Interferon-Linked Lipid and Bile Acid Imbalance Uncovered in Ankylosing Spondylitis in a Sibling-Controlled Multi-Omics Study
Abstract
1. Introduction
2. Results
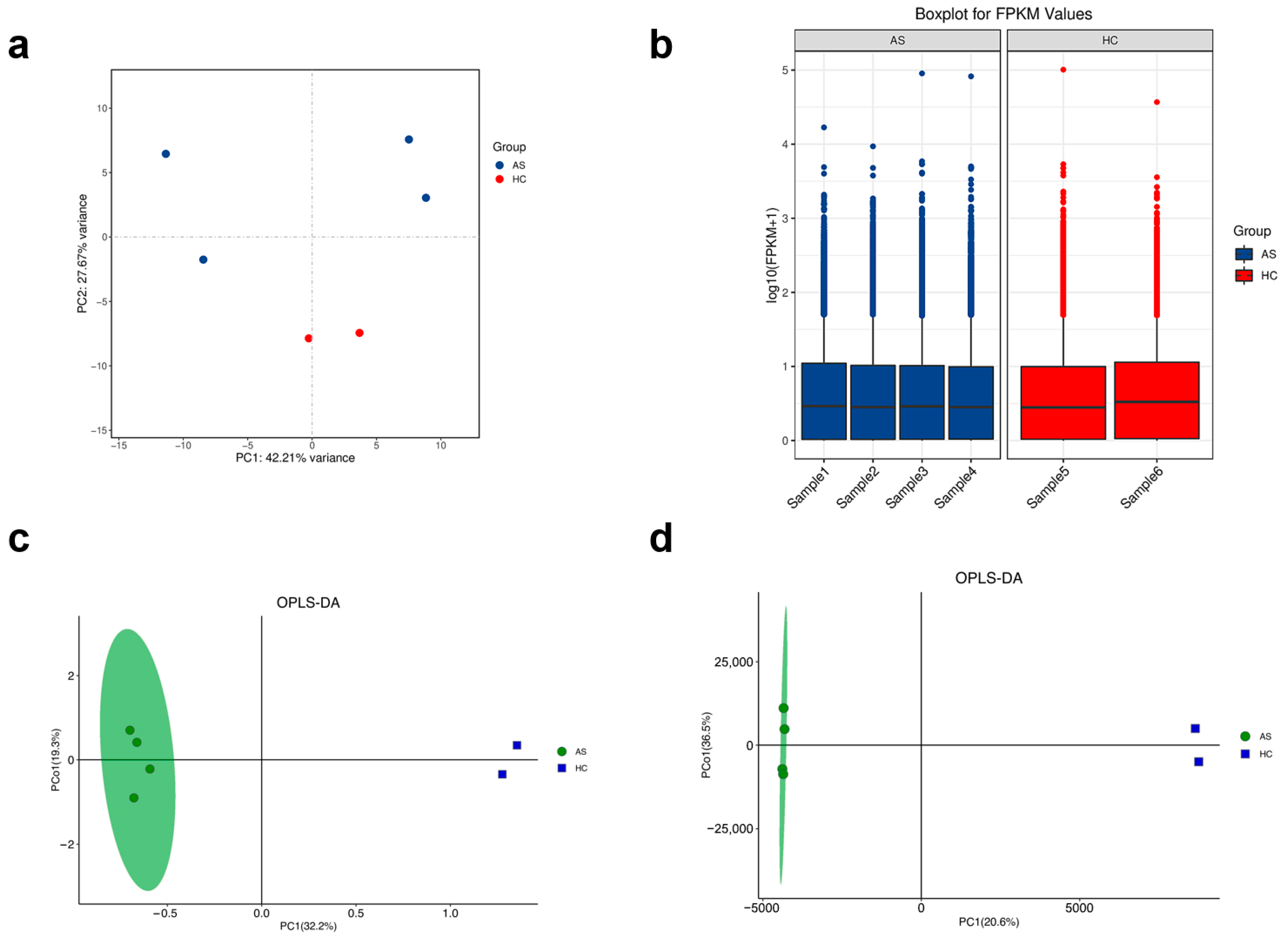
2.1. Global Quality Assessment and Sample Clustering
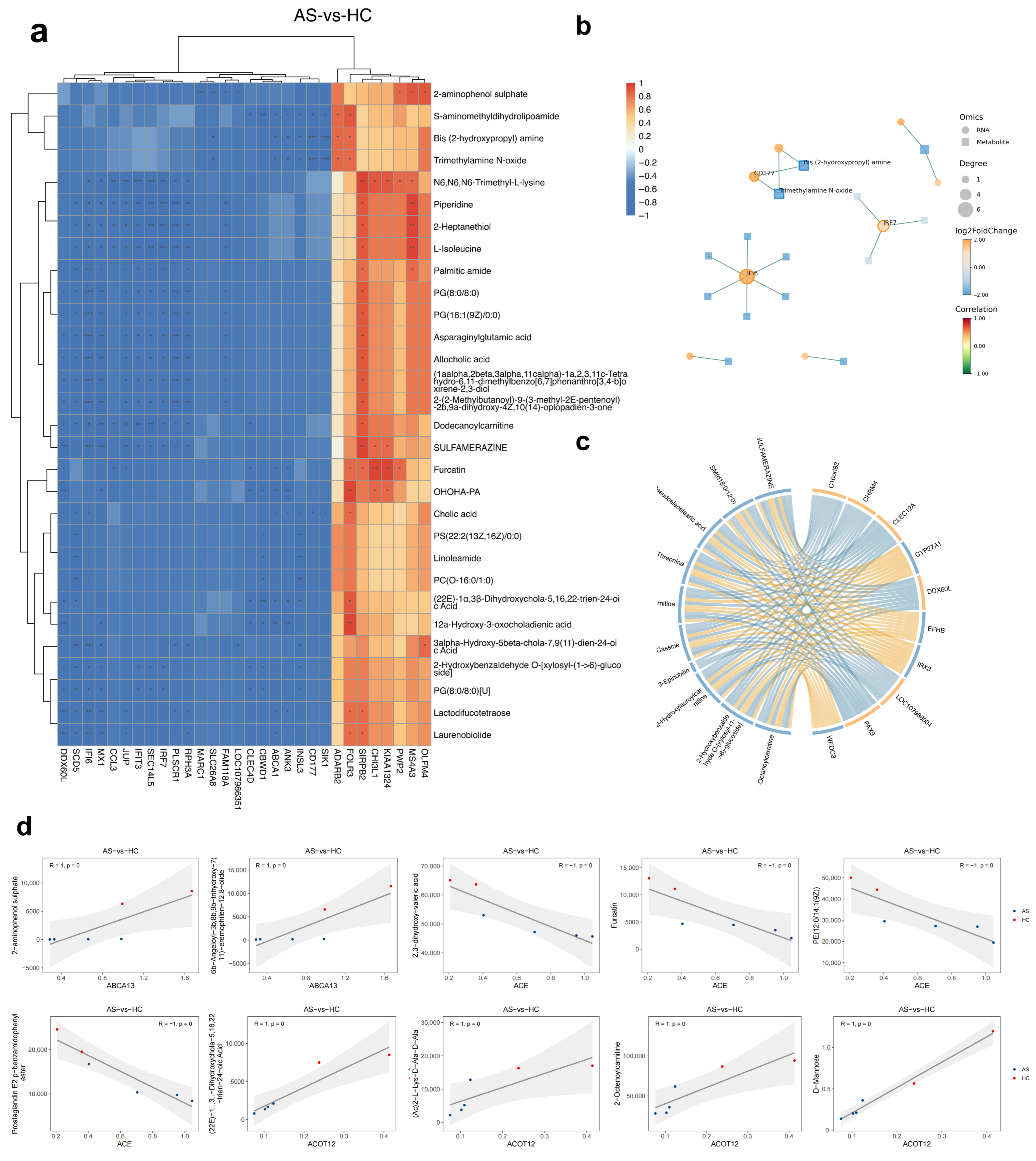
2.2. Transcriptomic Alterations Indicate an Interferon-Driven, Neutrophil-Skewed Immune Signature
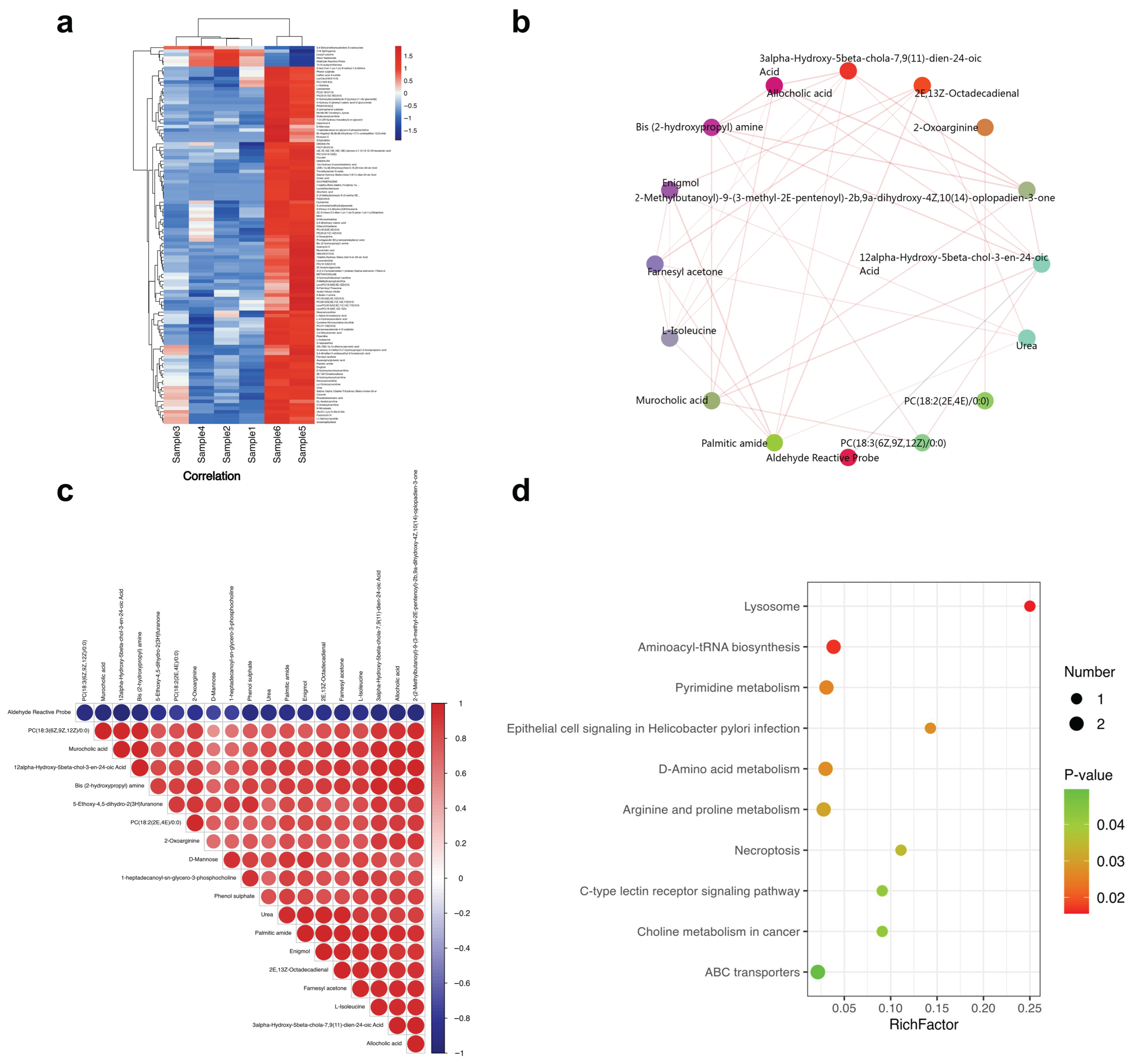
2.3. Combined GC-/LC-MS Analysis Highlights Bile Acid Dysregulation and Lipid Peroxidation Clusters
2.4. Multi-Omics Correlations Reveal Lipid Peroxidation and Bile Acid Hubs That Track Immune Activation
3. Discussion
3.1. A Systemic Type I Interferon/Neutrophil Program Dominates the AS PBMC Transcriptome
3.2. Suppression of Fatty Acid β-Oxidation and Accumulation of Lipid Peroxidation Products
3.3. Bile Acid Depletion as a Putative Metabolic Checkpoint in AS
3.4. Gene–Metabolite Hubs Suggest Novel Biomarkers and Therapeutic Avenues
3.5. Limitations and Future Directions
4. Conclusions
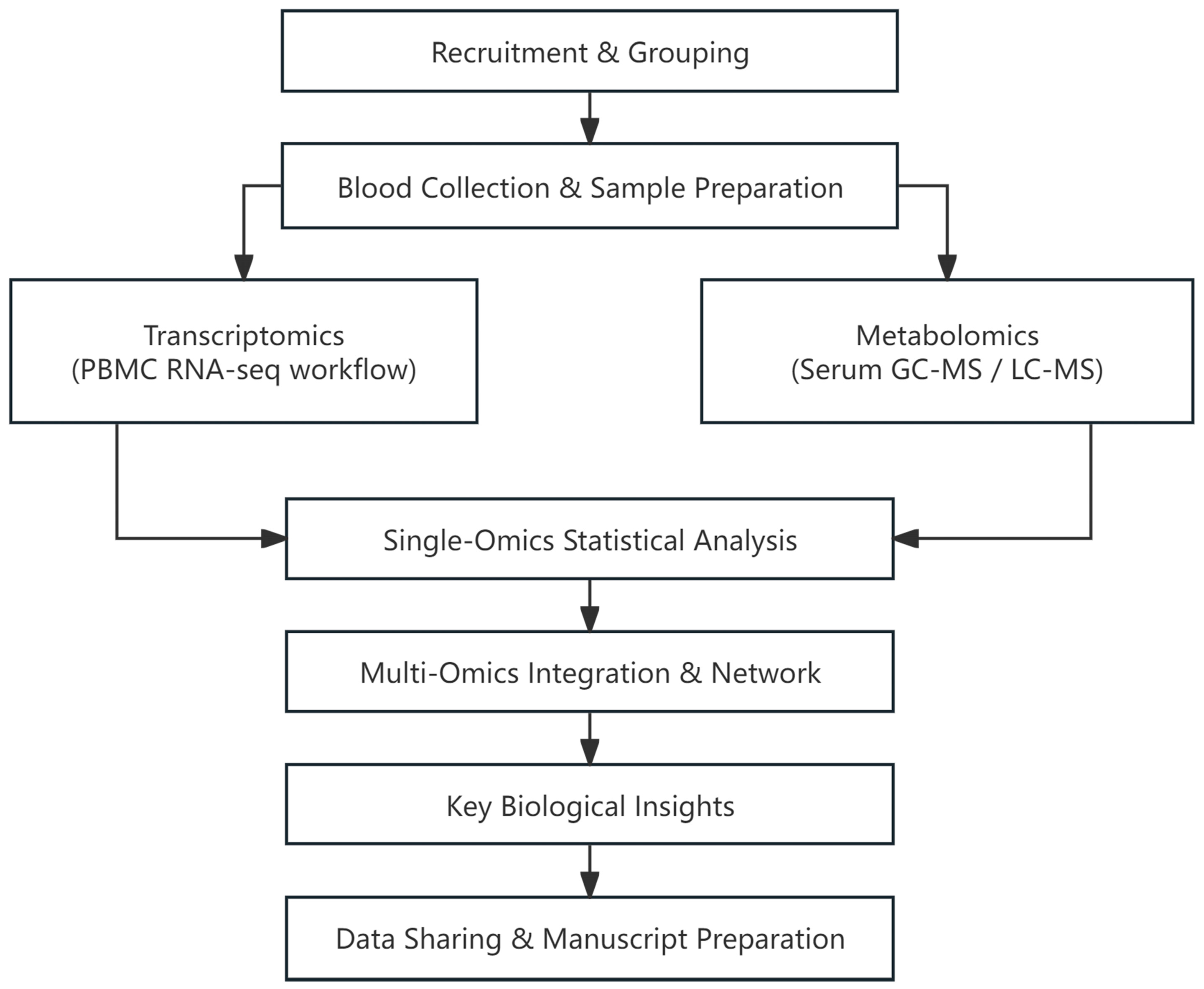
5. Materials and Methods
5.1. Study Design and Subjects
5.2. Clinical Assessment, Data Collection, and Ethics
5.3. RNA Extraction, Library Construction, and Sequencing
5.4. RNA Sequencing Analysis Process
5.5. Differential Gene Expression Analysis
5.6. Serum Metabolite Extraction and GC-MS Acquisition
5.7. Serum Metabolite Extraction and LC-MS Acquisition
5.8. Metabolomics Data Processing and Annotation
5.9. Gene Set Enrichment Analysis
5.10. Differential Feature Selection and Data Normalization
