Physiological and Transcriptome Analyses Reveal the Important Role of Microbial Fertilizer in the Response of Sugar Beet Seedlings to Saline-Alkali Stress
Abstract
1. Introduction
2. Results
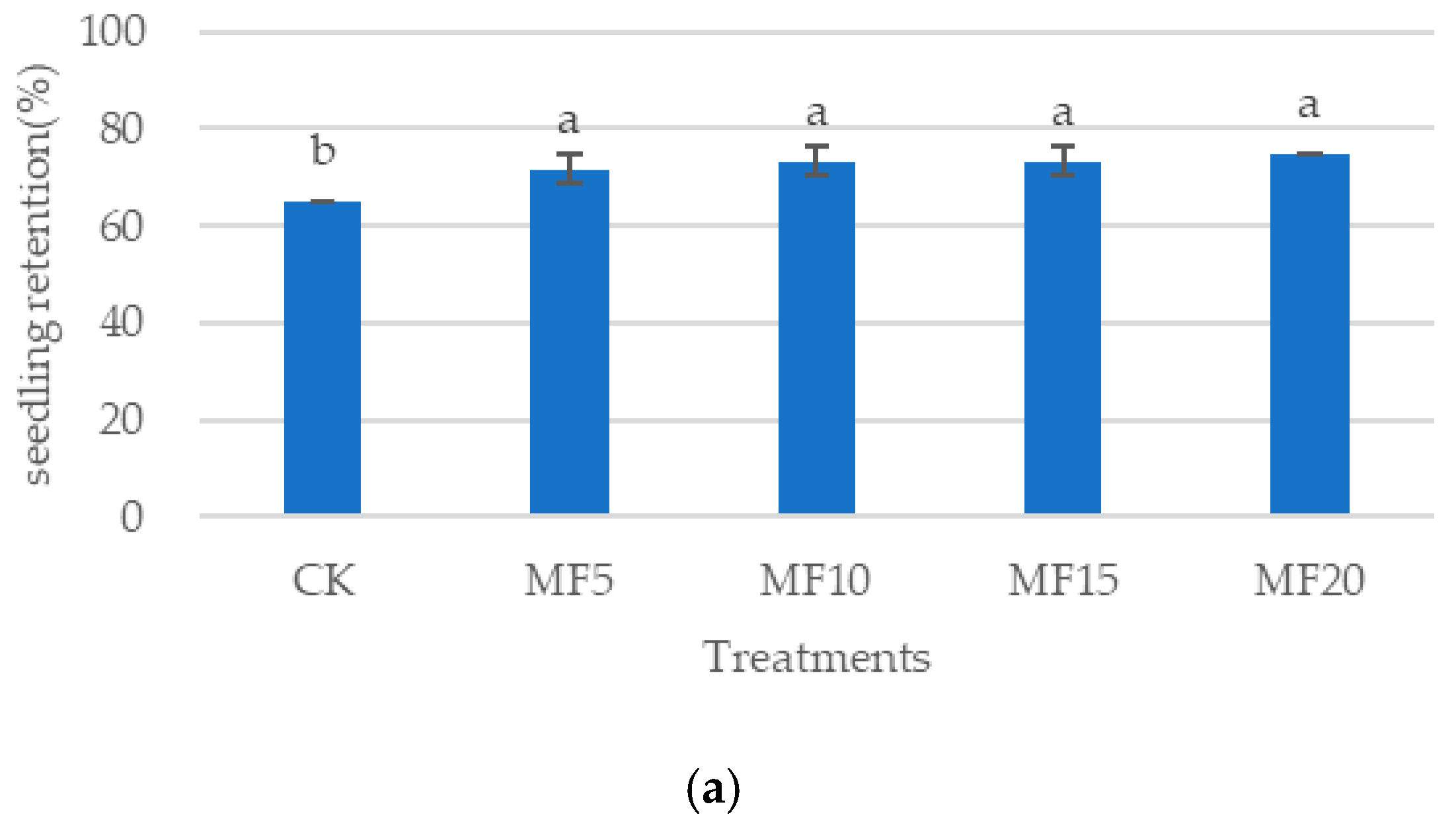
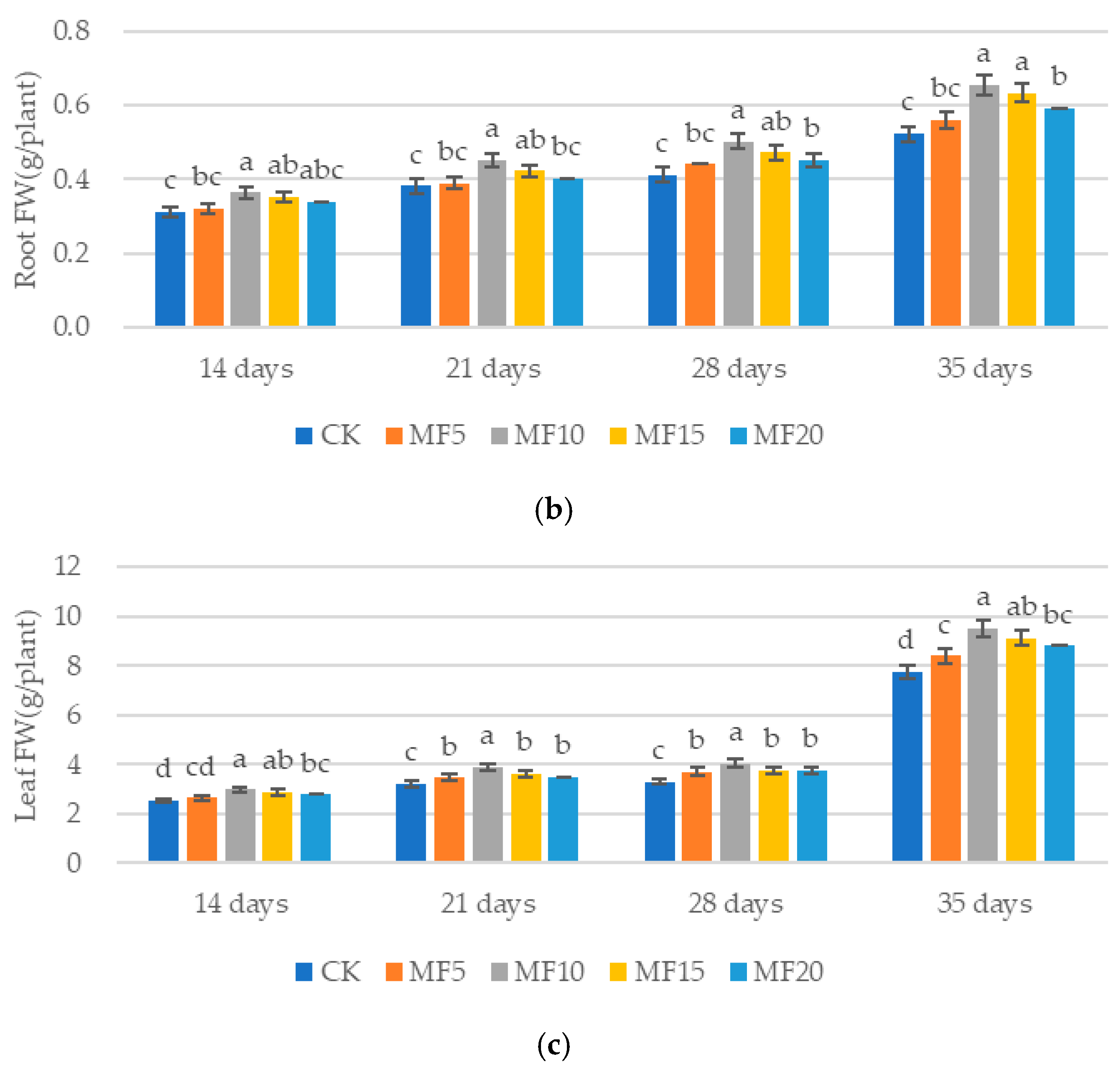
2.1. Impact of Microbial Fertilizer on the Growth of Saline–Alkali-Stressed Sugar Beet Seedlings
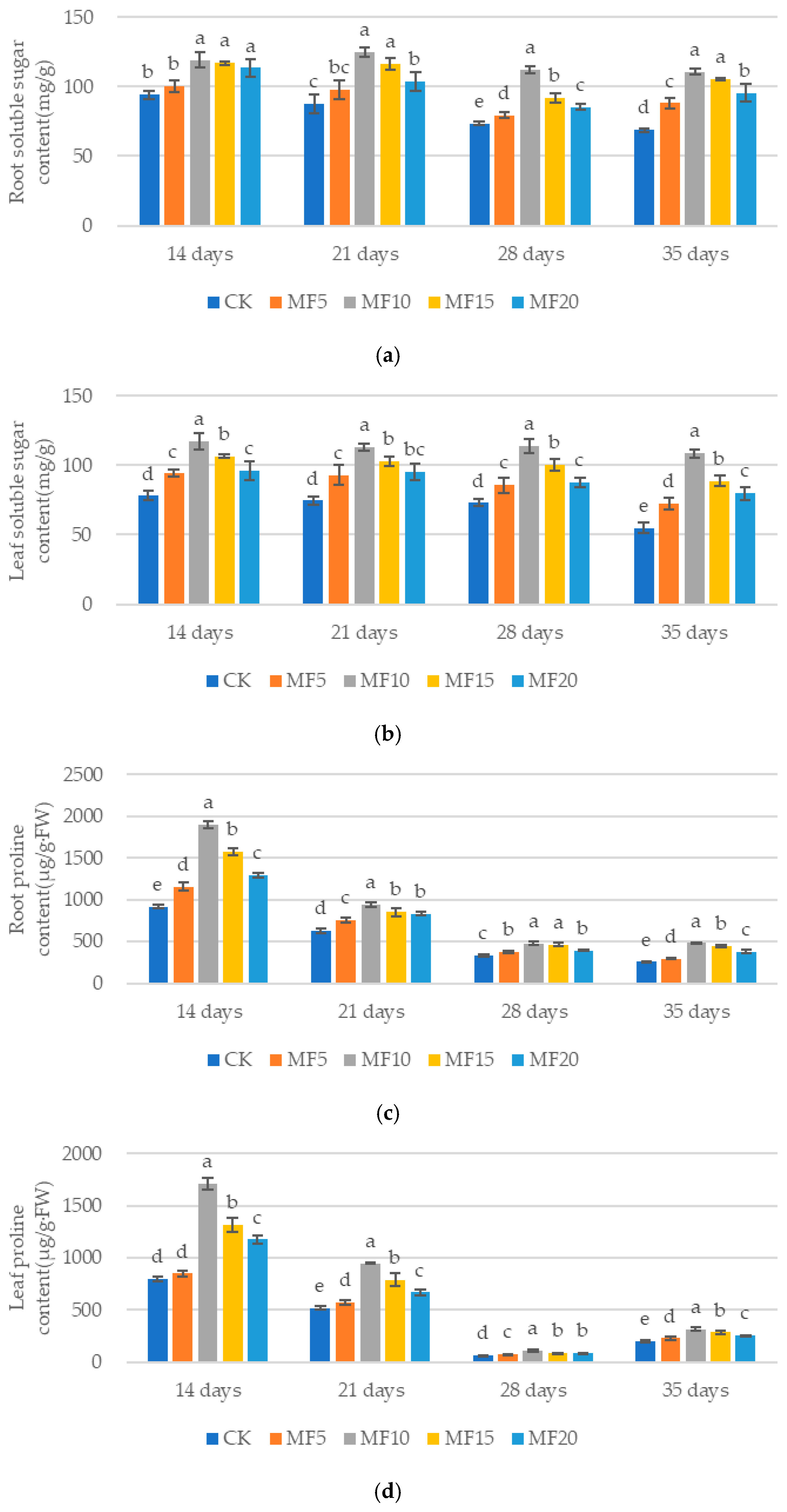
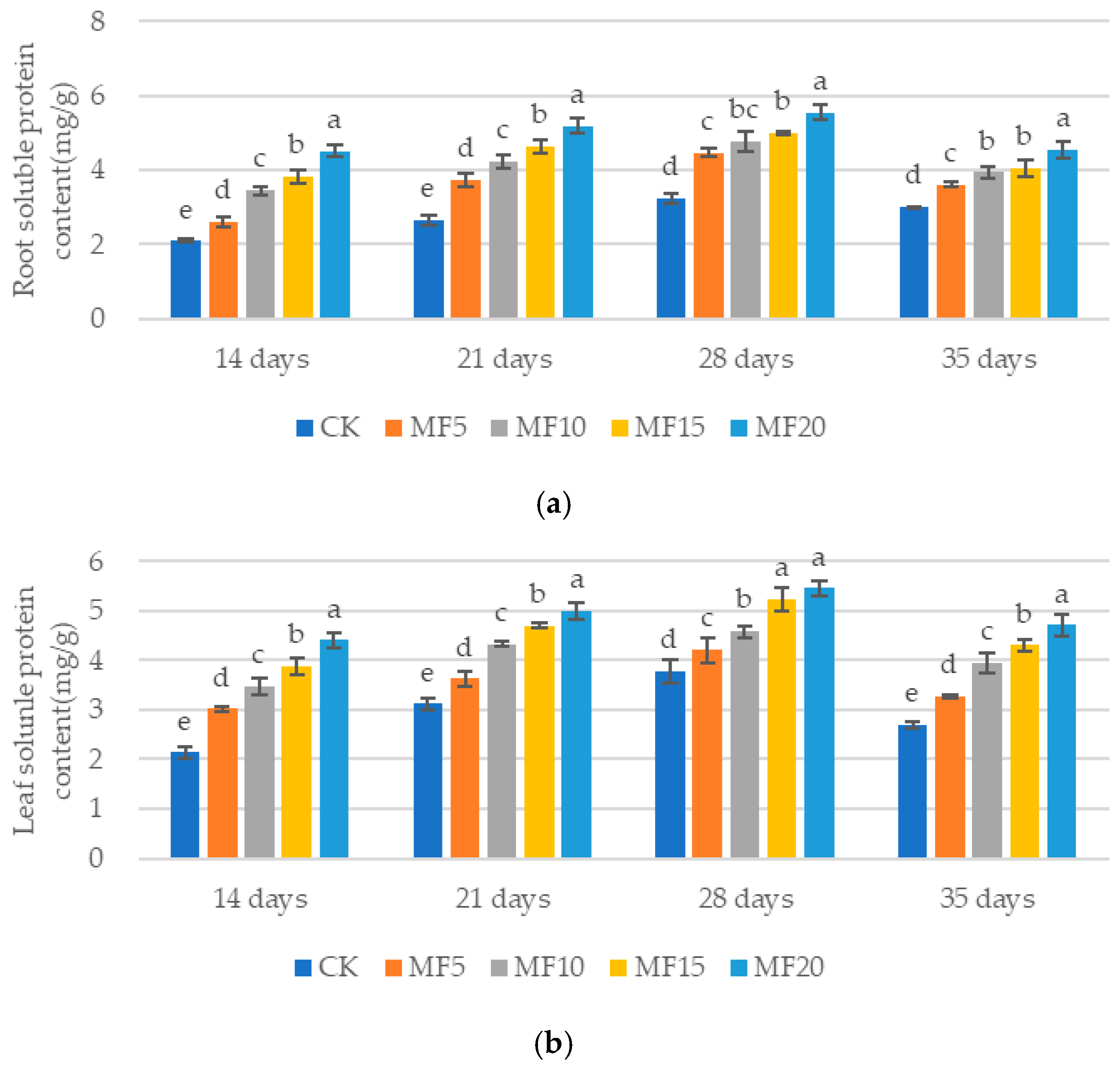
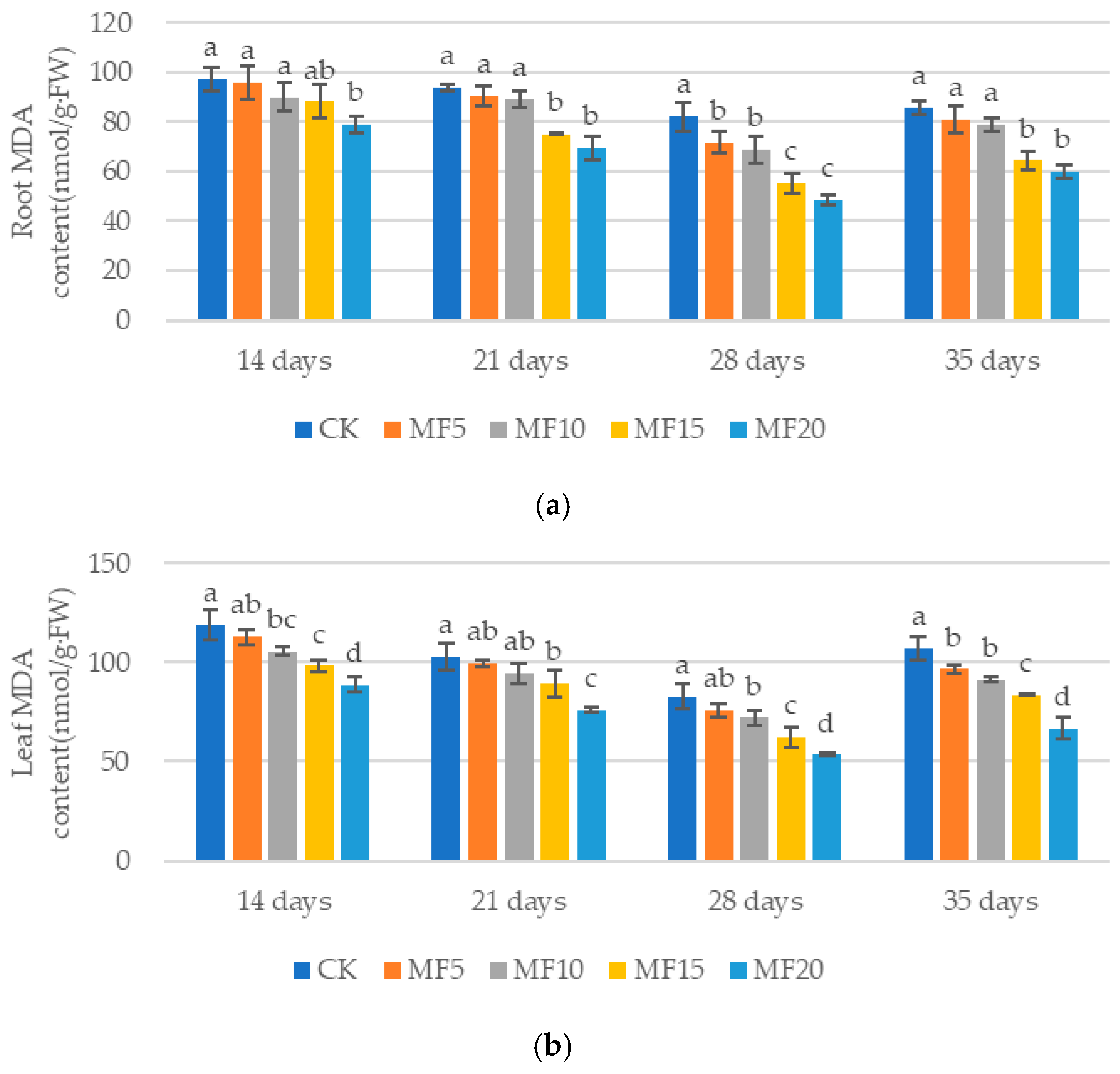
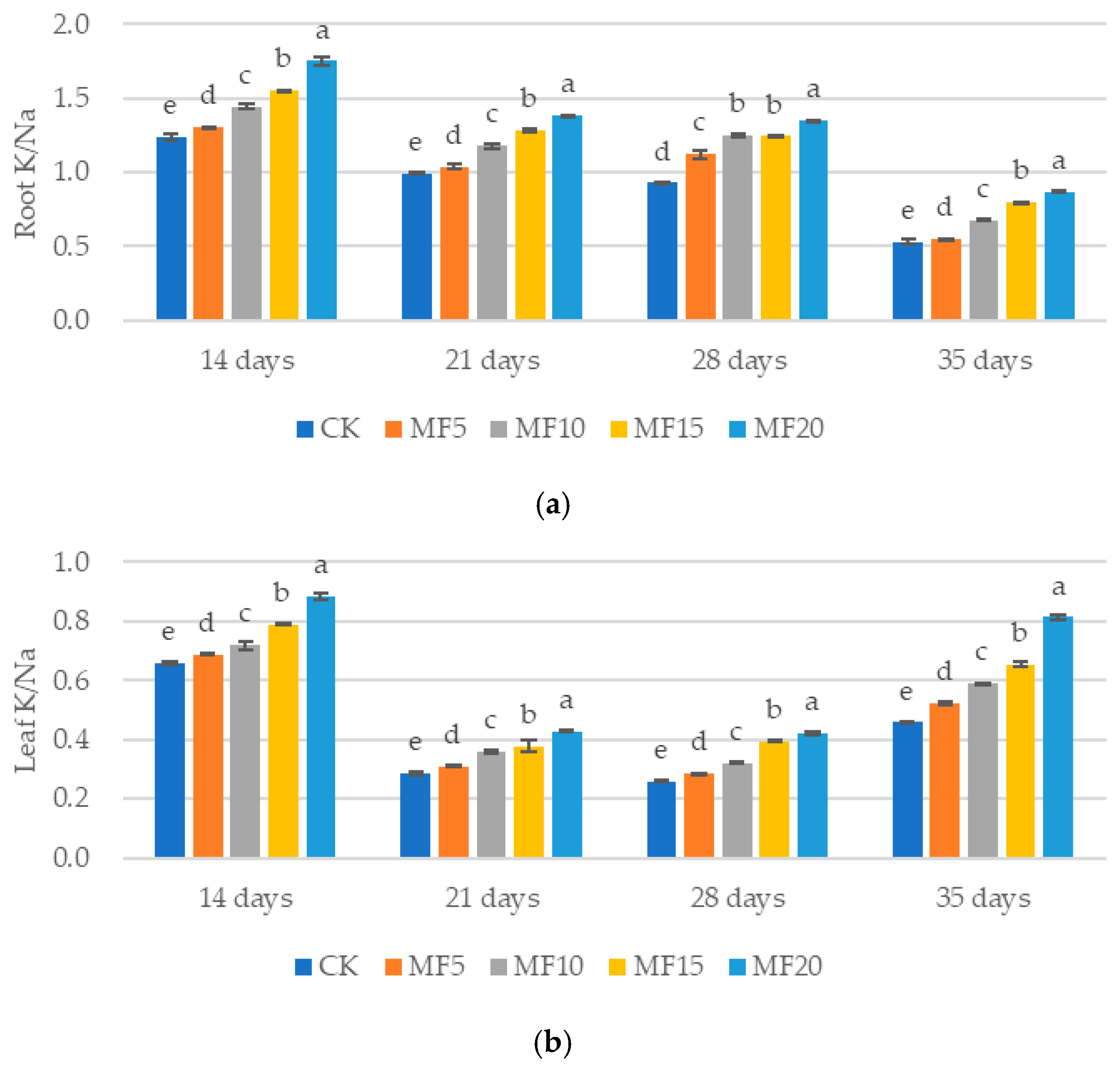
2.2. Impact of Microbial Fertilizer on Osmoregulatory Substances in Saline–Alkali-Stressed Sugar Beet Seedlings
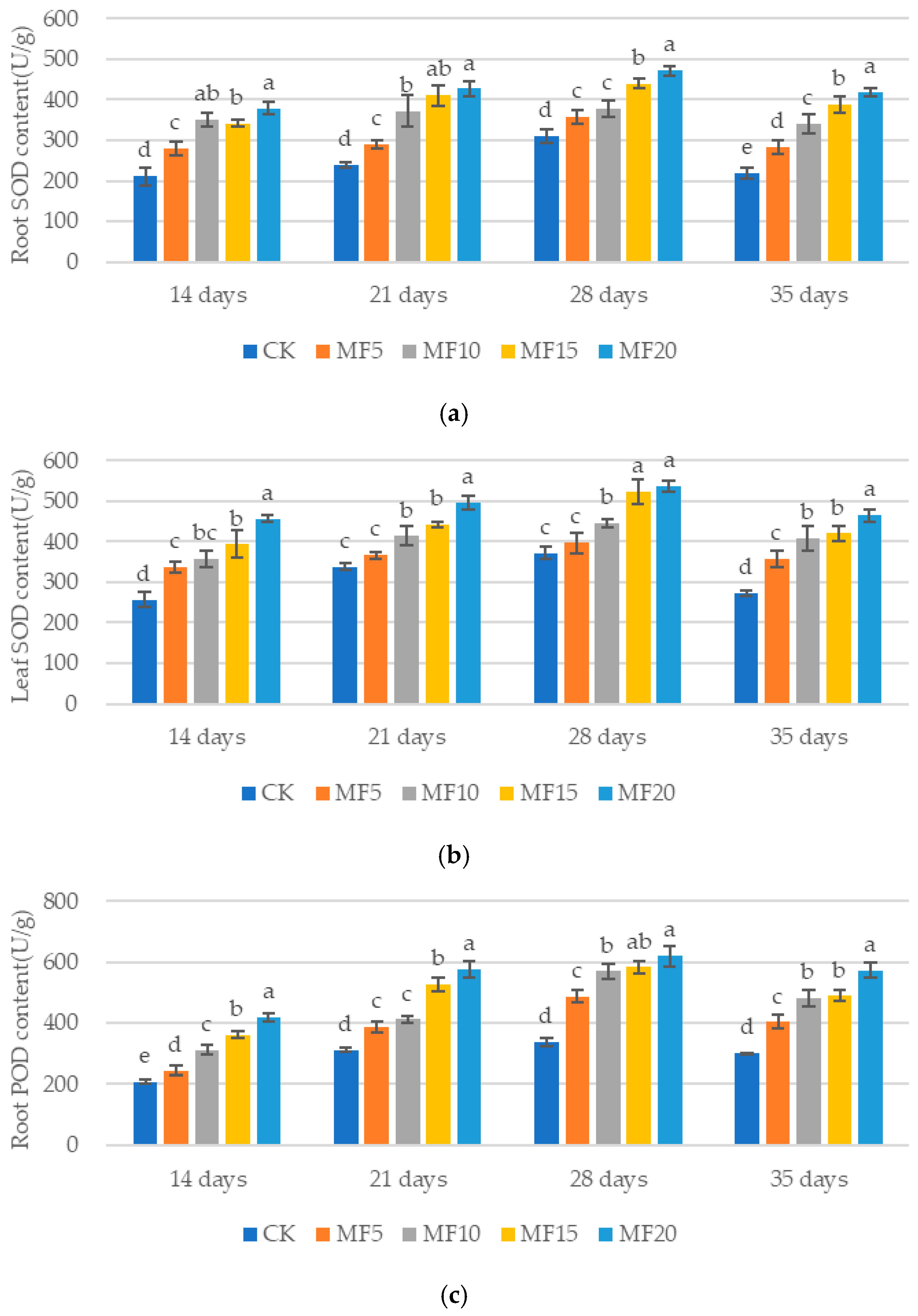
2.3. Impact of Microbial Fertilizer on Antioxidant Enzyme Activity in Saline–Alkali-Stressed Sugar Beet Seedlings
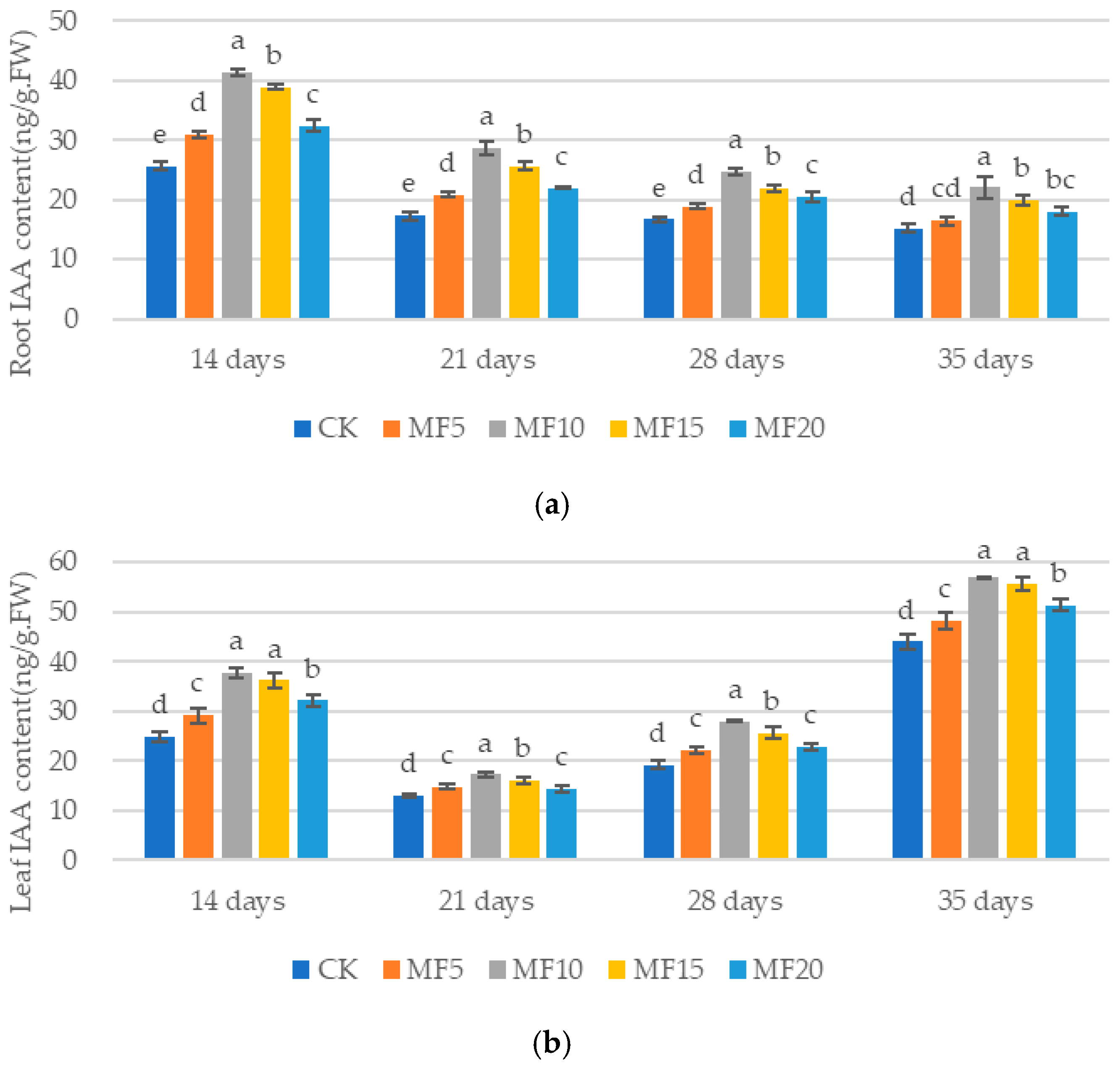
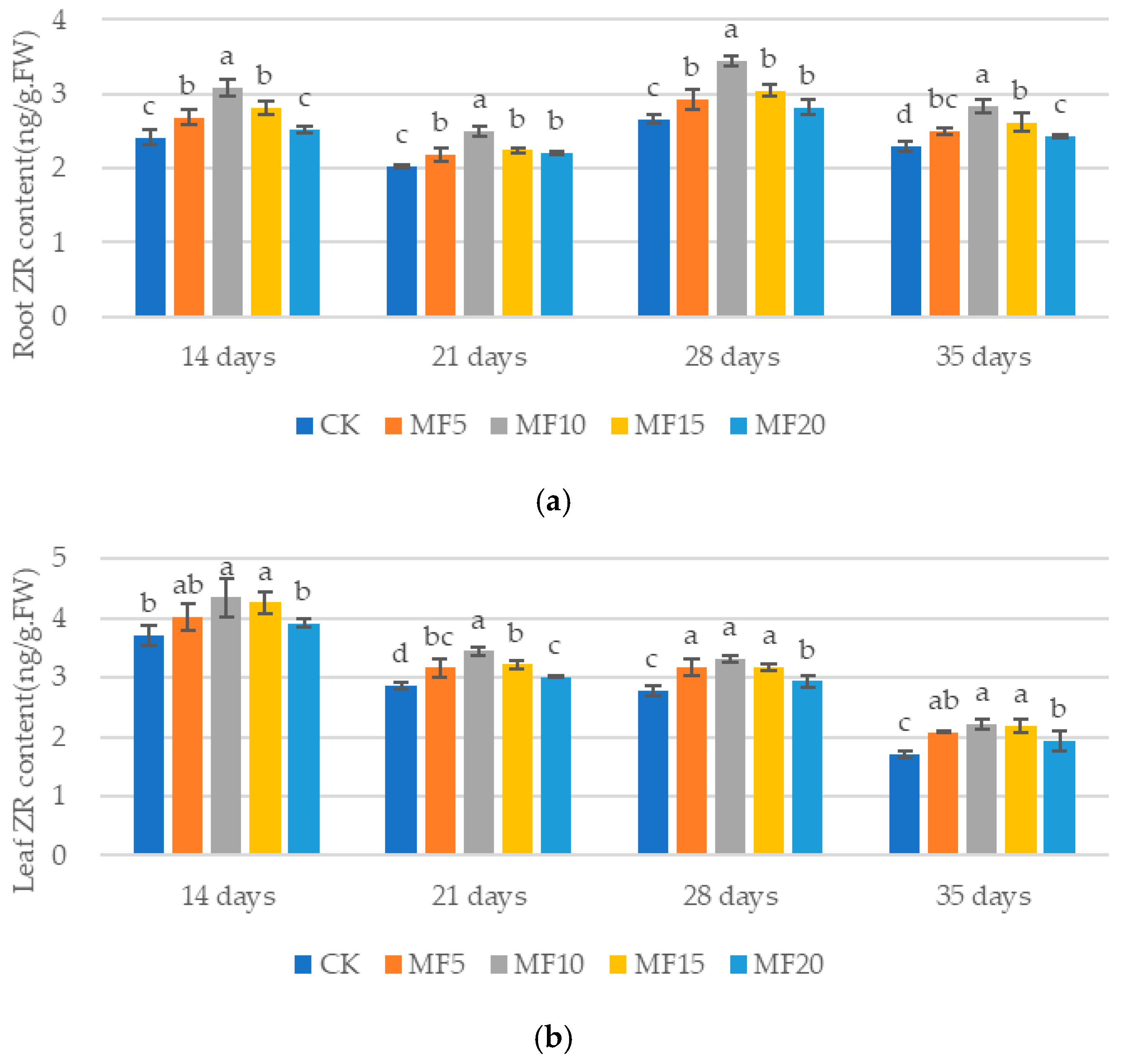
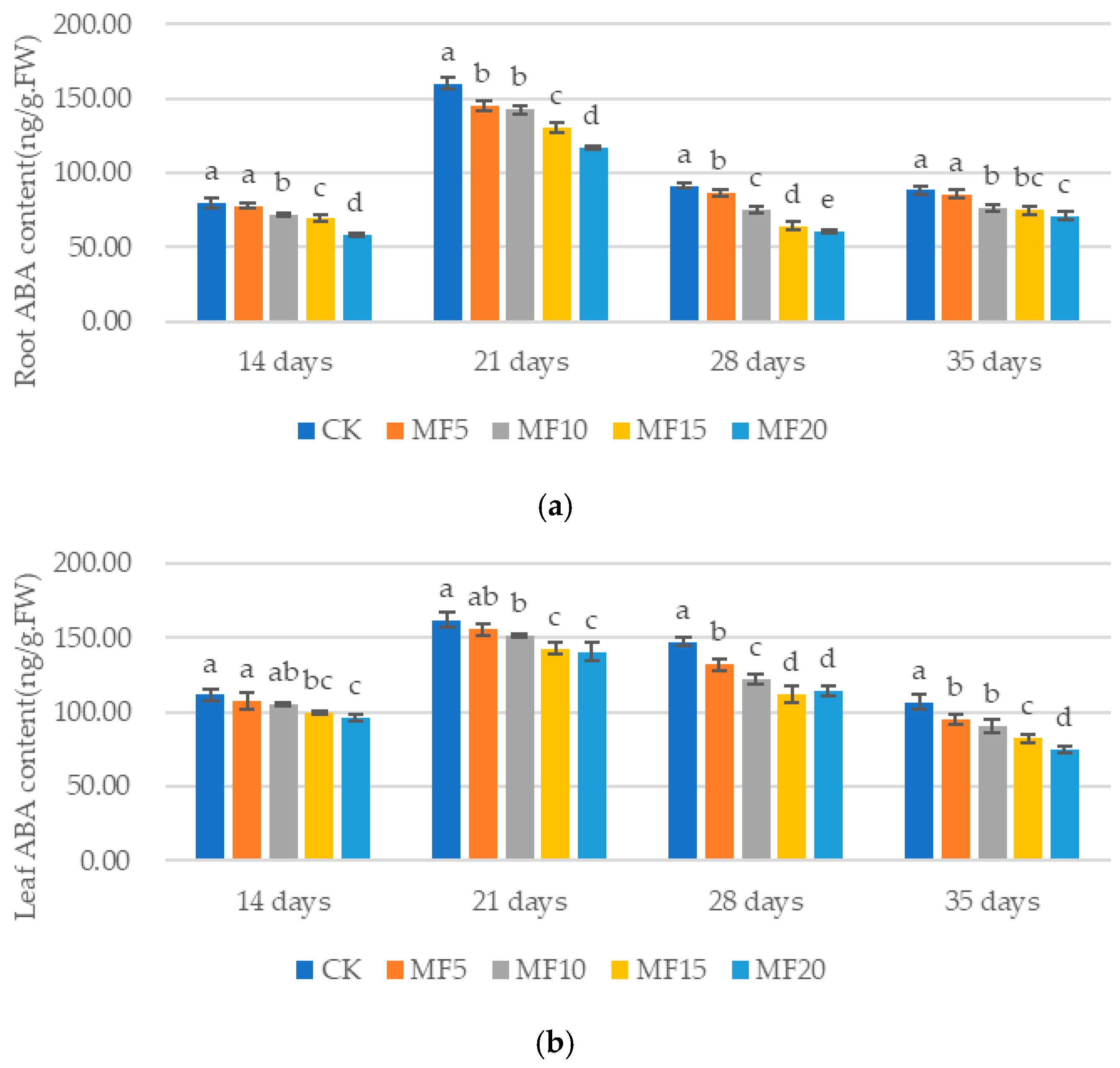
2.4. Impact of Microbial Fertilizer on Hormone Content in Saline–Alkali-Stressed Sugar BEET Seedlings
2.5. Transcriptome Sequencing and Alignment for Sugar Beet Seedlings
2.6. Analysis of Differentially Expressed Genes
2.7. Microbial Fertilization Regulates Important Genes in Saline–Alkali-Stressed Sugar Beet Seedlings
3. Discussion
3.1. Effect of Microbial Fertilizer on the Growth of Sugar Beet Seedlings Under Saline–Alkali Stress
3.2. Mechanisms of Microbial Fertilizers in Regulating the Roots of Sugar Beet Seedlings Under Saline–Alkali Stress
3.3. Mechanisms of Microbial Fertilizers in Regulating the Leaves of Sugar Beet Seedlings Under Saline–Alkali Stress
4. Materials and Methods
4.1. Plant Cultivation and Treatments
4.2. Sugar Beet Seeding Retention and Fresh Weight
4.3. Determination of Osmoregulatory Substances in Sugar Beet
4.4. Determination of Antioxidant Enzyme Levels in Sugar Beet
4.5. Measurement of IAA, GA, ZR and ABA
4.6. cDNA Library Preparation and RNA-Seq
4.7. Identification of Differentially Expressed Genes
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDA | Malonaldehyde |
| SOD | Superoxide dismutase |
| POD | Peroxidase |
| CAT | Catalase |
| IAA | Indole acetic acid |
| GA | Gibberellic acid |
| ZR | Zeatin riboside |
| ABA | Abscisic acid |
| FW | Fresh weight |
| ROS | Reactive oxygen species |
| AMF | Arbuscular mycorrhizal fungi |
| PGPRs | Plant growth-promoting rhizobacteria |
| JA | Jasmone acid |
