Research Progress on 35S rDNA and 5S rDNA in Sugarcane: Challenges and Prospects
Abstract
1. Introduction
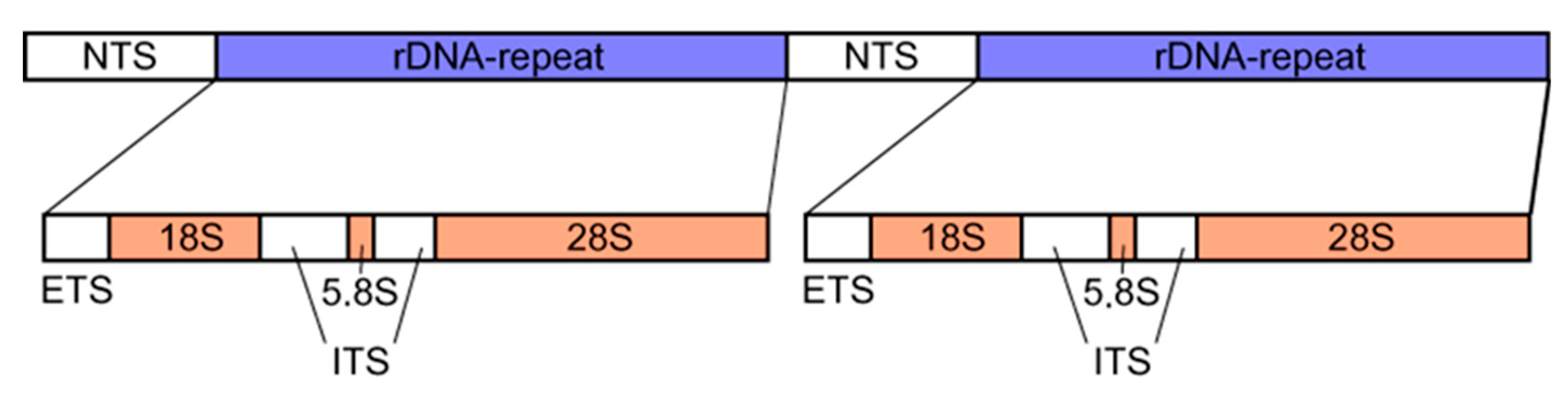
1.1. Composition and Characteristics of 35S rDNA and 5S rDNA
1.2. Development and Utilization of 35S rDNA and 5S rDNA in Plants Based on FISH
2. Research on the Phylogenetic Relationship and Evolutionary Relationship of rDNA in Sugarcane
2.1. Study on the Ploidy Relationship and Distribution of Sugarcane Using 35S rDNA and 5S rDNA
| Species | 5S Loci Num | 5S Position | 35S Loci Num | 35S Position | 2n | Ploidy |
|---|---|---|---|---|---|---|
| S. spontaneum [48] | 14 | interstitial | 10 | interstitial | 112 | 14 |
| S. spontaneum [48] | 12 | interstitial | 12 | interstitial | 96 | 12 |
| S. spontaneum [48] | 10 | interstitial | 10 | interstitial | 80 | 10 |
| S. spontaneum [48] | 8 | interstitial | 8 | interstitial | 64 | 8 |
| S. spontaneum [43] | 4 | (peri-)centromeric | 4 | satellite | 32 | 4 |
| S. officinarum [48] | 8 | interstitial | 8 | (sub-)terminal or satellite | 80 | 8 |
| S. robustum [24] | 6 | interstitial | 6 | (sub-)terminal or satellite | 60 | 6 |
| S. robustum [24] | 8 | interstitial | 8 | (sub-)terminal or satellite | 80 | 8 |
| S. arundinaceum [44] | 6 | interstitial | 6 | (sub-)terminal or satellite | 60 | 6 |
| Triticum aestivum [49] | 2 | (sub-)terminal | 6 | (sub-)terminal | 42 | 6 |
| Oryza glaberrima [50] | 2 | interstitial | 2 | (peri-)centromeric | 24 | 2 |
| Zea mays [51] | 2 | (sub-)terminal | 2 | satellite | 20 | 2 |
| Sorghum bicolor [52] | / | / | 2 | (peri-)centromeric | 20 | 2 |
2.2. Application of 35S rDNA and 5S rDNA in the Study of Sugarcane Evolution and Genetic Relationship
2.3. Application of 35S rDNA and 5S rDNA in Chromosome Composition and Genetics
3. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, Y.; Skinner, D.Z.; Liang, G.H.; Hulbert, S.H. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theor. Appl. Genet. 1994, 89, 26–32. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: Implications for biogeography and concerted evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 6813–6817. [Google Scholar] [CrossRef]
- Castilho, A.; Heslop-Harrison, J.S. Physical mapping of 5S and 18S-25S rDNA and repetitive DNA sequences in Aegilops umbellulata. Genome 1995, 38, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Kubis, S.; Schmidt, T.; Heslopharrison; Pat, J.S. Repetitive DNA Elements as a Major Component of Plant Genomes. Ann. Bot. 1998, 82, 45–55. [Google Scholar] [CrossRef]
- Flavell, R.B.; Bennett, M.D.; Smith, J.B.; Smith, D.B. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem. Genet. 1974, 12, 257–269. [Google Scholar] [CrossRef]
- Koornneef, M.; Fransz, P.; Jong, H.D.J. Cytogenetic tools for Arabidopsis thaliana. Chromosome Res. 2003, 11, 183–194. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Friebe, B.; Gill, B.S. Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome 1996, 39, 293–306. [Google Scholar] [CrossRef]
- Huang, Y. Chromosome Inheritance for the BC1 Progeny of Sugarcane and Erianthus arundinaceus Based on 45S rDNA-FISH and GISH. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2014. [Google Scholar]