5.11. Combined Transcriptome–Metabolome Analysis
5.12. Pathway Enrichment and KEGG Mapping
5.13. Statistical Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS | Ankylosing Spondylitis |
| HC | Healthy Controls |
| PBMCs | Peripheral Blood Mononuclear Cells |
| GC-MS | Gas Chromatography–Mass Spectrometry |
| LC-MS | Liquid Chromatography–Mass Spectrometry |
| mSASSS | Modified Stoke Ankylosing Spondylitis Spinal Score |
| ULN | Upper Limit of Normal |
| BMI | Body Mass Index |
| BASDAI | Bath Ankylosing Spondylitis Disease Activity Index |
| PCA | Principal Component Analysis |
| OPLS-DA | Orthogonal Partial Least Squares Discriminant Analysis |
| GSEA | Gene Set Enrichment Analysis |
| QC | Quality Control |
| MSI | Metabolomics Standards Initiative |
| DEM | Differentially Abundant Metabolite |
| DEGs | Differentially Expressed Genes |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Ritchlin, C.; Adamopoulos, I.E. Axial Spondyloarthritis: New Advances in Diagnosis and Management. BMJ 2021, 372, m4447. [Google Scholar] [CrossRef] [PubMed]
- Ortolan, A.; Webers, C.; Sepriano, A.; Falzon, L.; Baraliakos, X.; Landewé, R.B.; Ramiro, S.; van der Heijde, D.; Nikiphorou, E. Efficacy and Safety of Non-Pharmacological and Non-Biological Interventions: A Systematic Literature Review Informing the 2022 Update of the ASAS/EULAR Recommendations for the Management of Axial Spondyloarthritis. Ann. Rheum. Dis. 2023, 82, 142–152. [Google Scholar] [CrossRef] [PubMed]
- McGonagle, D.; David, P.; Macleod, T.; Watad, A. Predominant Ligament-Centric Soft-Tissue Involvement Differentiates Axial Psoriatic Arthritis from Ankylosing Spondylitis. Nat. Rev. Rheumatol. 2023, 19, 818–827. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Sieper, J. Ankylosing Spondylitis. Lancet 2007, 369, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Kenna, T.; Wordsworth, B.P. Genetics of Ankylosing Spondylitis—Insights into Pathogenesis. Nat. Rev. Rheumatol. 2015, 12, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Tam, L.-S.; Gu, J.; Yu, D. Pathogenesis of Ankylosing Spondylitis. Nat. Rev. Rheumatol. 2010, 6, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Mauro, D.; Thomas, R.; Guggino, G.; Lories, R.; Brown, M.A.; Ciccia, F. Ankylosing Spondylitis: An Autoimmune or Autoinflammatory Disease? Nat. Rev. Rheumatol. 2021, 17, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Li, Z.; Cao, K.-A.L. Biomarker Development for Axial Spondyloarthritis. Nat. Rev. Rheumatol. 2020, 16, 448–463. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.L.; Lejeune, A.; Chang, G.; Scher, J.U.; Koralov, S.B. Microbial-Derived Antigens and Metabolites in Spondyloarthritis. Semin. Immunopathol. 2021, 43, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Hedner, T.; Samulesson, O.; Währborg, P.; Wadenvik, H.; Ung, K.-A.; Ekbom, A. Nabumetone: Therapeutic Use and Safety Profile in the Management of Osteoarthritis and Rheumatoid Arthritis. Drugs 2004, 64, 2315–2343. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Tang, D.; Chen, D.; Zheng, F.; Huang, S.; Xu, Y.; Yu, H.; He, J.; Hong, X.; Yin, L.; et al. Advances in Applying of Multi-Omics Approaches in the Research of Systemic Lupus Erythematosus. Int. Rev. Immunol. 