References
- Haliu, B.; Mehari, H. Impacts of Soil Salinity/Sodicity on Soil-Water Relations and Plant Growth in Dry Land Areas: A Review. J. Nat. Sci. Res. 2021, 12, 1–10. [Google Scholar]
- Plant Breeding: Current and Future Views; Abdurakhmonov, I., Ed.; IntechOpen: London, UK, 2021. [Google Scholar]
- Sun, J.; Li, S.; Guo, H.; Hou, Z. Ion Homeostasis and Na+ Transport-Related Gene Expression in Two Cotton (Gossypium hirsutum L.) Varieties under Saline, Alkaline and Saline-Alkaline Stresses. PLoS ONE 2021, 16, e0256000. [Google Scholar] [CrossRef]
- Gao, Y.; Jin, Y.; Guo, W.; Xue, Y.; Yu, L. Metabolic and Physiological Changes in the Roots of Two Oat Cultivars in Response to Complex Saline-Alkali Stress. Front. Plant Sci. 2022, 13, 835414. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X. Response Mechanisms of Plants Under Saline-Alkali Stress. Front. Plant Sci. 2021, 12, 667458. [Google Scholar] [CrossRef]
- Otlewska, A.; Migliore, M.; Dybka-Stępień, K.; Manfredini, A.; Struszczyk-Świta, K.; Napoli, R.; Białkowska, A.; Canfora, L.; Pinzari, F. When Salt Meddles Between Plant, Soil, and Microorganisms. Front. Plant Sci. 2020, 11, 553087. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots Withstanding Their Environment: Exploiting Root System Architecture Responses to Abiotic Stress to Improve Crop Tolerance. Front. Plant Sci. 2016, 07, 1335. [Google Scholar] [CrossRef]
- Jin, C.-W.; Sun, Y.-L.; Cho, D.-H. Changes in Photosynthetic Rate, Water Potential, and Proline Content in Kenaf Seedlings under Salt Stress. Can. J. Plant Sci. 2012, 92, 311–319. [Google Scholar] [CrossRef]
- Reina-Sánchez, A.; Romero-Aranda, R.; Cuartero, J. Plant Water Uptake and Water Use Efficiency of Greenhouse Tomato Cultivars Irrigated with Saline Water. Agric. Water Manag. 2005, 78, 54–66. [Google Scholar] [CrossRef]
- Sun, Y.; Ou, Y.; Gao, Y.; Zhang, X.; He, Y.; Li, Y.; Yao, Y. Different Tolerance Mechanism to Alkaline Stresses between Populus Bolleana and Its Desert Relative Populus Euphratica. Plant Soil. 2018, 426, 349–363. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, M.; Wang, C.; Sun, H.; Qi, C.; Hou, L.; Wu, Z. Changes in osmolytes and related genes of different rice varieties under saline-alkali stress. J. Northwest AF Univ. (Nat. Sci. Ed.) 2016, 44, 39–47. [Google Scholar]