- Huang, Y.; Yu, F.; Li, X.; Luo, L.; Wu, J.; Yang, Y.; Deng, Z.; Chen, R.; Zhang, M. Comparative genetic analysis of the 45S rDNA intergenic spacers from three Saccharum species. PLoS ONE 2017, 12, e0183447. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A.; Rao, P.S.; Feldmann, P.; Grivet, L.; Islam-Faridi, N.; Taylor, P.; Glaszmann, J.C. Identification and characterisation of sugarcane intergeneric hybrids Saccharum officinarum × Erianthus arundinaceus, with molecular markers and DNA in situ hybridization. Theor. Appl. Genet. 1995, 91, 320–326. [Google Scholar] [CrossRef]
- Fransz, P.; Armstrong, S.; Alonso-Blanco, C.; Fischer, T.C.; Torres-Ruiz, R.A.; Jones, G. Cytogenetics for the model system Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 13, 867–876. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Z.; Lv, L. Chromosome Localization of 45S rDNA and 5S rDNA in Chinese Narcissus by Fluorescence in situ Hybridization. Chin. J. Trop. Crops 2008, 29, 618–621. [Google Scholar]
- Liao, J.; Yang, R.; Zhou, Y.; Tsujimoto, H. FISH analysis of 45S rDNA and 5S rDNA genes in Triticum polonicum L. and T. turgidum L. cv. Ailanmai. Hereditas 2007, 29, 449–454. [Google Scholar]
- Wang, Y.; Wang, C.; Jiang, Y.; Katz, L.A.; Gao, F.; Yan, Y. Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci. China. Life Sci. 2019, 62, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Lutzoni, F.; Wagner, P.; Reeb, V.; Zoller, S. Integrating ambiguously aligned regions of DNA sequences in phylogenetic analyses without violating positional homology. Syst. Biol. 2000, 49, 628–651. [Google Scholar] [CrossRef] [PubMed]
- Waminal, N.E.; Kim, H.H. Dual-color FISH karyotype and rDNA distribution analyses on four Cucurbitaceae species. Hortic. Environ. Biotechnol. 2012, 53, 49–56. [Google Scholar] [CrossRef]
- Xu, C.; Bie, T.; Wang, C.; Zhou, B.; Chen, P. Distribution of 45S rDNA sequence on chromosomes of Triticum aestivum and its relative species. Hereditas 2007, 29, 1126–1130. [Google Scholar]
- Baldwin, B.G.; Markos, S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: Congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogenetics Evol. 1998, 10, 449–463. [Google Scholar] [CrossRef]
- Thomas, H.M.; Harper, J.A.; Meredith, M.R.; Morgan, W.G.; Thomas, I.D.; Timms, E.; King, I.P. Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Res. 1996, 4, 486–490. [Google Scholar] [CrossRef]
- Huang, M.; Li, H.; Zhang, L.; Gao, F.; Wang, P.; Hu, Y.; Yan, S.; Zhao, L.; Zhang, Q.; Tan, J.; et al. Plant 45S rDNA clusters are fragile sites and their instability is associated with epigenetic alterations. PLoS ONE 2012, 7, e35139. [Google Scholar] [CrossRef]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Stults, D.M.; Killen, M.W.; Williamson, E.P.; Hourigan, J.S.; Vargas, H.D.; Arnold, S.M.; Moscow, J.A.; Pierce, A.J. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009, 69, 9096–9104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Chen, S.; Zhang, F.; Qi, F.; Guan, X. Localization of 45S and 5S rDNA sites and karyotype of Chrysanthemum and its related genera by fluorescent in situ hybridization. Biochem. Syst. 2015, 62, 164–172. [Google Scholar]