2020, 39, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Kreitmaier, P.; Katsoula, G.; Zeggini, E. Insights from Multi-Omics Integration in Complex Disease Primary Tissues. Trends Genet. TIG 2023, 39, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.J.; Maksymowych, W.P. Beyond the TNF-α Inhibitors: New and Emerging Targeted Therapies for Patients with Axial Spondyloarthritis and Their Relation to Pathophysiology. Drugs 2018, 78, 1397–1418. [Google Scholar] [CrossRef] [PubMed]
- Gasparotto, M.; Franco, C.; Zanatta, E.; Ghirardello, A.; Zen, M.; Iaccarino, L.; Fabris, B.; Doria, A.; Gatto, M. The Interferon in Idiopathic Inflammatory Myopathies: Different Signatures and New Therapeutic Perspectives. A Literature Review. Autoimmun. Rev. 2023, 22, 103334. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Laurent-Rolle, M.; Grove, T.L.; Hsu, J.C.-C. Interferon-Stimulated Genes and Immune Metabolites as Broad-Spectrum Biomarkers for Viral Infections. Viruses 2025, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, K.Z.; Wilhelm, T.R.; Ulff-Møller, C.J.; Jacobsen, S. Cluster of Highly Expressed Interferon-Stimulated Genes Associate More with African Ancestry than Disease Activity in Patients with Systemic Lupus Erythematosus. A Systematic Review of Cross-Sectional Studies. Transl. Res. J. Lab. Clin. Med. 2021, 238, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, Y.-H.; Liu, B.; Yang, H.-X.; Li, G.