- Wang, H.; Takano, T.; Liu, S. Screening and Evaluation of Saline–Alkaline Tolerant Germplasm of Rice (Oryza sativa L.) in Soda Saline–Alkali Soil. Agronomy 2018, 8, 205. [Google Scholar] [CrossRef]
- Sakamoto, A.; Murata, N. Genetic Engineering of Glycinebetaine Synthesis in Plants: Current Status and Implications for Enhancement of Stress Tolerance. J. Exp. Bot. 2000, 51, 81–88. [Google Scholar] [CrossRef]
- Adem, G.D.; Roy, S.J.; Zhou, M.; Bowman, J.P.; Shabala, S. Evaluating Contribution of Ionic, Osmotic and Oxidative Stress Components towards Salinity Tolerance in Barley. BMC Plant Biol. 2014, 14, 113. [Google Scholar] [CrossRef]
- Bhanbhro, N.; Xiao, B.; Han, L.; Lu, H.; Wang, H.; Yang, C. Adaptive Strategy of Allohexaploid Wheat to Long-Term Salinity Stress. BMC Plant Biol. 2020, 20, 210. [Google Scholar] [CrossRef]
- Sawatdee, S.; Jarunglumlert, T.; Pavasant, P.; Sakihama, Y.; Flood, A.E.; Prommuak, C. Effect of Mixed Light Emitting Diode Spectrum on Antioxidants Content and Antioxidant Activity of Red Lettuce Grown in a Closed Soilless System. BMC Plant Biol. 2023, 23, 351. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Ji, K.; Mo, W.; Wang, L.; Chen, L.; Gao, L.; Gong, W.; Yuan, D. Dynamics of Sugars, Endogenous Hormones, and Oil Content during the Development of Camellia oleifera Fruit. Botany 2021, 99, 515–529. [Google Scholar] [CrossRef]
- Zhu, X.; Li, X.; Jiu, S.; Zhang, K.; Wang, C.; Fang, J. Analysis of the Regulation Networks in Grapevine Reveals Response to Waterlogging Stress and Candidate Gene-Marker Selection for Damage Severity. R. Soc. Open Sci. 2018, 5, 172253. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Jiang, T.; Xu, Y.; Zong, L. Advances in Researches on Microbial Inoculants on Promoting Plant Growth. Plant Nu-Trition Fertil. Sci. 1999, 19, 2–10. [Google Scholar]
- Jia, Y.; Liao, Z.; Chew, H.; Wang, L.; Lin, B.; Chen, C.; Lu, G.; Lin, Z. Effect of Pennisetum Giganteum z.x.Lin Mixed Nitrogen-Fixing Bacterial Fertilizer on the Growth, Quality, Soil Fertility and Bacterial Community of Pakchoi (Brassica chinensis L.). PLoS ONE 2020, 15, e0228709. [Google Scholar] [CrossRef]
- Yan, N.; Wang, W.; Mi, T.; Zhang, X.; Li, X.; Du, G. Enhancing Tomato Growth and Soil Fertility under Salinity Stress Using Halotolerant Plant Growth-Promoting Rhizobacteria. Plant Stress 2024, 14, 100638. [Google Scholar] [CrossRef]