- D’Hont, A.; Ison, D.; Alix, K.; Roux, C.; Glaszmann, J.C. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome Biol. Evol. 1998, 41, 221–225. [Google Scholar] [CrossRef]
- Piperidis, G.; Christopher, M.J.; Carroll, B.J.; Berding, N.; D’Hont, A. Molecular contribution to selection of intergeneric hybrids between sugarcane and the wild species Erianthus arundinaceus. Genome 2000, 43, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Acevedo, R.; Moreno Díaz de la Espina, S.; Jouve, N.; de la Torre, C. Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J. Exp. Bot. 2004, 55, 847–854. [Google Scholar] [CrossRef]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, Z.; Yan, T.; Lin, Q.; Wang, Y.; Huang, W.; Huang, Y.; Li, Z.; Yu, Q.; Wang, J.; et al. Comprehensively Characterizing the Cytological Features of Saccharum spontaneum by the Development of a Complete Set of Chromosome-Specific Oligo Probes. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, X.; Tian, L.; Chen, L.; Yu, W. Identification of peanut (Arachis hypogaea) chromosomes using a fluorescence in situ hybridization system reveals multiple hybridization events during tetraploid peanut formation. New Phytol. 2016, 211, 1424–1439. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, W.; Hao, L.; Wang, X.; Ma, Y.; Liu, Y. Analysis of karyotype and 45S rDNA locus of purple grain wheat. Biotechnology 2020, 30, 242–246. [Google Scholar]
- Zhang, T.; Jian, H.; Tian, M.; Wang, Q.; Zhang, H.; Yan, H.; Qiu, X.; Tang, K. Physical Location of 45S rDNA and 5S rDNA in the Genomes of Three Wild Rose Species. Acta Hortic. Sin. 2014, 41, 994–1000. [Google Scholar]
- Wang, J.; Roe, B.; Macmil, S.; Yu, Q.; Murray, J.E.; Tang, H.; Chen, C.; Najar, F.; Wiley, G.; Bowers, J.; et al. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genom. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Kole, C. Pulses, Sugar and Tuber Crops; Genome Mapping and Molecular Breeding in Plants; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007; Volume 3. [Google Scholar]
- Evans, D.L.; Joshi, S.V. Complete chloroplast genomes of Saccharum spontaneum, Saccharum officinarum and Miscanthus floridulus (Panicoideae: Andropogoneae) reveal the plastid view on sugarcane origins. Syst. Biodivers. 2016, 14, 548–571. [Google Scholar] [CrossRef]
- Amalraj, V.A.; Balasundaram, N. On the Taxonomy of the Members of ‘Saccharum Complex’. Genet. Resour. Crop Evol. 2006, 53, 35–41. [Google Scholar] [CrossRef]
- Ming, R. Sugarcane improvement through breeding and biotechnology. Plant Breed. Rev. 2006, 27, 15–118. [Google Scholar]
- Zhang, J.; Zhang, Q.; Li, L.; Tang, H.; Zhang, Q.; Chen, Y.; Arrow, J.; Zhang, X.; Wang, A.; Miao, C.; et al. Recent polyploidization events in three Saccharum founding species. Plant Biotechnol. J. 2019, 17, 264–274. [Google Scholar] [CrossRef]
- Lloyd Evans, D.; Joshi, S.V.; Wang, J. Whole chloroplast genome and gene locus phylogenies reveal the taxonomic placement and relationship of Tripidium (Panicoideae: Andropogoneae) to sugarcane. BMC Evol. Biol. 2019, 19, 33. [Google Scholar] [CrossRef]