-T.; Zhou, H.-L.; Wang, Y.-S. Role of Peroxisomes in the Pathogenesis and Therapy of Renal Fibrosis. Metab. Clin. Exp. 2025, 166, 156173. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gu, Y.; Li, L.; Liu, T.; Song, X.; Sun, Y.; Cao, X.; Wang, B.; Jiang, K.; Cao, H. Bile Acid-Gut Microbiota Axis in Inflammatory Bowel Disease: From Bench to Bedside. Nutrients 2021, 13, 3143. [Google Scholar] [CrossRef] [PubMed]
- Christofides, A.; Konstantinidou, E.; Jani, C.; Boussiotis, V.A. The Role of Peroxisome Proliferator-Activated Receptors (PPAR) in Immune Responses. Metab. Clin. Exp. 2021, 114, 154338. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Log2FC | Fold Change | Adjusted p-Value (BH-FDR) | Regulation |
|---|---|---|---|---|
| SIK1 | 3.34 | 10.16 | 5.86 × 10−7 | Up |
| TMEM191B | 2.89 | 7.40 | 7.00 × 10−5 | Up |
| LOC107986351 | 2.63 | 6.19 | 4.59 × 10−8 | Up |
| CD177 | 2.49 | 5.62 | 3.95 × 10−10 | Up |
| IFI6 | 2.42 | 5.35 | 1.71 × 10−6 | Up |
| RPH3A | 2.16 | 4.48 | 3.08 × 10−8 | Up |
| MX1 | 2.03 | 4.08 | 4.42 × 10−6 | Up |
| SCD5 | 2.01 | 4.04 | 4.37 × 10−10 | Up |
| IFIT3 | 2.01 | 4.02 | 2.94 × 10−5 | Up |
| OSM | 1.76 | 3.38 | 4.89 × 10−5 | Up |
| MS4A3 | −1.33 | 0.40 | 3.65 × 10−5 | Down |
| PTPRF | −1.37 | 0.39 | 1.24 × 10−4 | Down |
| CAMP | −1.73 | 0.30 | 5.13 × 10−5 | Down |
| CHI3L1 | −2.16 | 0.22 | 2.16 × 10−6 | Down |
| OLFM4 | −2.33 | 0.20 | 7.52 × 10−6 | Down |
| FOLR3 | −2.82 | 0.14 | 1.28 × 10−21 | Down |
| PWP2 | −2.95 | 0.13 | 9.85 × 10−13 | Down |
| MYO18B | −3.08 | 0.12 | 1.61 × 10−4 | Down |
| TBC1D3 | −4.64 | 0.04 | 4.05 × 10−5 | Down |
| ADARB2 | −6.62 | 0.01 | 1.87 × 10−6 | Down |
| Data Class | Metabolites | VIP | Fold Change | Adjusted p-Value (BH-FDR) | Regulation |
|---|---|---|---|---|---|
| GC | D-Mannose | 8.49 | 0.26 | 3.38 × 10−2 | Down |
| GC | Urea | 6.57 | 0.77 | 2.34 × 10−2 | Down |
| GC | L-Alpha-aminobutyric acid | 1.03 | 0.47 | 4.07 × 10−2 | Down |
| LC | Leucyl-leucine | 1.14 | 3.27 | 3.96 × 10−2 | Up |
| LC | Hexyl heptanoate | 1.58 | 3.18 | 1.99 × 10−2 | Up |
| LC | Aldehyde reactive probe | 3.69 | 3.08 | 1.31 × 10−2 | Up |
| LC | Tri-N-acetylchitotriose | 2.19 | 2.97 | 1.46 × 10−2 | Up |
| LC | 3,4-Dehydrothiomorpholine-3-carboxylate | 1.57 | 2.24 | 2.76 × 10−2 | Up |
| LC | C16 Sphinganine | 2.92 | 1.33 | 4.69 × 10−2 | Up |
| LC | 2-(2-methylbutanoyl)-9-(3-methyl-2E-pentenoyl)-2b,9a-dihydroxy-4Z,10(14)-oplopadien-3-one | 3.22 | 0.02 | 2.59 × 10−4 | Down |
| LC | 6b-Angeloyl-3b,8b,9b-trihydroxy-7(11)-eremophilen-12,8-olide | 1.14 | 0.02 | 4.37 × 10−3 | Down |
| LC | PG(8:0/8:0) | 2.22 | 0.02 | 2.51 × 10−4 | Down |
| LC | (1aalpha,2beta,3alpha,11calpha)-1a,2,3,11c-Tetrahydro-6,11-dimethylbenzo [6,7]phenanthro [3,4-b]oxirene-2,3-diol | 1.64 | 0.01 | 1.09 × 10−4 | Down |
| LC | 2-Aminophenol sulphate | 1.06 | 0.01 | 4.19 × 10−4 | Down |
| LC | PG(8:0/8:0) [U] | 1.07 | 0.00 | 2.87 × 10−5 | Down |
| LC | Lactodifucotetraose | 1.22 | 0.00 | 1.54 × 10−4 | Down |
| LC | Niveusin C | 2.67 | 0.00 | 9.92 × 10−3 | Down |
| LC | SM(d18:0/12:0) | 1.47 | 0.00 | 4.73 × 10−3 | Down |
| LC | 3-Epinobilin | 1.53 | 0.00 | 1.71 × 10−2 | Down |