- Wu, G.-Q.; Feng, R.-J.; Wang, S.-M.; Wang, C.-M.; Bao, A.-K.; Wei, L.; Yuan, H.-J. Co-Expression of Xerophyte Zygophyllum Xanthoxylum ZxNHX and ZxVP1-1 Confers Enhanced Salinity Tolerance in Chimeric Sugar Beet (Beta vulgaris L.). Front. Plant Sci. 2015, 6, 581. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-R.; Zhao, J.-N.; Zhuang, Y.-L.; Zhao, L.-H.; Wang, C.; Yang, X.-F.; Wang, Y.-B.; Li, C.-F. Effects of Spraying Allantoin at Different Stages on Inorganic Nitrogen Assimilation, Endogenous Hormones, Yield, and Quality of Sugar Beet in Saline-Alkali Land. Sugar Tech 2024, 26, 1283–1296. [Google Scholar] [CrossRef]
- Abbasi, Z.; Arzani, A.; Majidi, M.M.; Rajabi, A.; Jalali, A. Genetic Analysis of Sugar Yield and Physiological Traits in Sugar Beet under Salinity Stress Conditions. Euphytica 2019, 215, 99. [Google Scholar] [CrossRef]
- Zou, C.; Sang, L.; Gai, Z.; Wang, Y.; Li, C. Morphological and Physiological Responses of Sugar Beet to Alkaline Stress. Sugar Technol. 2018, 20, 202–211. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Ghouili, E.; Abid, G.; Hogue, R.; Jeanne, T.; D’Astous-Pagé, J.; Sassi, K.; Hidri, Y.; M’Hamed, H.C.; Somenahally, A.; Xue, Q.; et al. Date Palm Waste Compost Application Increases Soil Microbial Community Diversity in a Cropping Barley (Hordeum vulgare L.) Field. Biology 2023, 12, 546. [Google Scholar] [CrossRef]
- Verbon, E.H.; Liberman, L.M. Beneficial Microbes Affect Endogenous Mechanisms Controlling Root Development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Boughattas, S.; Hu, S.; Oh, S.-H.; Sa, T. A Meta-Analysis of Arbuscular Mycorrhizal Effects on Plants Grown under Salt Stress. Mycorrhiza 2014, 24, 611–625. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B.; Patel, S. Combination of Endophytic and Rhizospheric Plant Growth Promoting Rhizobacteria in Oryza Sativa Shows Higher Accumulation of Osmoprotectant against Saline Stress. Acta Physiol. Plant. 2011, 33, 797–802. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Yang, W.; Liu, W.; Wang, Y.; Wang, Q.; Zhao, Y. Identification of Whirly Transcription Factors in Triticeae Species and Functional Analysis of TaWHY1-7D in Response to Osmotic Stress. Front. Plant Sci. 2023, 14, 1297228. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, Y.; Zhang, F. Screening and Identification of Salt-Tolerant Plant Growth-Promoting Bacteria and Its Promo-tion Effect on Wheat Seedling under Salt Stress. J. Triticeae Crops 2020, 40, 110–117. [Google Scholar]
- Sritongon, N.; Boonlue, S.; Mongkolthanaruk, W.; Jogloy, S.; Riddech, N. The Combination of Multiple Plant Growth Promotion and Hydrolytic Enzyme Producing Rhizobacteria and Their Effect on Jerusalem Artichoke Growth Improvement. Sci. Rep. 2023, 13, 5917. [Google Scholar] [CrossRef]
- Kim, K.; Jang, Y.-J.; Lee, S.-M.; Oh, B.-T.; Chae, J.-C.; Lee, K.-J. Alleviation of Salt Stress by Enterobacter Sp. EJ01 in Tomato and Arabidopsis Is Accompanied by Up-Regulation of Conserved Salinity Responsive Factors in Plants. Mol. Cells 2014, 37, 109–117. [Google Scholar] [CrossRef]
- Li, W.; Meng, R.; Liu, Y.; Chen, S.; Jiang, J.; Wang, L.; Zhao, S.; Wang, Z.; Fang, W.; Chen, F.; et al. Heterografted Chrysanthemums Enhance Salt Stress Tolerance by Integrating Reactive Oxygen Species, Soluble Sugar, and Proline. Hortic. Res. 2022, 9, uhac073. [Google Scholar] [CrossRef]
- Wang, X.; Ma, J.; He, F.; Wang, L.; Zhang, T.; Liu, D.; Xu, Y.; Li, F.; Feng, X. A Study on the Functional Identification of Overexpressing Winter Wheat Expansin Gene TaEXPA7-B in Rice under Salt Stress. Int. J. Mol. Sci. 2024, 25, 7707. [Google Scholar] [CrossRef] [PubMed]
- Giri, B.; Kapoor, R.; Mukerji, K.G. Improved Tolerance of Acacia Nilotica to Salt Stress by Arbuscular Mycorrhiza, Glomus Fasciculatum May Be Partly Related to Elevated K/Na Ratios in Root and Shoot Tissues. Microb. Ecol. 2007, 54, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Bharti, N.; Pandey, S.S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant Growth Promoting Rhizobacteria Dietzia Natronolimnaea Modulates the Expression of Stress Responsive Genes Providing Protection of Wheat from Salinity Stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B. PGPR Regulate Caspase-like Activity, Programmed Cell Death, and Antioxidant Enzyme Activity in Paddy under Salinity. Physiol. Mol. Biol. Plants 2014, 20, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.; Abd_Allah, E.F. Plant Hormones as Key Regulators in Plant-Microbe Interactions Under Salt Stress. In Plant Microbiome: Stress Response; Egamberdieva, D., Ahmad, P., Eds.; Microorganisms for Sustainability; Springer: Singapore, 2018; Volume 5, pp. 165–182. [Google Scholar]