- Pachakkil, B.; Terajima, Y.; Ohmido, N.; Ebina, M.; Irei, S.; Hayashi, H.; Takagi, H. Cytogenetic and agronomic characterization of intergeneric hybrids between Saccharum spp. hybrid and Erianthus arundinaceus. Sci. Rep. 2019, 9, 1748. [Google Scholar] [CrossRef]
- Pan, Y.B.; Burner, D.M.; Legendre, B.L. An assessment of the phylogenetic relationship among sugarcane and related taxa based on the nucleotide sequence of 5S rRNA intergenic spacers. Genetica 2000, 108, 285–295. [Google Scholar] [CrossRef]
- Yu, F.; Chai, J.; Li, X.; Yu, Z.; Yang, R.; Ding, X.; Wang, Q.; Wu, J.; Yang, X.; Deng, Z. Chromosomal Characterization of Tripidium arundinaceum Revealed by Oligo-FISH. Int. J. Mol. Sci. 2021, 22, 8539. [Google Scholar] [CrossRef]
- Li, S.; Wang, X.; Yang, Q. The Ploidy Identification of Saccharum spontaneum Collected from Myanmar Based on 5S rDNA-FISH Localization. Mol. Plant Breed. 2018, 16, 1229–1235. [Google Scholar]
- Ha, S.; Moore, P.H.; Heinz, D.; Kato, S.; Ohmido, N.; Fukui, K. Quantitative chromosome map of the polyploid Saccharum spontaneum by multicolor fluorescence in situ hybridization and imaging methods. Plant Mol. Biol. 1999, 39, 1165–1173. [Google Scholar] [CrossRef]
- Wu, J.; Huang, Y.; Wang, Q.; Ling, Q.; Deng, Z.; Li, Q.; Chen, R. Physical Mapping of rDNA on the Chromosomes of Erianthus arundinaceus and Determination of Basic Chromosome Number. Sugarcane Canesugar 2018, 2, 1–5. [Google Scholar]
- Besse, P.; Taylor, G.; Carroll, B.; Berding, N.; Burner, D.; Mcintyre, C.L. Assessing genetic diversity in a sugarcane germplasm collection using an automated AFLP analysis. Genetica 1998, 104, 143. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, J.; Wang, P.; Lin, Y.; Fu, C.; Deng, Z.; Wang, Q.; Li, Q.; Chen, R.; Zhang, M. Characterization of Chromosome Inheritance of the Intergeneric BC2 and BC3 Progeny between Saccharum spp. and Erianthus arundinaceus. PLoS ONE 2015, 10, e0133722. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, Y.; Lin, Y.; Fu, C.; Liu, S.; Deng, Z.; Li, Q.; Huang, Z.; Chen, R.; Zhang, M. Unexpected inheritance pattern of Erianthus arundinaceus chromosomes in the intergeneric progeny between Saccharum spp. and Erianthus arundinaceus. PLoS ONE 2014, 9, e110390. [Google Scholar] [CrossRef] [PubMed]
- D’Hont, A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet. Genome Res. 2005, 109, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Gerlach, W.L.; Dennis, E.S.; Swift, H.; Peacock, W.J. Molecular and chromosomal organization of DNA sequences coding for the ribosomal RNAs in cereals. Chromosoma 1980, 78, 293–311. [Google Scholar] [CrossRef]
- Ohmido, N.; Fukui, K. Cytological studies of African cultivated rice, Oryza glaberrima. TAG. Theor. Appl. genetics. Theor. Und Angew. Genet. 1995, 91, 212–217. [Google Scholar] [CrossRef]
- Han, Y.H.; Li, L.J.; Song, Y.C.; Li, Z.Y.; Xiong, Z.Y.; Li, D.Y. Physical mapping of the 5S and 45S rDNA in teosintes. Hereditas 2002, 137, 16–19. [Google Scholar] [CrossRef]
- Sang, Y.; Liang, G.H. Comparative physical mapping of the 18S-5.8S-26S rDNA in three sorghum species. Genome 2000, 43, 918–922. [Google Scholar] [CrossRef]
- Maoyin, S. Physical Mapping of the 45S and 5S rDNA of Cultivated Buckwheat. J. Plant Genet. Resour. 2013, 014, 317–321. [Google Scholar]