| Number | Group | F/M | Age | Surgery History | HLA-B27 | mSASSS | BASDAI |
|---|---|---|---|---|---|---|---|
| Sample 1 | AS | M | 49 | YES | + | 32 | 4.5 |
| Sample 2 | AS | M | 51 | NO | + | 10 | 2.3 |
| Sample 3 | AS | M | 40 | YES | + | 45 | 5.1 |
| Sample 4 | AS | M | 43 | NO | + | 12 | 2.6 |
| Sample 5 | HC | M | 41 | NO | − | − | − |
| Sample 6 | HC | M | 41 | NO | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Huang, Y.; Guo, Z.; Sun, J.; Zheng, G. Interferon-Linked Lipid and Bile Acid Imbalance Uncovered in Ankylosing Spondylitis in a Sibling-Controlled Multi-Omics Study. Int. J. Mol. Sci. 2025, 26, 7919. https://doi.org/10.3390/ijms26167919
Wang Z, Huang Y, Guo Z, Sun J, Zheng G. Interferon-Linked Lipid and Bile Acid Imbalance Uncovered in Ankylosing Spondylitis in a Sibling-Controlled Multi-Omics Study. International Journal of Molecular Sciences. 2025; 26(16):7919. https://doi.org/10.3390/ijms26167919
Chicago/Turabian StyleWang, Ze, Yi Huang, Ziyu Guo, Jianhua Sun, and Guoquan Zheng. 2025. "Interferon-Linked Lipid and Bile Acid Imbalance Uncovered in Ankylosing Spondylitis in a Sibling-Controlled Multi-Omics Study" International Journal of Molecular Sciences 26, no. 16: 7919. https://doi.org/10.3390/ijms26167919
APA StyleWang, Z., Huang, Y., Guo, Z., Sun, J., & Zheng, G. (2025). Interferon-Linked Lipid and Bile Acid Imbalance Uncovered in Ankylosing Spondylitis in a Sibling-Controlled Multi-Omics Study. International Journal of Molecular Sciences, 26(16), 7919. https://doi.org/10.3390/ijms26167919