- Chen, X.; Liu, K.; Xuan, M.; Shao, J.; Zhang, R. Screening and identification of plant growth-promoting rhizobacteria to enhance salt stress tolerance of crops and their effects in field experiment. J. Nanjing Agric. Univ. 2020, 43, 452–459. [Google Scholar]
- Deng, C.; Deng, Y.; Wang, B.; Yang, X. Gas Chromatography–Mass Spectrometry Method for Determination of Phenylalanine and Tyrosine in Neonatal Blood Spots. J. Chromatogr. B 2002, 780, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Bruce, S.J.; Guy, P.A.; Rezzi, S.; Ross, A.B. Quantitative Measurement of Betaine and Free Choline in Plasma, Cereals and Cereal Products by Isotope Dilution LC-MS/MS. J. Agric. Food Chem. 2010, 58, 2055–2061. [Google Scholar] [CrossRef] [PubMed]



| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Error_ Rate | Q20 | Q30 | GC (%) |
|---|---|---|---|---|---|---|---|---|
| CKR141 | 47753286 | 7.16 G | 46780204 | 7.02 G | 0.03 | 97.22 | 92.43 | 42.94 |
| CKR142 | 47589848 | 7.14 G | 46715122 | 7.01 G | 0.03 | 97.31 | 92.74 | 43.01 |
| CKR143 | 41420298 | 6.21 G | 40502964 | 6.08 G | 0.03 | 97.15 | 92.30 | 42.96 |
| MFR141 | 45841150 | 6.88 G | 44684554 | 6.70 G | 0.03 | 97.35 | 92.77 | 42.93 |
| MFR142 | 42508768 | 6.38 G | 41146434 | 6.17 G | 0.03 | 97.31 | 92.61 | 42.74 |
| MFR143 | 47536994 | 7.13 G | 46153018 | 6.92 G | 0.03 | 97.11 | 92.18 | 42.88 |
| CKR351 | 46944566 | 7.04 G | 45633760 | 6.85 G | 0.03 | 97.24 | 92.47 | 42.38 |
| CKR352 | 49176016 | 7.38 G | 48067686 | 7.21 G | 0.03 | 97.30 | 92.54 | 41.97 |
| CKR353 | 42532576 | 6.38 G | 41035740 | 6.16 G | 0.03 | 97.26 | 92.53 | 41.52 |
| MFR351 | 41906002 | 6.29 G | 40858146 | 6.13 G | 0.03 | 97.02 | 91.93 | 42.01 |
| MFR352 | 46465650 | 6.97 G | 45025034 | 6.75 G | 0.03 | 97.14 | 92.21 | 42.09 |
| MFR353 | 43744572 | 6.56 G | 43043216 | 6.46 G | 0.03 | 97.39 | 92.85 | 42.78 |
| CKL141 | 47319126 | 7.10 G | 46530012 | 6.98 G | 0.03 | 97.38 | 92.69 | 43.61 |
| CKL142 | 44370914 | 6.66 G | 42726458 | 6.41 G | 0.03 | 97.45 | 92.93 | 43.38 |
| CKL143 | 45810000 | 6.87 G | 44462518 | 6.67 G | 0.03 | 97.61 | 93.31 | 43.70 |
| MFL141 | 46487394 | 6.97 G | 45105062 | 6.77 G | 0.03 | 97.59 | 93.19 | 43.68 |
| MFL142 | 48043430 | 7.21 G | 46680844 | 7.00 G | 0.03 | 97.04 | 92.14 | 43.68 |
| MFL143 | 51087790 | 7.66 G | 49202892 | 7.38 G | 0.03 | 97.33 | 92.64 | 43.12 |
| CKL351 | 46409334 | 6.96 G | 45000052 | 6.75 G | 0.03 | 97.48 | 92.95 | 42.79 |
| CKL352 | 45967052 | 6.90 G | 44912846 | 6.74 G | 0.03 | 97.00 | 92.04 | 42.61 |
| CKL353 | 46424230 | 6.96 G | 45259438 | 6.79 G | 0.03 | 97.45 | 92.89 | 42.69 |
| MFL351 | 41423172 | 6.21 G | 40703336 | 6.11 G | 0.03 | 97.25 | 92.42 | 42.42 |
| MFL352 | 45310110 | 6.80 G | 43811570 | 6.57 G | 0.03 | 97.26 | 92.42 | 42.52 |
| MFL353 | 47306530 | 7.10 G | 46569176 | 6.99 G | 0.03 | 97.19 | 92.19 | 42.62 |
| Comparison | All Differentially Expressed Genes | Upregulated Genes | Downregulated Genes |
|---|---|---|---|
| MFR14 vs. CKR14 | 388 | 82 | 306 |
| MFR35 vs. CKR35 | 171 | 107 | 64 |
| MFL14 vs. CKL14 | 102 | 79 | 23 |
| MFL35 vs. CKL35 | 418 | 175 | 243 |
| Treatment | Gene Classification | Gene ID | Gene Name | Gene Description (Swissprot Database) | Regulation Pattern |
|---|---|---|---|---|---|
| MFR14 vs. CKR14 | Osmoregulation | IMABv01g023760 | ABCG25 | ABC transporter G family member 25 | Up |
| IMABv02g032324 | At5g20260 | Probable glycosyltransferase | Up | ||
| novel.664 | SWEET4 | Bidirectional sugar transporter | Up | ||
| IMABv08g029340 | ABCG32 | ABC transporter G family member 32 | Up | ||
| IMABv07g015513 | At3g07620 | Probable glycosyltransferase | Up | ||
| IMABv05g000506 | BASS3 | Probable sodium/metabolite cotransporter | Down | ||
| IMABv04g008626 | IRKI | IRK-interacting protein | Up | ||
| novel.3487 | SECA2 | Protein translocase subunit | Up | ||
| Antioxidant | IMABv09g020441 | GSVIVT00023967001 | Peroxidase 4 | Down | |
| Signal transduction | IMABv05g000249 | GA2OX2 | Gibberellin 2-beta-dioxygenase | Down | |
| IMABv05g000235 | GA2OX2 | Gibberellin 2-beta-dioxygenase 2 | Down | ||
| IMABv08g029383 | D14 | Strigolactone esterase | Up | ||
| IMABv04g006193 | NCED2 | 9-cis-Epoxycarotenoid dioxygenase | Down | ||
| MFR35 vs. CKR35 | Osmoregulation | IMABv06g019546 | HAK17 | Probable potassium transporter 17 | Up |
| IMABv03g009139 | UGT85A24 | 7-Deoxyloganetin glucosyltransferase | Up | ||
| IMABv05g000475 | At5g26710 | Glutamate-tRNA ligase cytoplasmic | Up | ||
| Antioxidant | IMABv06g020044 | APX3 | L-ascorbate peroxidase 3 | Up | |
| novel.2533 | APX3 | L-ascorbate peroxidase 3 | Up | ||
| IMABv09g021299 | APX1 | L-ascorbate peroxidase 1 | Up | ||
| Signal transduction | IMABv08g028764 | ABP19A | Auxin-binding protein | Up | |
| novel.1826 | ERF098 | Ethylene-responsive transcription factor | Up | ||
| MFL14 vs. CKL14 | Osmoregulation | IMABv03g00921 | AVT1A | Amino acid transporter | Up |