- Mizuochi, H.; Marasek, A.; Okazaki, K. Molecular cloning of Tulipa fosteriana rDNA and subsequent FISH analysis yields cytogenetic organization of 5S rDNA and 45S rDNA in T. gesneriana and T. fosteriana. Euphytica 2007, 155, 235–248. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Chen, J. Application of ITS sequences of nuclear rDNA in phylogenetic and evolutionary studies of angiosperms. J. Syst. Evol. 1999, 37, 407–416. [Google Scholar]
- Poczai, P.; Hyvönen, J. Nuclear ribosomal spacer regions in plant phylogenetics: Problems and prospects. Mol. Biol. Rep. 2010, 37, 1897–1912. [Google Scholar] [CrossRef]
- Wang, X.; Deng, Z.; Hong, D. The Systematic Position of Beesia: Evidence from ITS (nrDNA) Sequence Analysis. Acta Phytotaxon. Sin. 1998, 36, 403–410. [Google Scholar]
- Ainouche, M.L.; Bayer, R.J. On the origins of the tetraploid Bromus species (section Bromus, Poaceae): Insights from internal transcribed spacer sequences of nuclear ribosomal DNA. Genome 1997, 40, 730–743. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Baldwin, B.G.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S. The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny. Ann. Mo. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Chen, H.; Fan, Y.; Xiangyu, J.; Cai, Q.; Zhang, Y. Phylogenetic Relationships of Saccharum and Related Species Inferred from Sequence Analysis of the nrDNA ITS Region. Acta Agron. Sin. 2003, 29, 379–385. [Google Scholar]
- Hodkinson, T.R.; Chase, M.W.; Lledó, M.D.; Salamin, N.; Renvoize, S.A. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J. Plant Res. 2002, 115, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.G.; Yu, F.; Huang, Y.J.; Sun, L.; Li, X.T.; Yang, S.; Chen, K.; Huang, F.; Zeng, K.; Zhang, M.Q.; et al. Characterization analysis of the 35S rDNA intergenic spacers in Erianthus arundinaceus. Gene 2019, 694, 63–70. [Google Scholar] [CrossRef]
- Lee, H.I.; Younis, A.; Hwang, Y.J.; Kang, Y.I.; Lim, K.B. Molecular cytogenetic analysis and phylogenetic relationship of 5S and 45S ribosomal DNA in sinomartagon Lilium species by fluorescence in situ hybridization (FISH). Hortic. Environ. Biotechnol. 2014, 55, 514–523. [Google Scholar] [CrossRef]
- Gruendler, P.; Unfried, I.; Pascher, K.; Schweizer, D. rDNA intergenic region from Arabidopsis thaliana. Structural analysis, intraspecific variation and functional implications. J. Mol. Biol. 1991, 221, 1209–1222. [Google Scholar] [CrossRef]
- Volkov, R.A.; Bachmair, A.; Panchuk, I.I.; Kostyshyn, S.S.; Schweizer, D. 25S-18S rDNA intergenic spacer of Nicotiana sylvestris (Solanaceae): Primary and secondary structure analysis. Plant Syst. Evol. 1999, 218, 89–97. [Google Scholar] [CrossRef]
- Chai, J.; Luo, L.; Yu, Z.; Lei, J.; Zhang, M.; Deng, Z. Repetitive Sequence Barcode Probe for Karyotype Analysis in Tripidium arundinaceum. Int. J. Mol. Sci. 2022, 23, 6726. [Google Scholar] [CrossRef]
- Mahelka, V.; Kopecky, D.; Baum, B.R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol. Biol. Evol. 2013, 30, 2065–2086. [Google Scholar] [CrossRef] [PubMed]
- Li, X.T.; Huang, F.; Chai, J.; Wang, