Optimized tDR Sequencing Reveals Diversity and Heterogeneity in tRNA-Derived Fragment Landscapes in Mouse Tissues
Abstract
1. Background
2. Results
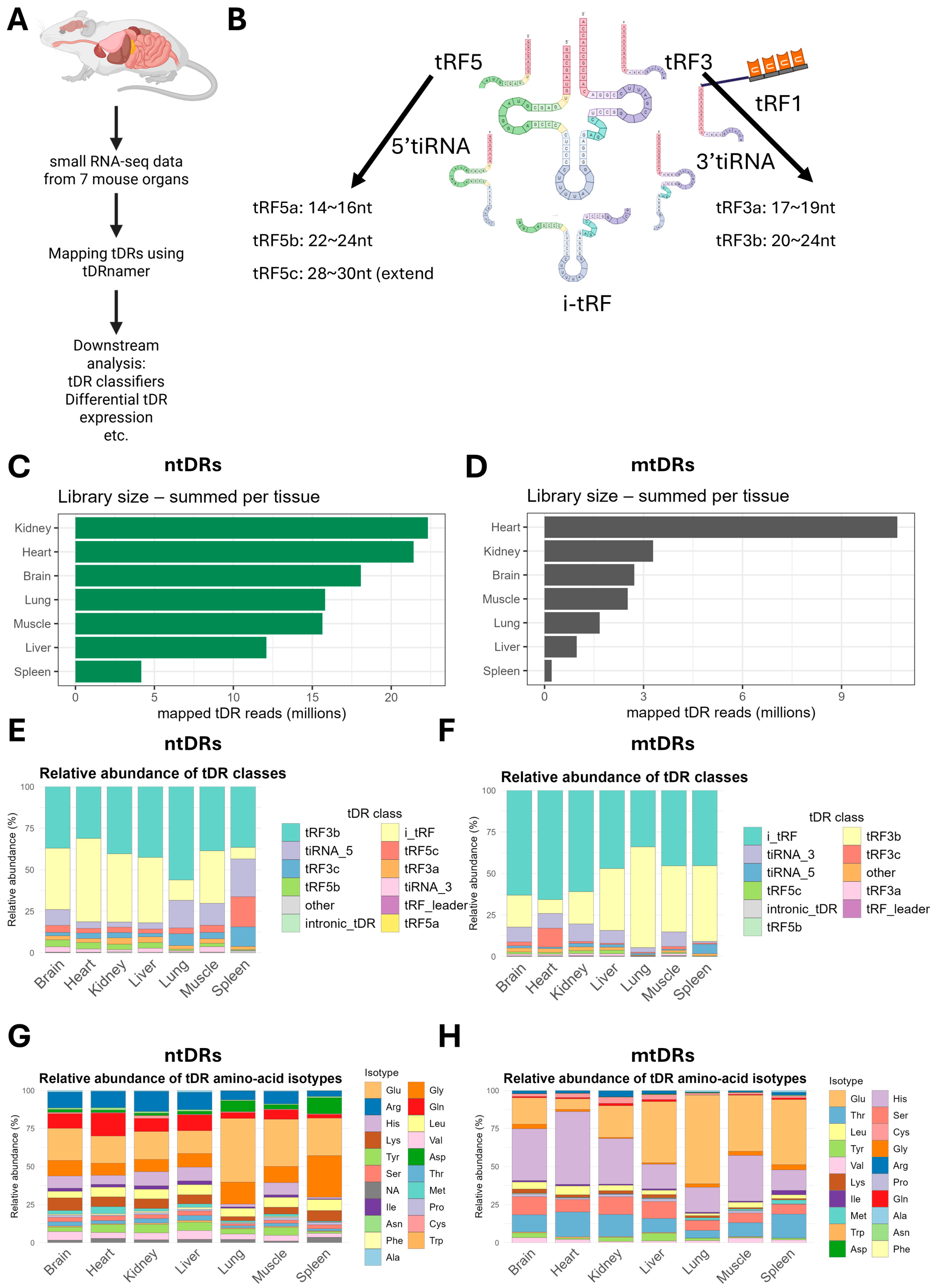
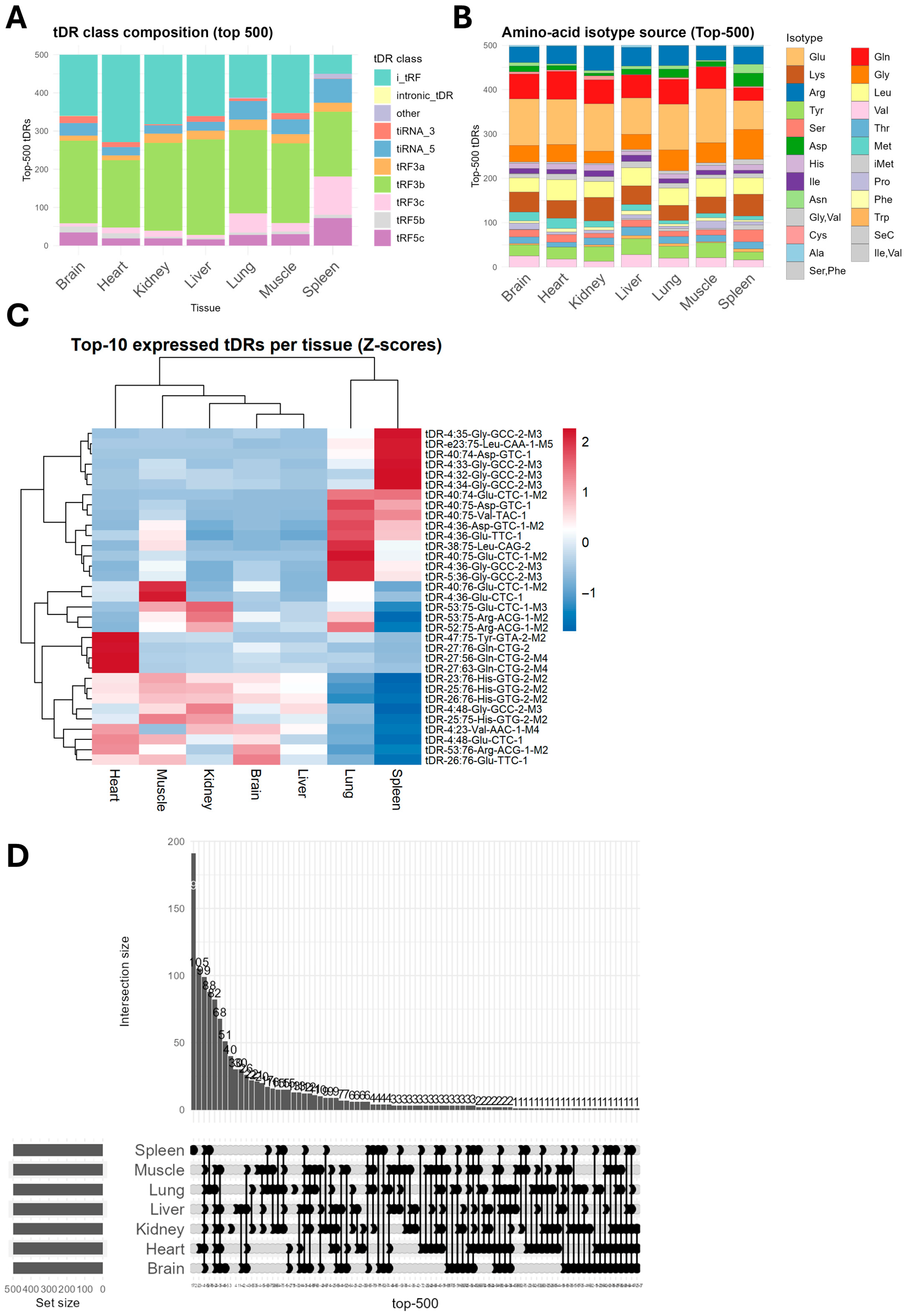
2.1. Global Patterns of tDRs Expression in Seven Mouse Tissues
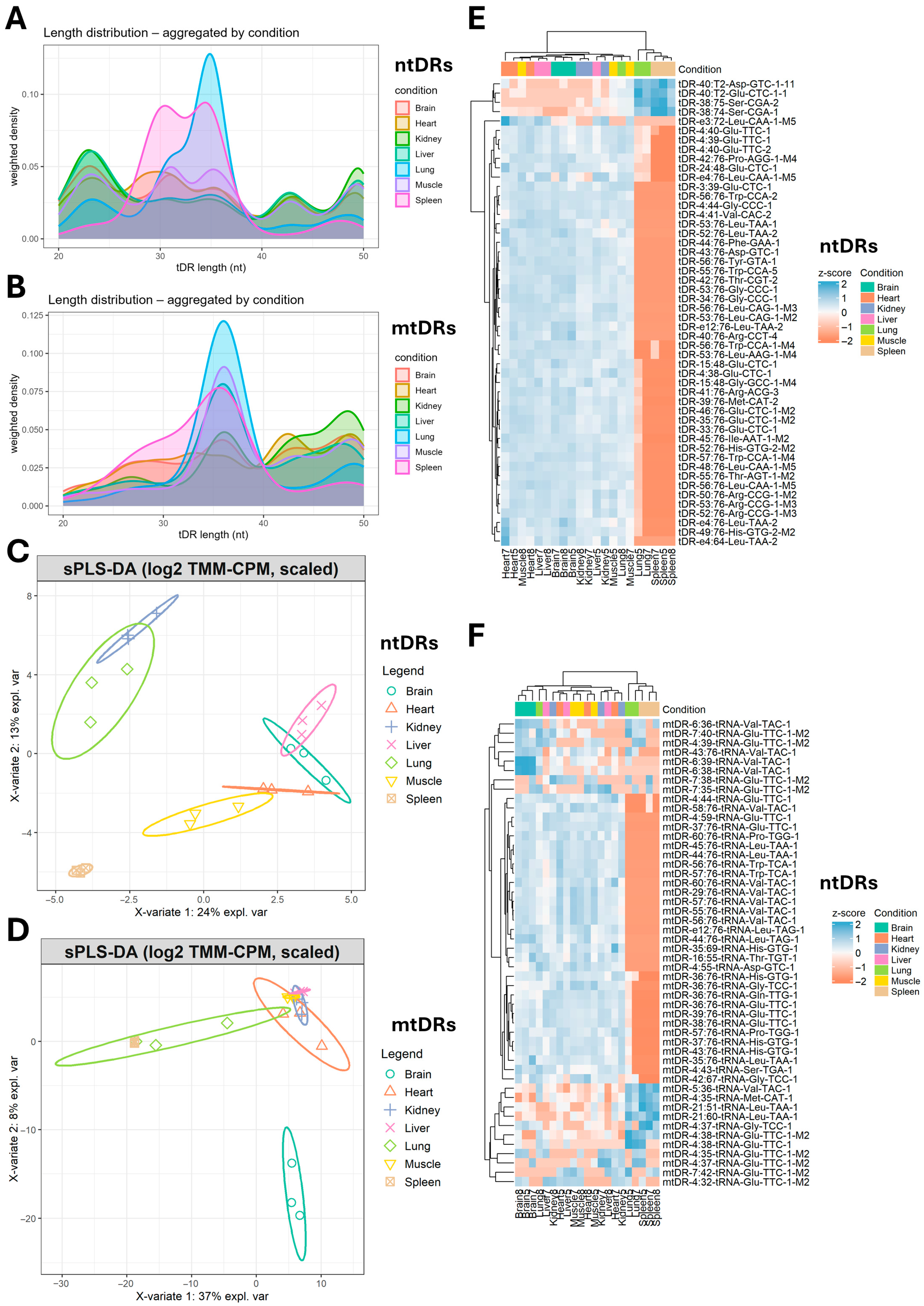
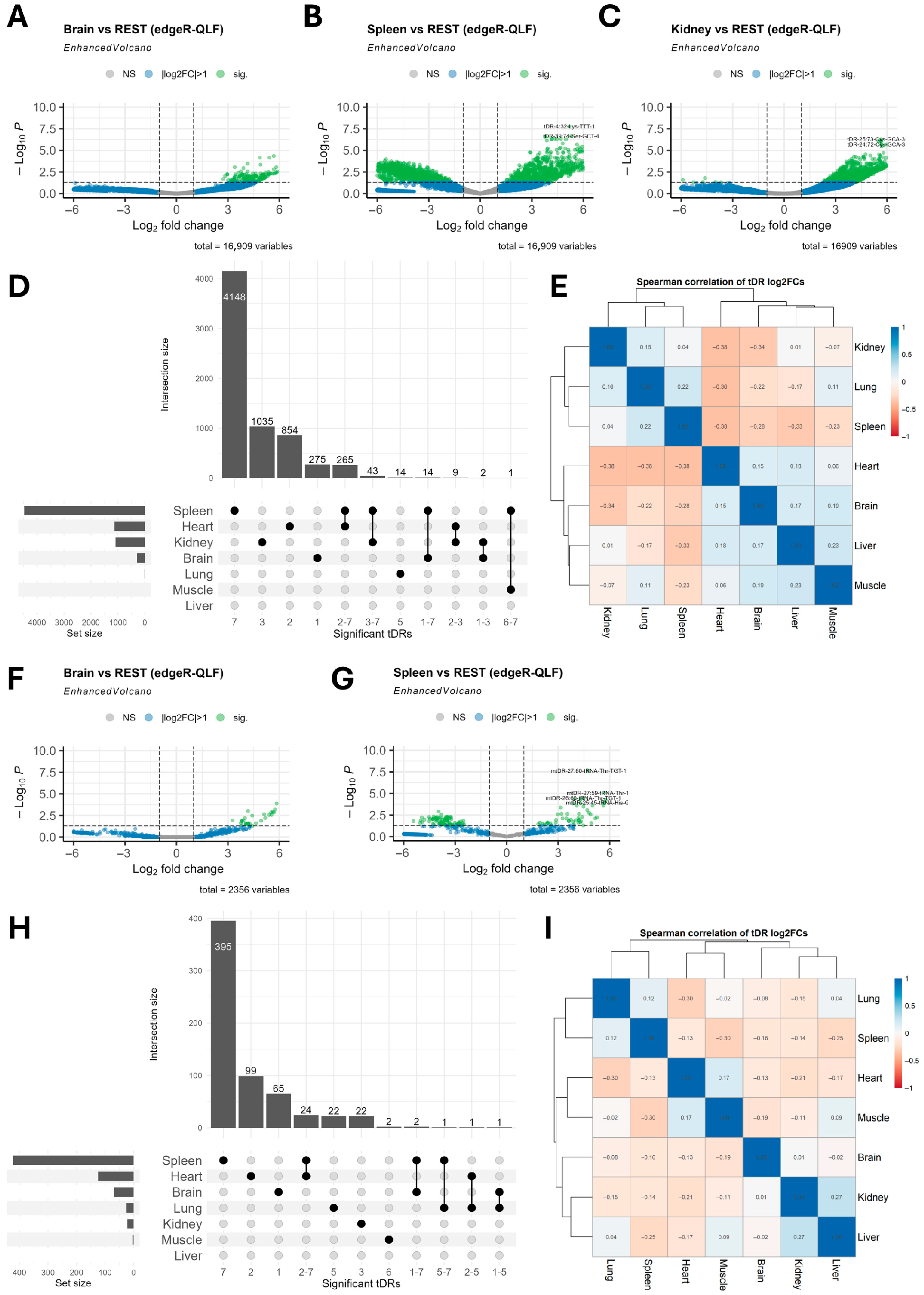
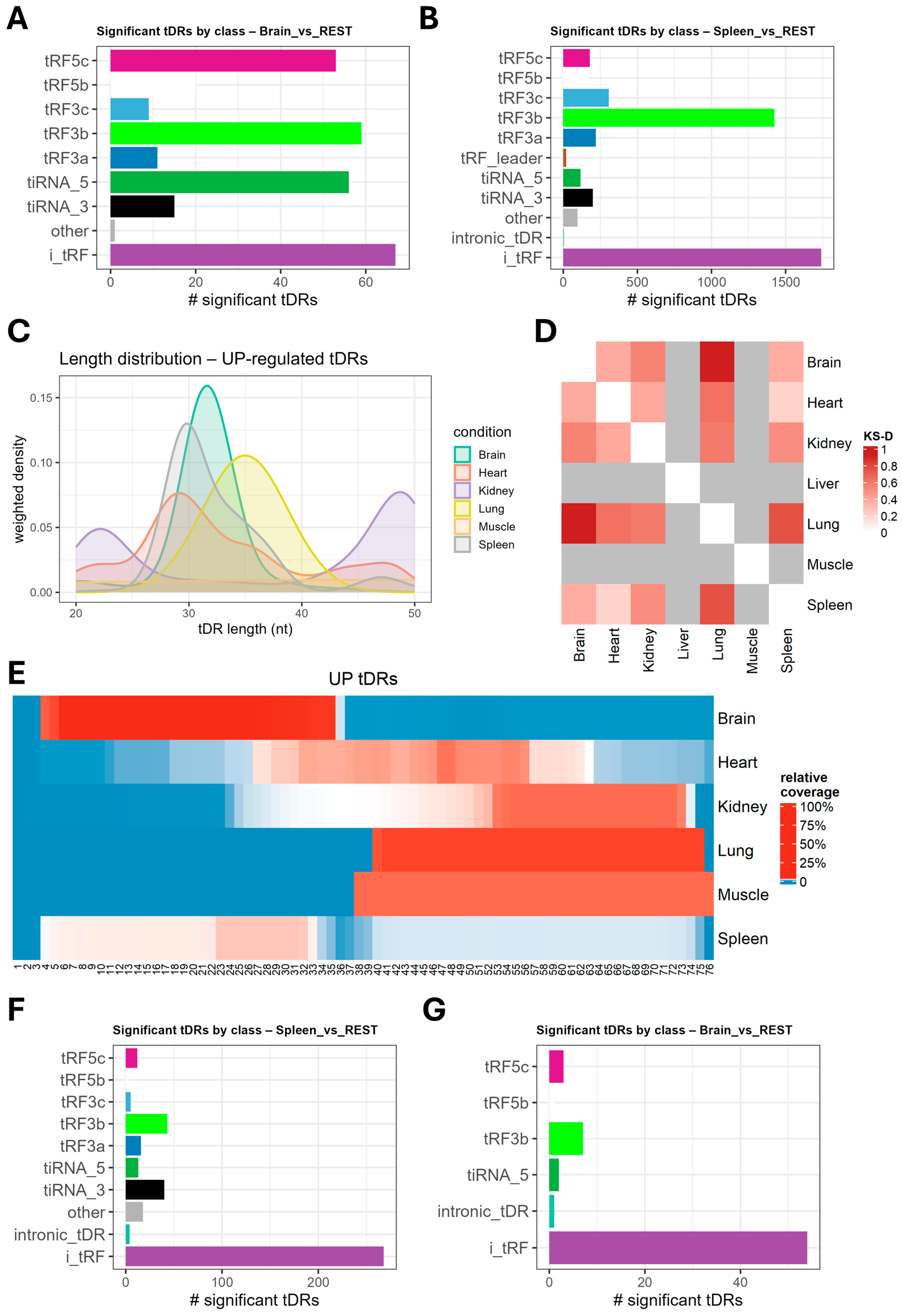
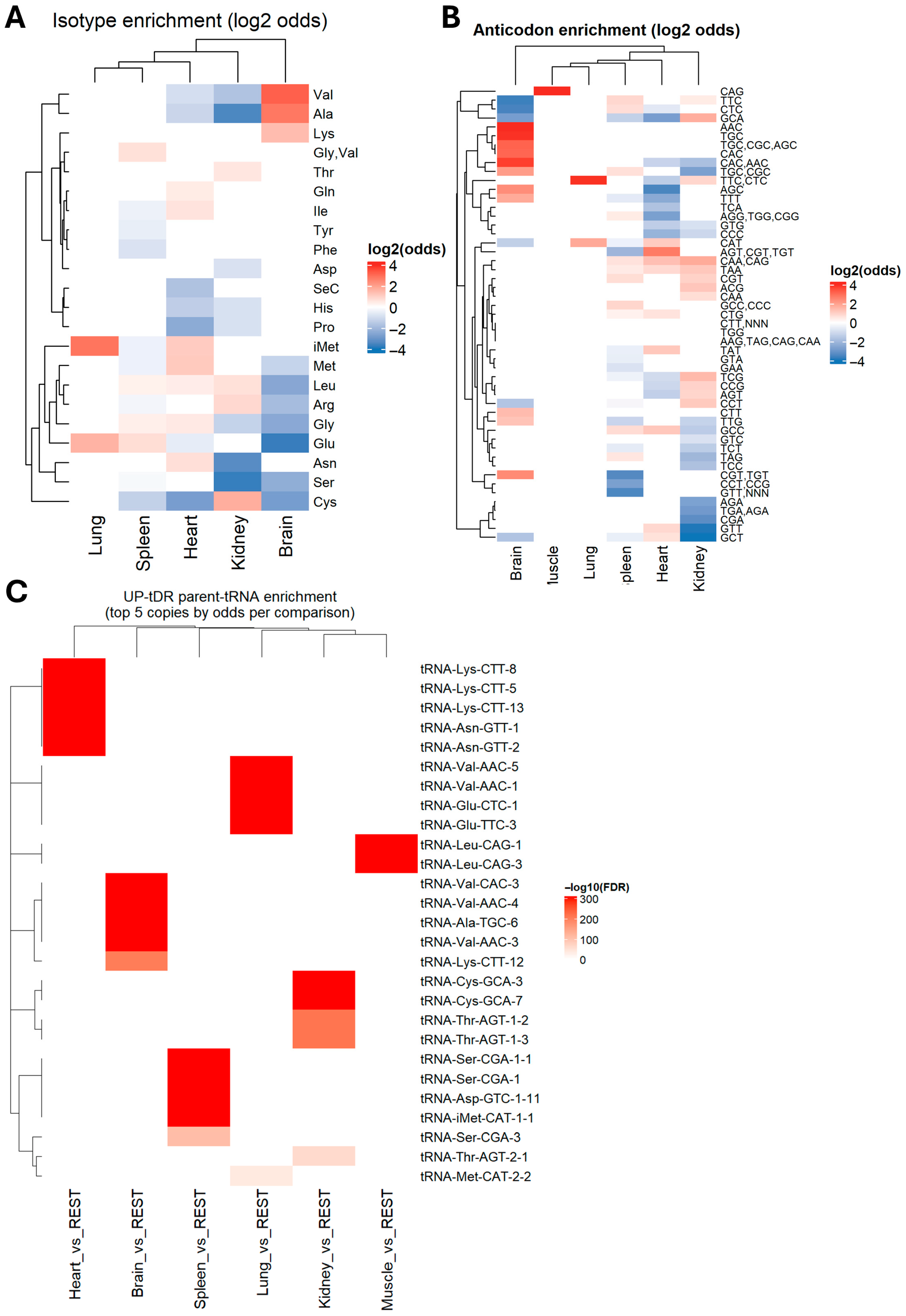
2.2. tDRs Show Tissue-Specific Enrichment Patterns
2.3. tDRs Enriched in Different Tissues Have Unique tRNA Origins
2.4. Highly Expressed Tissue-Resident tDRs Are Heterogeneous
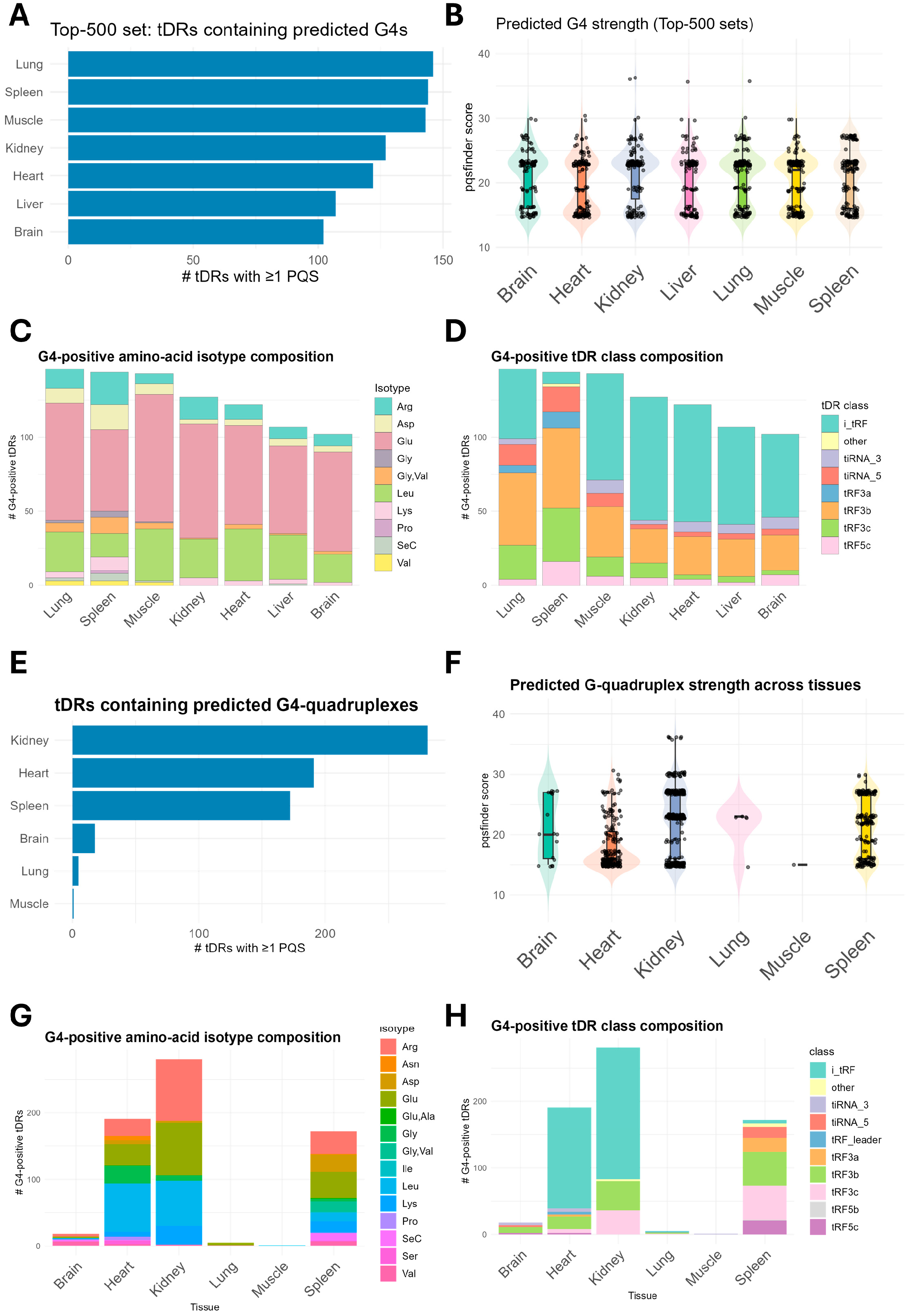
2.5. G4 Quadruplexes Forming tDRs Are Enriched in the Spleen
2.6. Sequencing Methods Omitting RNA Deacetylation Skew Towards 5′tiRNA Fragments
3. Discussion
4. Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kuhle, B.; Chen, Q.; Schimmel, P. tRNA renovatio: Rebirth through fragmentation. Mol. Cell 2023, 83, 3953–3971. [Google Scholar] [CrossRef] [PubMed]
- Muthukumar, S.; Li, C.T.; Liu, R.J.; Bellodi, C. Roles and regulation of tRNA-derived small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2024, 25, 359–378. [Google Scholar] [CrossRef]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef]
- Rashad, S.; Niizuma, K.; Tominaga, T. tRNA cleavage: A new insight. Neural Regen. Res. 2020, 15, 47–52. [Google Scholar] [CrossRef]
- Akiyama, Y.; Ivanov, P. tRNA-derived RNAs: Biogenesis and roles in translational control. Wiley Interdiscip. Rev. RNA 2023, 14, e1805. [Google Scholar] [CrossRef]
- Rashad, S.; Tominaga, T.; Niizuma, K. The cell and stress-specific canonical and noncanonical tRNA cleavage. J. Cell Physiol. 2021, 236, 3710–3724. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Manning, A.C.; Bagi, A.; Yang, X.; Gokulnath, P.; Spanos, M.; Howard, J.; Chan, P.P.; Sweeney, T.; Kitchen, R.; et al. Distinct Stress-Dependent Signatures of Cellular and Extracellular tRNA-Derived Small RNAs. Adv. Sci. 2022, 9, e2200829. [Google Scholar] [CrossRef]
- Yue, T.; Zhan, X.; Zhang, D.; Jain, R.; Wang, K.W.; Choi, J.H.; Misawa, T.; Su, L.; Quan, J.; Hildebrand, S.; et al. SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity. Science 2021, 372, eaba4220. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Malone, D.; Gao, X.; Wang, J.Y.J.; David, M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018, 25, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ji, Q.; Chaturvedi, J.; Morales, M.; Mao, Y.; Meng, X.; Dong, L.; Deng, J.; Qian, S.B.; Xiang, Y. Human SAMD9 is a poxvirus-activatable anticodon nuclease inhibiting codon-specific protein synthesis. Sci. Adv. 2023, 9, eadh8502. [Google Scholar] [CrossRef]
- Di Fazio, A.; Schlackow, M.; Pong, S.K.; Alagia, A.; Gullerova, M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022, 50, 1734–1752. [Google Scholar] [CrossRef]
- Rashad, S.; Han, X.; Sato, K.; Mishima, E.; Abe, T.; Tominaga, T.; Niizuma, K. The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol. 2020, 17, 1092–1103. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Wang, X.; Matuszek, Z.; Huang, Y.; Parisien, M.; Dai, Q.; Clark, W.; Schwartz, M.H.; Pan, T. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 2018, 24, 1305–1313. [Google Scholar] [CrossRef]
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206. [Google Scholar] [CrossRef]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.; Valdmanis, P.N.; Kim, H.K. Emerging roles of tRNA-derived small RNAs in cancer biology. Exp. Mol. Med. 2023, 55, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Yao, W.; Ni, X.S.; Yao, M.D.; Bai, W.; Yang, T.J.; Zhang, Z.R.; Li, X.M.; Jiang, Q.; Yan, B. Hyperglycemia-regulated tRNA-derived fragment tRF-3001a propels neurovascular dysfunction in diabetic mice. Cell Rep. Med. 2023, 4, 101209. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, K.A.; Hu, G.F. Mechanism and Function of Angiogenin in Hematopoietic Malignancy. Zhongguo Sheng Wu Hua Xue Yu Fen Zi Sheng Wu Xue Bao 2015, 31, 1267–1275. [Google Scholar] [PubMed]
- Goncalves, K.A.; Silberstein, L.; Li, S.; Severe, N.; Hu, M.G.; Yang, H.; Scadden, D.T.; Hu, G.F. Angiogenin Promotes Hematopoietic Regeneration by Dichotomously Regulating Quiescence of Stem and Progenitor Cells. Cell 2016, 166, 894–906. [Google Scholar] [CrossRef]
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef]
- Bayazit, M.B.; Jacovetti, C.; Cosentino, C.; Sobel, J.; Wu, K.; Brozzi, F.; Rodriguez-Trejo, A.; Stoll, L.; Guay, C.; Regazzi, R. Small RNAs derived from tRNA fragmentation regulate the functional maturation of neonatal β cells. Cell Rep. 2022, 40, 111069. [Google Scholar] [CrossRef]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960. [Google Scholar] [CrossRef]
- Kim, H.K.; Xu, J.; Chu, K.; Park, H.; Jang, H.; Li, P.; Valdmanis, P.N.; Zhang, Q.C.; Kay, M.A. A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice. Cell Rep. 2019, 29, 3816–3824.e3814. [Google Scholar] [CrossRef]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef]
- Lyons, S.M.; Gudanis, D.; Coyne, S.M.; Gdaniec, Z.; Ivanov, P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017, 8, 1127. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, W.; Liu, K.; Jiang, Z.; Han, Y.; Jin, H.; Zhang, L.; Shen, W.; Jia, S.; Sun, Q.; et al. 5′ Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 2021, 7, eabh0494. [Google Scholar] [CrossRef]
- Schorn, A.J.; Gutbrod, M.J.; LeBlanc, C.; Martienssen, R. LTR-Retrotransposon Control by tRNA-Derived Small RNAs. Cell 2017, 170, 61–71.e11. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Muthukumar, S.; Cieśla, M.; Todisco, G.; Ngoc, P.C.T.; Madej, M.; Munita, R.; Fazio, S.; Ekström, S.; Mortera-Blanco, T.; et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat. Cell Biol. 2022, 24, 299–306. [Google Scholar] [CrossRef]
- Ganser, L.R.; Kelly, M.L.; Herschlag, D.; Al-Hashimi, H.M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019, 20, 474–489. [Google Scholar] [CrossRef]
- Madrer, N.; Vaknine-Treidel, S.; Zorbaz, T.; Tzur, Y.; Bennett, E.R.; Drori, P.; Suissa, N.; Greenberg, D.S.; Lerner, E.; Soreq, E.; et al. Pre-symptomatic Parkinson’s disease blood test quantifying repetitive sequence motifs in transfer RNA fragments. Nat. Aging 2025, 5, 868–882. [Google Scholar] [CrossRef]
- Dhahbi, J.M.; Spindler, S.R.; Atamna, H.; Yamakawa, A.; Boffelli, D.; Mote, P.; Martin, D.I. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 2013, 14, 298. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Hu, W.; Huang, Y.; Lv, Y.; Ji, D.; Chen, C.; Yang, B.; Zhang, C.; Liang, Y.; Zhang, H.; et al. A Novel tsRNA, m(7)G-3′ tiRNA Lys(TTT), Promotes Bladder Cancer Malignancy Via Regulating ANXA2 Phosphorylation. Adv. Sci. 2024, 11, e2400115. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Honda, S.; Jing, Y.; Palazzo, J.; Kirino, Y.; Rigoutsos, I. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 2015, 6, 24797–24822. [Google Scholar] [CrossRef]
- La Ferlita, A.; Alaimo, S.; Nigita, G.; Distefano, R.; Beane, J.D.; Tsichlis, P.N.; Ferro, A.; Croce, C.M.; Pulvirenti, A. tRFUniverse: A comprehensive resource for the interactive analyses of tRNA-derived ncRNAs in human cancer. iScience 2024, 27, 108810. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Tan, D.; Zhang, X.; Yan, M.; Zhang, Y.; Franklin, R.; Shahbazi, M.; Mackinlay, K.; Liu, S.; et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 2021, 23, 424–436. [Google Scholar] [CrossRef]
- Wang, H.; Huang, R.; Li, L.; Zhu, J.; Li, Z.; Peng, C.; Zhuang, X.; Lin, H.; Shi, S.; Huang, P. CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov. 2021, 7, 25. [Google Scholar] [CrossRef]
- Isakova, A.; Fehlmann, T.; Keller, A.; Quake, S.R. A mouse tissue atlas of small noncoding RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 25634–25645. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Zhu, L.; Guo, Z.; Liu, W.; Zhang, J.; Zeng, Z.; Wu, Q.; Cheng, J.; Fu, X.; Jin, Y.; et al. tsRBase: A comprehensive database for expression and function of tsRNAs in multiple species. Nucleic Acids Res. 2021, 49, D1038–D1045. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef]
- Liu, Z.; Kim, H.K.; Xu, J.; Jing, Y.; Kay, M.A. The 3′tsRNAs are aminoacylated: Implications for their biogenesis. PLoS Genet. 2021, 17, e1009675. [Google Scholar] [CrossRef]
- Ando, D.; Rashad, S.; Begley, T.J.; Endo, H.; Aoki, M.; Dedon, P.C.; Niizuma, K. Decoding Codon Bias: The Role of tRNA Modifications in Tissue-Specific Translation. Int. J. Mol. Sci. 2025, 26, 706. [Google Scholar] [CrossRef]
- Hu, J.F.; Yim, D.; Ma, D.; Huber, S.M.; Davis, N.; Bacusmo, J.M.; Vermeulen, S.; Zhou, J.; Begley, T.J.; DeMott, M.S.; et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat. Biotechnol. 2021, 39, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Cozen, A.E.; Quartley, E.; Holmes, A.D.; Hrabeta-Robinson, E.; Phizicky, E.M.; Lowe, T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 2015, 12, 879–884. [Google Scholar] [CrossRef]
- Holmes, A.D.; Chan, P.P.; Chen, Q.; Ivanov, P.; Drouard, L.; Polacek, N.; Kay, M.A.; Lowe, T.M. A standardized ontology for naming tRNA-derived RNAs based on molecular origin. Nat. Methods 2023, 20, 627–628. [Google Scholar] [CrossRef] [PubMed]
- Pinkard, O.; McFarland, S.; Sweet, T.; Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 2020, 11, 4104. [Google Scholar] [CrossRef]
- Sanadgol, N.; König, L.; Drino, A.; Jovic, M.; Schaefer, M.R. Experimental paradigms revisited: Oxidative stress-induced tRNA fragmentation does not correlate with stress granule formation but is associated with delayed cell death. Nucleic Acids Res. 2022, 50, 6919–6937. [Google Scholar] [CrossRef]
- Hon, J.; Martínek, T.; Zendulka, J.; Lexa, M. pqsfinder: An exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics 2017, 33, 3373–3379. [Google Scholar] [CrossRef]
- Winek, K.; Lobentanzer, S.; Nadorp, B.; Dubnov, S.; Dames, C.; Jagdmann, S.; Moshitzky, G.; Hotter, B.; Meisel, C.; Greenberg, D.S.; et al. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc. Natl. Acad. Sci. USA 2020, 117, 32606–32616. [Google Scholar] [CrossRef]
- Gao, L.; Behrens, A.; Rodschinka, G.; Forcelloni, S.; Wani, S.; Strasser, K.; Nedialkova, D.D. Selective gene expression maintains human tRNA anticodon pools during differentiation. Nat. Cell Biol. 2024, 26, 100–112. [Google Scholar] [CrossRef]
- Lyons, S.M.; Kharel, P.; Akiyama, Y.; Ojha, S.; Dave, D.; Tsvetkov, V.; Merrick, W.; Ivanov, P.; Anderson, P. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 2020, 48, 6223–6233. [Google Scholar] [CrossRef] [PubMed]
- Boon, N.J.; Oliveira, R.A.; Körner, P.R.; Kochavi, A.; Mertens, S.; Malka, Y.; Voogd, R.; van der Horst, S.E.M.; Huismans, M.A.; Smabers, L.P.; et al. DNA damage induces p53-independent apoptosis through ribosome stalling. Science 2024, 384, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, L.; Lun, A.T.L.; Baldoni, P.L.; Smyth, G.K. edgeR v4: Powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. Nucleic Acids Res. 2025, 53, gkaf018. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ando, D.; Rashad, S.; Niizuma, K. Optimized tDR Sequencing Reveals Diversity and Heterogeneity in tRNA-Derived Fragment Landscapes in Mouse Tissues. Int. J. Mol. Sci. 2025, 26, 8772. https://doi.org/10.3390/ijms26188772
Ando D, Rashad S, Niizuma K. Optimized tDR Sequencing Reveals Diversity and Heterogeneity in tRNA-Derived Fragment Landscapes in Mouse Tissues. International Journal of Molecular Sciences. 2025; 26(18):8772. https://doi.org/10.3390/ijms26188772
Chicago/Turabian StyleAndo, Daisuke, Sherif Rashad, and Kuniyasu Niizuma. 2025. "Optimized tDR Sequencing Reveals Diversity and Heterogeneity in tRNA-Derived Fragment Landscapes in Mouse Tissues" International Journal of Molecular Sciences 26, no. 18: 8772. https://doi.org/10.3390/ijms26188772
APA StyleAndo, D., Rashad, S., & Niizuma, K. (2025). Optimized tDR Sequencing Reveals Diversity and Heterogeneity in tRNA-Derived Fragment Landscapes in Mouse Tissues. International Journal of Molecular Sciences, 26(18), 8772. https://doi.org/10.3390/ijms26188772





