. [Google Scholar] [CrossRef]
- Newman, A.M.; Arshad, M. The Role of Probiotics, Prebiotics and Synbiotics in Combating Multidrug-Resistant Organisms. Clin. Ther. 2020, 42, 1637–1648. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Barberio, D.; Microbiome Therapeutics Innovation Group. Navigating Regulatory and Analytical Challenges in Live Biotherapeutic Product Development and Manufacturing. Front. Microbiomes 2024, 3, 1441290. [Google Scholar] [CrossRef]
- Denkel, L.A.; Gastmeier, P.; Reichardt, C.; Köck, R.; Vehreschild, M.; Seifert, H.; de With, K.; Kern, W.V.; Weichert, S.; Knobloch, J.K. Can probiotics trigger a paradigm shift for cleaning healthcare? Antimicrob. Resist. Infect. Control. 2024, 13, 101. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. Enhancing Mechanistic Research on Precision Probiotics (PAR-25-211). NIH Grants Funding 2024. Available online: https://grants.nih.gov/grants/guide/pa-files/PAR-25-211.html (accessed on 30 August 2025).
- del Olmo, M.; Andreu, C. Current Status of the Application of Antimicrobial Peptides and Their Conjugated Derivatives. Molecules 2025, 30, 3070. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Fang, D.; Li, F.; Wang, Z.; Liu, Y. Vitamin B6 Resensitizes mcr-Carrying Gram-Negative Bacteria to Colistin. Commun. Biol. 2025, 8, 459. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a Therapeutic Tool to Combat Microbial Resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef]
- Arnold, A.; McLellan, S.; Stokes, J.M. How AI Can Help Us Beat AMR. npj Antimicrob. Resist. 2025, 3, 18. [Google Scholar] [CrossRef]
- Li, Y.; Cui, X.; Yang, X.; Liu, G.; Zhang, J. Artificial Intelligence in Predicting Pathogenic Microorganisms’ Antimicrobial Resistance: Challenges, Progress, and Prospects. Front. Cell. Infect. Microbiol. 2024, 14, 1482186. [Google Scholar] [CrossRef]
- Valavarasu, S.; Sangu, Y.; Mahapatra, T. Prediction of Antibiotic Resistance from Antibiotic Susceptibility Testing Results from Surveillance Data Using Machine Learning. Sci. Rep. 2025, 15, 30509. [Google Scholar] [CrossRef]
- Lin, B.; Yan, S.; Zhen, B. A Machine Learning Method for Predicting Molecular Antimicrobial Activity. Sci. Rep. 2025, 15, 6559. [Google Scholar] [CrossRef]
- Renz, J.; Dauda, K.A.; Aga, O.N.L.; Diaz-Uriarte, R.; Löhr, I.H.; Blomberg, B.; Johnston, I.G. Evolutionary Accumulation Modelling in AMR: Machine Learning to Infer and Predict Evolutionary Dynamics of Multi-Drug Resistance. arXiv 2024, arXiv:2411.00219. [Google Scholar] [CrossRef]
- López-Cortés, X.A.; Manríquez-Troncoso, J.M.; Hernández-García, R.; Peralta, D. MSDeepAMR: Antimicrobial Resistance Prediction Based on Deep Neural Networks and Transfer Learning. Front. Microbiol. 2024, 15, 1361795. [Google Scholar] [CrossRef]
- Zhou, G.; Janarthanan, S.; Lu, Y.; Hu, P. CL-MFAP: A Contrastive Learning-Based Multimodal Foundation Model for Molecular Property Prediction and Antibiotic Screening. arXiv 2025, arXiv:2502.11001. [Google Scholar] [CrossRef]
- Theodosiou, A.A.; Read, R.C. Artificial intelligence, machine learning and deep learning: Potential resources for the infection clinician. J. Infect. 2023, 87, 287–294. [Google Scholar] [CrossRef]
- Kim, J.I.; Maguire, F.; Tsang, K.K.; Gouliouris, T.; Peacock, S.J.; McAllister, T.A.; McArthur, A.G.; Beiko, R.G. Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective. Clin. Microbiol. Rev. 2022, 35, e00179-21. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Chen, L.; Kreiswirth, B.N.; Clancy, C.J. In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem-susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob. Agents Chemother. 2017, 61, e00079-17. [Google Scholar] [CrossRef] [PubMed]
- Cannatelli, A.; Giani, T.; D’Andrea, M.M.; Di Pilato, V.; Arena, F.; Conte, V.; Tryfinopoulou, K.; Vatopoulos, A.; Rossolini, G.M. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014, 58, 5696–5703. [Google Scholar] [CrossRef]
- Sargianou, M.; Stathopoulos, P.; Vrysis, C.; Tzvetanova, I.D.; Falagas, M.E. New β-Lactam/β-Lactamase Inhibitor Combination Antibiotics. Pathogens 2025, 14, 307. [Google Scholar] [CrossRef]
- El-Sokkary, R.; Erdem, H.; Kullar, R.; Pekok, A.U.; Amer, F.; Grgić, S.; Carevic, B.; El-Kholy, A.; Liskova, A.; Özdemir, M.; et al. Self-Reported Antibiotic Stewardship and Infection Control Measures from 57 Intensive Care Units: An International ID-IRI Survey. J. Infect. Public Health 2022, 15, 950–954. [Google Scholar] [CrossRef]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Aidara-Kane, A.; Andremont, A.; Collignon, P.; Angulo, F.J.; Conly, J.M.; Minato, Y.; Silbergeld, E.K.; McEwen, S.A.; Balkhy, H.; Friedman, C.; et al. World Health Organization guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob. Resist. Infect. Control. 2018, 7, 7. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report; WHO: Geneva, Switzerland, 2022; Available online: https://www.who.int/publications/i/item/9789240062702 (accessed on 30 August 2025).
- Do, P.C.; Assefa, Y.A.; Batikawai, S.M.; Reid, S.A. Strengthening Antimicrobial Resistance Surveillance Systems: A Scoping Review. BMC Infect. Dis. 2023, 23, 593. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Khong, D.T.; Le, H.V.; Tran, H.T.; Phan, Q.N.; Le, H.T.T.; Kawahara, R.; Yamamoto, Y. Quantitative analysis of colistin-resistant Escherichia coli in retail meat from local Vietnamese markets. Biomed. Res. Int. 2021, 2021, 6678901. [Google Scholar] [CrossRef] [PubMed]
- Talat, A.; Usmani, A.; Khan, A.U. Detection of E. coli IncX1 Plasmid-Mediated mcr-5.1 Gene in an Indian Hospital Sewage Water Using Shotgun Metagenomic Sequencing: A First Report. Microbiol. Drug Resist. 2022, 28, 759–764. [Google Scholar] [CrossRef]
- Trung, N.V.; Matamoros, S.; Carrique-Mas, J.J.; Nghia, N.H.; Nhung, N.T.; Chieu, T.T.B.; Mai, H.H.; van Rooijen, W.; Campbell, J.; Wagenaar, J.A.; et al. Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerg. Infect. Dis. 2017, 23, 529–532. [Google Scholar] [CrossRef]
- Schmitt, H.; Blaak, H.; Kemper, M.; de Roda Husman, A.M. Wastewater Based Surveillance of AMR in the Netherlands; Rijksinstituut voor Volksgezondheid en Milieu (RIVM): Bilthoven, The Netherlands, 2023; Available online: https://www.rivm.nl/sites/default/files/2023-02/1.%20rioolwatersurveillance%20SNIV%20dag%20%28H.%20Schmitt%29.pdf (accessed on 28 July 2025).
- Miftode, I.-L.; Leca, D.; Miftode, R.-S.; Roşu, F.; Plesca, C.; Loghin, I.; Timpau, A.S.; Mitu, I.; Mititiuc, I.; Dorneanu, O.; et al. The Clash of the Titans: COVID-19, Carbapenem-Resistant Enterobacterales, and First mcr-1-Mediated Colistin Resistance in Humans in Romania. Antibiotics 2023, 12, 324. [Google Scholar] [CrossRef]
- Maciuca, I.E.; Cummins, M.L.; Cozma, A.P.; Rimbu, C.M.; Guguianu, E.; Panzaru, C.; Licker, M.; Szekely, E.; Flonta, M.; Djordjevic, S.P.; et al. Genetic Features of mcr-1-Mediated Colistin Resistance in CMY-2-Producing Escherichia coli from Romanian Poultry. Front. Microbiol. 2019, 10, 2267. [Google Scholar] [CrossRef]
- Schaumburg, F.; Sertic, S.M.; Correa-Martinez, C.; Mellmann, A.; Köck, R.; Becker, K. Acquisition and colonization dynamics of antimicrobial-resistant bacteria during international travel: A prospective cohort study. Clin. Microbiol. Infect. 2019, 25, 1287.e1–1287.e7. [Google Scholar] [CrossRef]
- Mistry, K.; Thumbi, D.; Li, X.R.; Charlebois, A.; Avery, B.P.; Deckert, A.E.; Cormier, A.C.; Murphy, C.; Kearney, A.; Campbell, J.; et al. One Health Study of Mobile Colistin Resistance (mcr) in Salmonella enterica in Canada, 2017–2022. Microbiol. Spectr. 2025, 13, e02156-24. [Google Scholar] [CrossRef]
- Wang, P.; Smith, A.L. Emergence of mobile colistin resistance genes within Los Angeles County wastewater. Environ. Sci. Technol. Lett. 2023, 10, 299–305. [Google Scholar] [CrossRef]
- De La Cadena, E.; Mahecha, M.; Velandia, A.M.; García-Betancur, J.C.; Rojas, L.J.; Porras, J.; Pallares, C.; Villegas, M.V. Identification of mcr-1 Genes and Characterization of Resistance Mechanisms to Colistin in Escherichia coli Isolates from Colombian Hospitals. Antibiotics 2023, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Lentz, S.A.M.; Dalmolin, T.V.; Barth, A.L.; Martins, A.F. mcr-1 gene in Latin America: How is it disseminated among humans, animals, and the environment? Front. Public Health 2021, 9, 648940. [Google Scholar] [CrossRef]
- Bastidas-Caldes, C.; de Waard, J.H.; Salgado, M.S.; Villacís, M.J.; Coral-Almeida, M.; Yamamoto, Y.; Calvopiña, M. Worldwide Prevalence of mcr-mediated Colistin-Resistance Escherichia coli in Isolates of Clinical Samples, Healthy Humans, and Livestock—A Systematic Review and Meta-Analysis. Pathogens 2022, 11, 659. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Otter, J.A.; Mutters, N.T.; Tacconelli, E.; Gikas, A.; Holmes, A.H. Controversies in guidelines for the control of multidrug-resistant Gram-negative bacteria in EU hospitals. Clin. Microbiol. Infect. 2015, 21, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, A.M.; Janice, J.; Hjerde, E.; Simonsen, G.S.; Hanssen, A.-M. Shotgun-Metagenomics Based Prediction of Antibiotic Resistance and Virulence Determinants in Staphylococcus aureus from Periprosthetic Tissue on Blood Culture Bottles. Sci. Rep. 2021, 11, 20848. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yin, X.; Xu, X.; Zhang, T. Species-Resolved Profiling of Antibiotic Resistance Genes in Complex Metagenomes through Long-Read Overlapping with Argo. Nat. Commun. 2025, 16, 1744. [Google Scholar] [CrossRef]
- Rajput, V.; Pramanik, R.; Nannaware, K.; Shah, P.; Bhalerao, A.; Jain, N.; Shashidhara, L.S.; Kamble, S.; Dastager, S.; Dharne, M. Metagenomics-Based Longitudinal Monitoring of Antibiotic Resistome and Microbiome in the Inlets of Wastewater Treatment Plants in an Indian Megacity. Sci. Total Environ. 2025, 986, 179691. [Google Scholar] [CrossRef]
- Naylor, N.R.; Hasso-Agopsowicz, M.; Kim, C.; Ma, Y.; Frost, I.; Abbas, K.; Aguilar, G.; Fuller, N.; Robotham, J.V.; Jit, M. The Global Economic Burden of Antibiotic-Resistant Infections and the Potential Impact of Bacterial Vaccines: A Modelling Study. BMJ Glob. Health 2025, 10, e016249. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, M.; Sameni, F.; Bostanshirin, N.; Yaslianifard, S.; Khosravi-Dehaghi, N.; Nasiri, M.J.; Goudarzi, M.; Hashemi, A.; Hajikhani, B. Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 2022, 29, 444–461. [Google Scholar] [CrossRef] [PubMed]
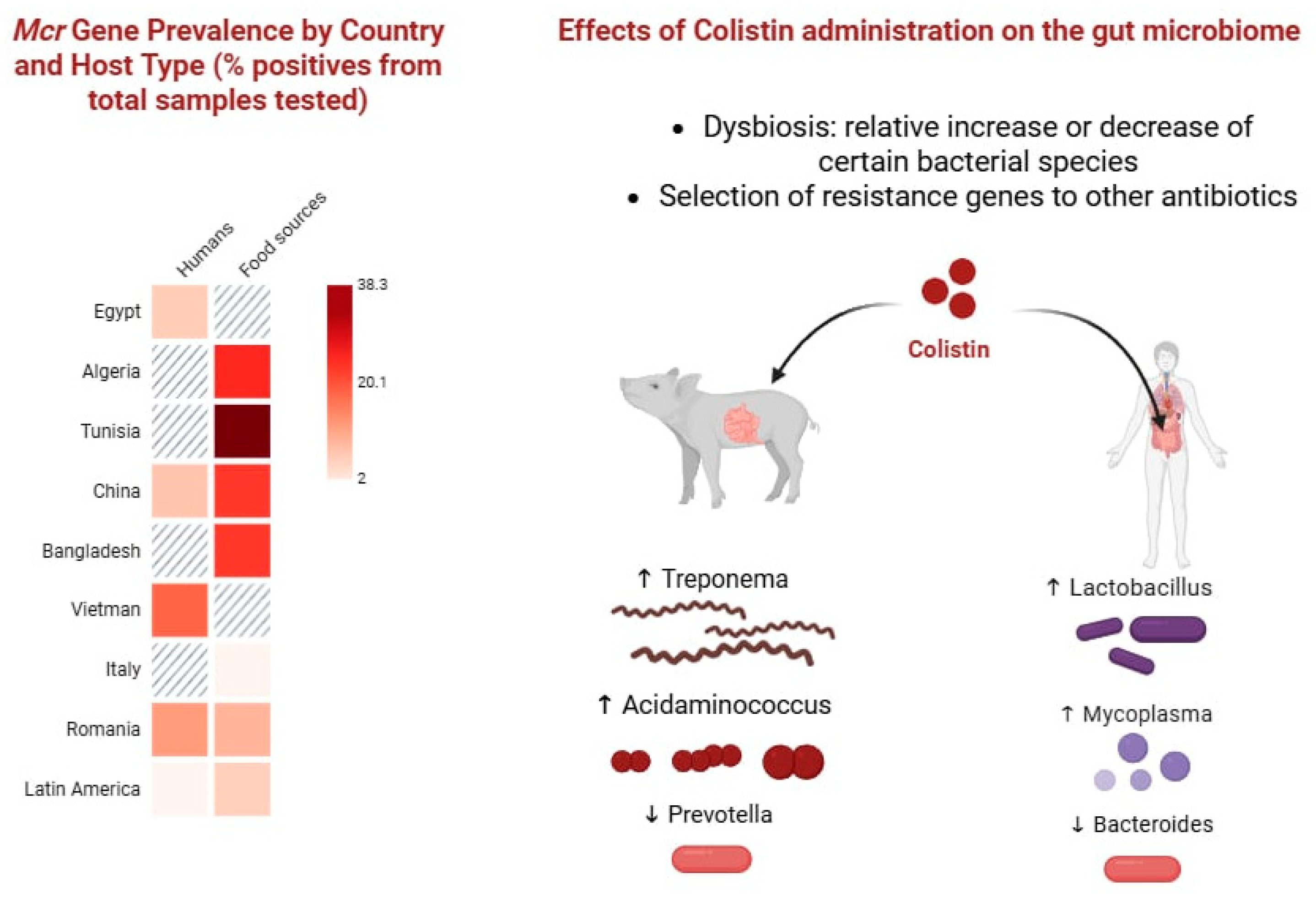
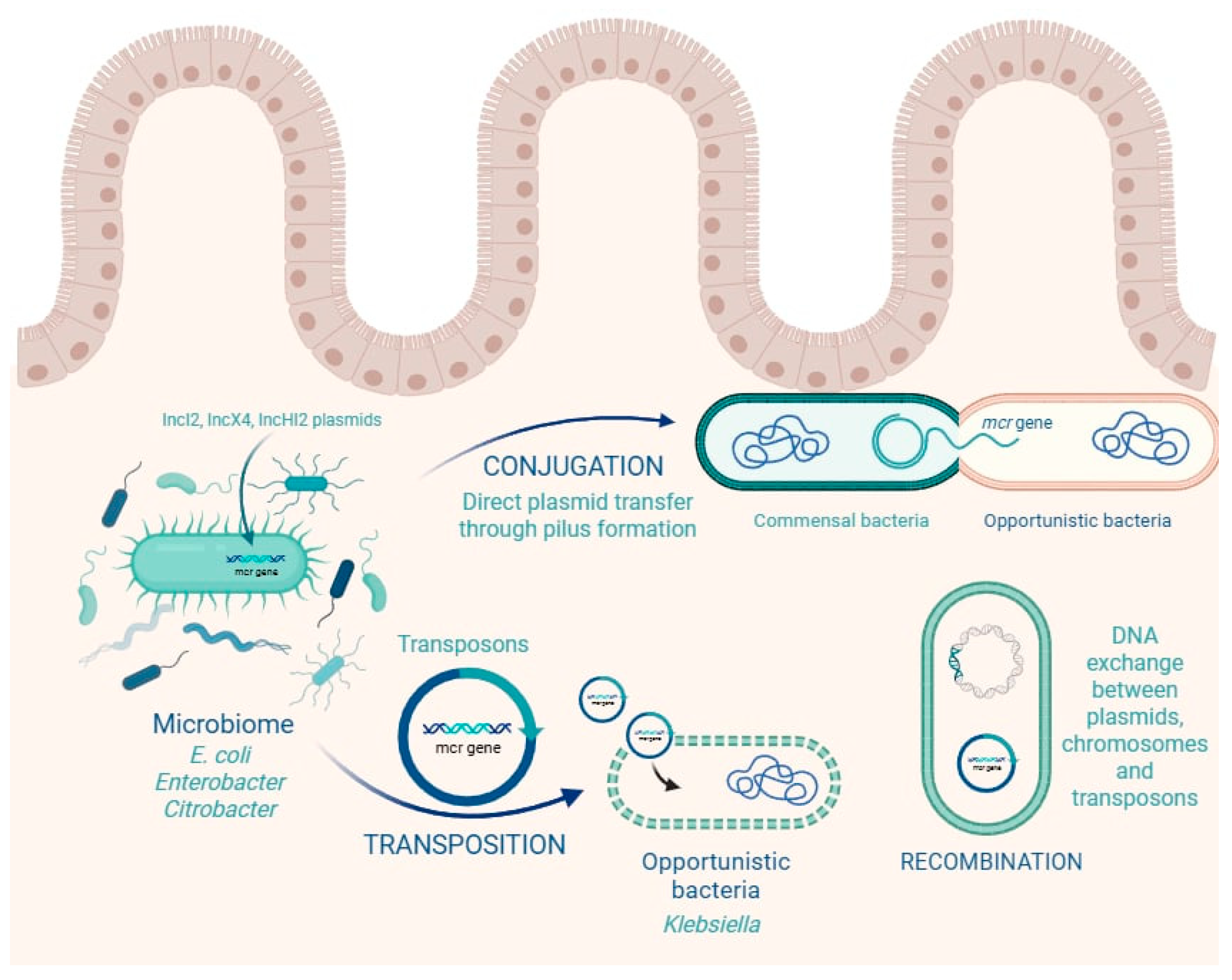
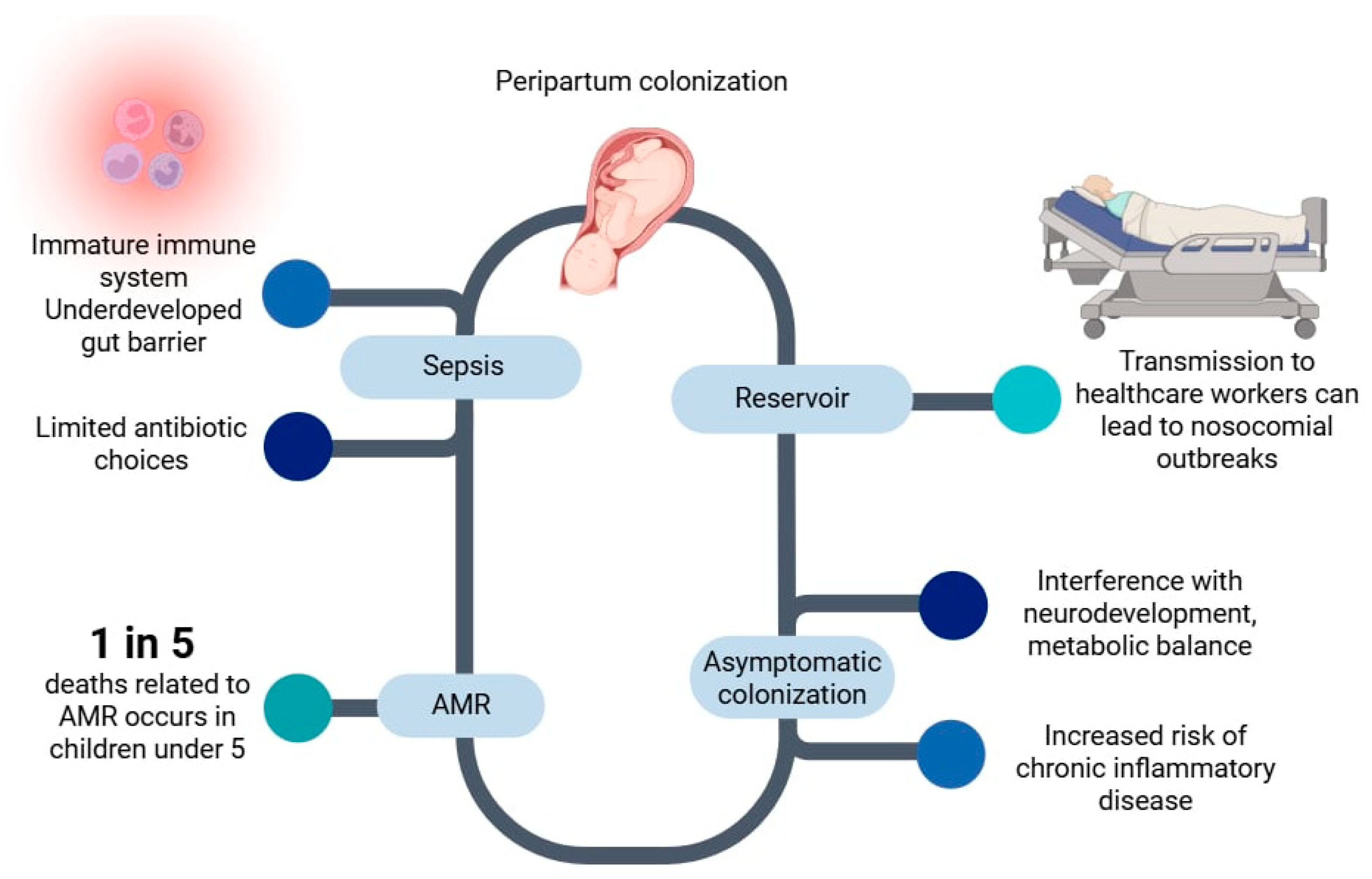
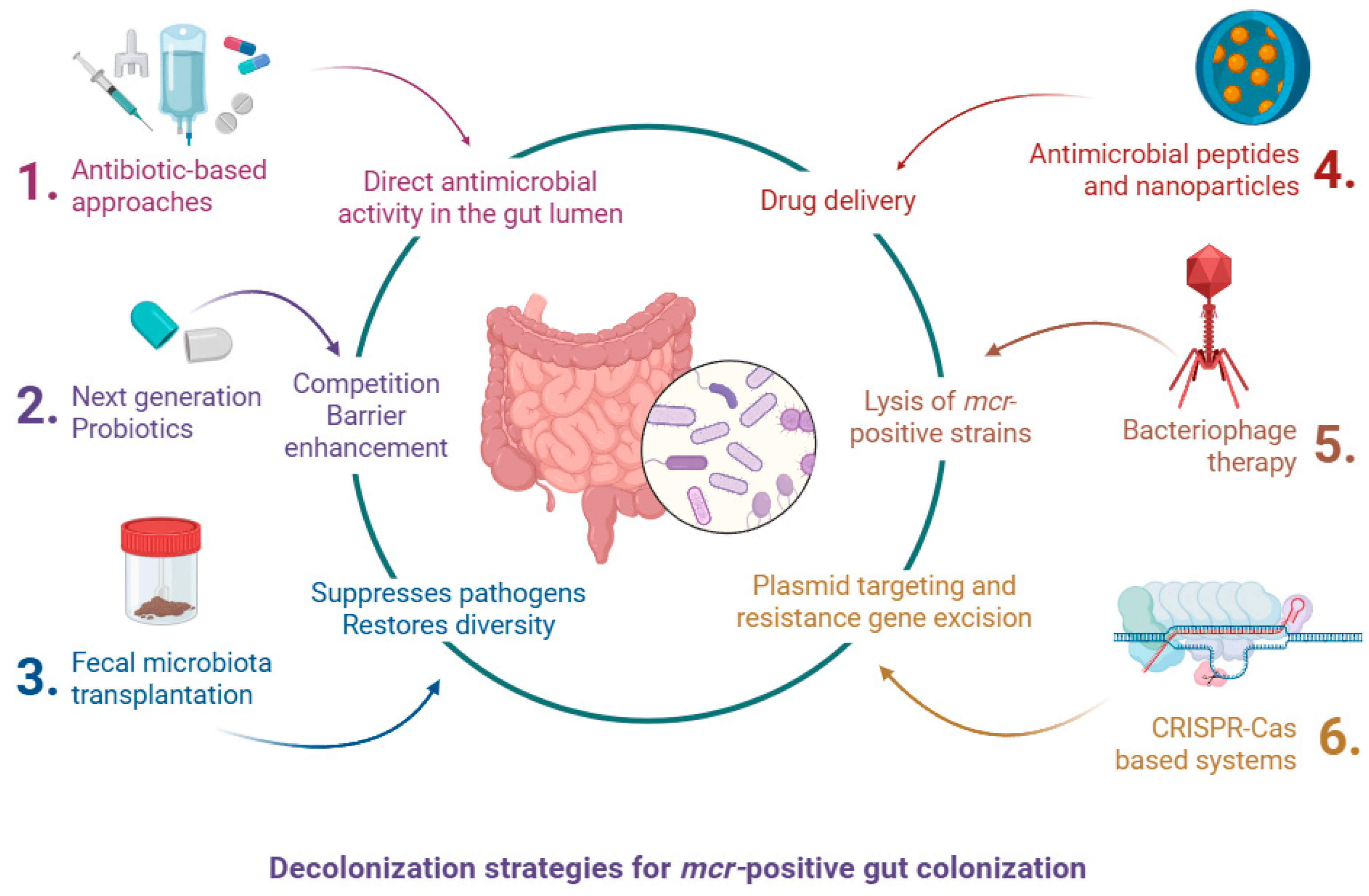
| Gene | Year and Country of First Report | Typical Hosts and Sources | Plasmid/Location | Horizontal Transfer Efficiency | Colistin MIC Impact | Notes |
|---|---|---|---|---|---|---|
| mcr-1 | 2015, China (E. coli from pigs and meat) | E. coli, Klebsiella Enterobacterales | IncI2, IncX4, IncHI2 | High—highly conjugative on diverse plasmid backbone widely disseminated | 2–8 mg/L [17] | Most globally prevalent; food/clinical link |
| mcr-2 | 2016, Belgium (pigs) | E. coli | IncX4 | Efficient but less prevalent | Similar to mcr-1, commonly 4–8 mg/L in E. coli [18,19] | Limited global spread |
| mcr-3 | 2017, China (pig feces) | E. coli, Salmonella, Aeromonas | IncHI2, IncP, IncFII | Moderate, some chromosomal insertions | 4–8 mg/L in E. coli, may be higher in other hosts (e.g., Aeromonas) [20,21] | Environmental reservoirs important |
| mcr-4 | 2017, Italy (swine feces) | Salmonella enterica, E. coli | IncHI2, ColE-like | Transferable | 4–8 mg/L [22] | Sporadic |
| mcr-5 | 2017, Germany (animal feces, food) | Salmonella, E. coli | IncX1, chromosomal | Integrated into chromosomes; conjugation possible but less efficient. | ~8 mg/L in Salmonella Paratyphi B ~4 mg/L in other isolates [23] | Detected mostly in food chain |
| mcr-6 | 2017, UK (pig) | Moraxella spp. | Chromosomal | Not plasmid-borne Chromosomally located in Moraxella; no evidence of efficient conjugative transfer. | Low–moderate (1–2 mg/L) [24] | Rare |
| mcr-7 | 2018, China (chicken) | K. pneumoniae | IncI1 | Transferable Experimentally shown to mobilize between Enterobacterales. | 4–8 mg/L [25] | Limited distribution |
| mcr-8 | 2018, China (cattle, humans) | K. pneumoniae, Raoultella | IncFII(K), IncHI2 | Efficient in K. pneumoniae Demonstrated conjugation | 8–16 mg/L in K. pneumoniae; 16 mg/L have been reported for mcr-8.1 [26,27] | Clinical outbreaks in Asia |
| mcr-9 | 2019, USA (human/animal/food/environment) | Serratia, Morganella | IncHI2, IncFII | Conjugative, inducible (qseB/qseC regulation) | frequently “silent” with MIC ≤ 2 mg/L unless induced; upon induction, MIC may increase [28,29] | Widespread, but variable phenotypic resistance |
| mcr-10 | 2020, China (clinical Enterobacter roggenkampii) | Enterobacter spp., Klebsiella | IncFII(K) | Transferable Conjugation demonstrated | ~4 mg/L (E. roggenkampii), but broader ranges from 4 up to >16–128 mg/L in Enterobacter spp.; inducibility has been observed [30,31] | Emerging, low prevalence |
| Strengths | Weaknesses | Opportunities | Threats |
|---|---|---|---|