| Antioxidant | IMABv02g030636 | APX6 | Putative L-ascorbate peroxidase 6 | Up | |
| IMABv02g031865 | PER52 | Peroxidase 52 | Up | ||
| Signal transduction | IMABv05g003391 | ERF5 | Ethylene-responsive transcription factor 5 | Down | |
| novel.3195 | JOX4 | Jasmonate-induced oxygenase 4 | Up | ||
| MFL35 vs. CKL35 | Osmoregulation | IMABv07g0148989 | ABCG28 | ABC transporter G family member 28 | Up |
| Antioxidant | IMABv05g002263 | PER44 | Peroxidase 44 | Down | |
| Signal transduction | IMABv08g028833 | ERF017 | Ethylene-responsive transcription factor | Down | |
| IMABv01g024418 | CKX5 | Cytokinin dehydrogenase 5 | Down | ||
| IMABv03g008822 | RALF | Rapid alkalinization factor | Down | ||
| IMABv05g004284 | ERF061 | Ethylene-responsive transcription factor | Down | ||
| IMABv08g028764 | ABP19A | Auxin-binding protein | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.; Han, K.; Guo, X.; Tian, L.; Jian, C.; Su, W.; Wei, Z.; Zhang, P.; Li, Y.; Ren, H.; et al. Physiological and Transcriptome Analyses Reveal the Important Role of Microbial Fertilizer in the Response of Sugar Beet Seedlings to Saline-Alkali Stress. Int. J. Mol. Sci. 2025, 26, 8840. https://doi.org/10.3390/ijms26188840
Huang C, Han K, Guo X, Tian L, Jian C, Su W, Wei Z, Zhang P, Li Y, Ren H, et al. Physiological and Transcriptome Analyses Reveal the Important Role of Microbial Fertilizer in the Response of Sugar Beet Seedlings to Saline-Alkali Stress. International Journal of Molecular Sciences. 2025; 26(18):8840. https://doi.org/10.3390/ijms26188840
Chicago/Turabian StyleHuang, Chunyan, Kang Han, Xiaoxia Guo, Lu Tian, Caiyuan Jian, Wenbin Su, Zhigang Wei, Peng Zhang, Yinghao Li, Huimin Ren, and et al. 2025. "Physiological and Transcriptome Analyses Reveal the Important Role of Microbial Fertilizer in the Response of Sugar Beet Seedlings to Saline-Alkali Stress" International Journal of Molecular Sciences 26, no. 18: 8840. https://doi.org/10.3390/ijms26188840
APA StyleHuang, C., Han, K., Guo, X., Tian, L., Jian, C., Su, W., Wei, Z., Zhang, P., Li, Y., Ren, H., Song, J., Wang, L., Zhang, Y., & Li, Z. (2025). Physiological and Transcriptome Analyses Reveal the Important Role of Microbial Fertilizer in the Response of Sugar Beet Seedlings to Saline-Alkali Stress. International Journal of Molecular Sciences, 26(18), 8840. https://doi.org/10.3390/ijms26188840