| Established AMR surveillance platforms (e.g., GLASS, national veterinary monitoring programs) provide a ready framework into which mcr surveillance can be embedded across clinical, veterinary, and environmental compartments. | Sparse randomized controlled trials for decolonization strategies; heterogeneous designs limiting inference | Incorporation of wastewater-based metagenomics and high-throughput sequencing pipelines could enable near real-time monitoring of mcr gene circulation, improving early detection of resistance hotspots. | Co-selection pressures from non-polymyxin antibiotics, disinfectants, and heavy metals sustaining mcr despite reduced colistin use |
| Implementation of antimicrobial stewardship policies and colistin-use restrictions in multiple regions demonstrates feasibility of coordinated policy-driven interventions. | Limited laboratory capacity and lack of standardized susceptibility testing methods | Phase-out of colistin in agriculture, vaccination and biosecurity measures in livestock, and improved sanitation infrastructure | Global dissemination of high-risk clones and plasmids via travel, trade, and wildlife vectors |
| Increasing accessibility of sequencing and bioinformatics tools supports comprehensive resistome characterization and comparative genomics across sectors. | Data fragmentation between human, veterinary, and environmental sectors impede integrated responses | Network-informed targeting of hub taxa or plasmid backbones driving dissemination | Economic, regulatory, and ethical challenges may slow translation of microbiome-based, phage, or CRISPR-driven decolonization approaches into clinical or agricultural practice. |
| Growing recognition of One Health approaches in policy frameworks | Incomplete epidemiological data from LMICs creates geographic blind spots in global risk maps | Development of standardized trial protocols for horizontal gene transfer inhibition, microbiota restoration, and decolonization therapies could generate actionable clinical evidence. | Risk of delayed global coordination leading to entrenched resistance reservoirs |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miftode, I.-L.; Vâţă, A.; Miftode, R.-Ş.; Oancea, A.F.; Pasăre, M.-A.; Parângă, T.G.; Miftode, E.G.; Mititiuc, I.L.; Radu, V.D. The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure. Int. J. Mol. Sci. 2025, 26, 8899. https://doi.org/10.3390/ijms26188899
Miftode I-L, Vâţă A, Miftode R-Ş, Oancea AF, Pasăre M-A, Parângă TG, Miftode EG, Mititiuc IL, Radu VD. The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure. International Journal of Molecular Sciences. 2025; 26(18):8899. https://doi.org/10.3390/ijms26188899
Chicago/Turabian StyleMiftode, Ionela-Larisa, Andrei Vâţă, Radu-Ştefan Miftode, Alexandru Florinel Oancea, Maria-Antoanela Pasăre, Tudoriţa Gabriela Parângă, Egidia Gabriela Miftode, Irina Luanda Mititiuc, and Viorel Dragoş Radu. 2025. "The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure" International Journal of Molecular Sciences 26, no. 18: 8899. https://doi.org/10.3390/ijms26188899
APA StyleMiftode, I.-L., Vâţă, A., Miftode, R.-Ş., Oancea, A. F., Pasăre, M.-A., Parângă, T. G., Miftode, E. G., Mititiuc, I. L., & Radu, V. D. (2025). The Gut Microbiome and Colistin Resistance: A Hidden Driver of Antimicrobial Failure. International Journal of Molecular Sciences, 26(18), 8899. https://doi.org/10.3390/ijms26188899











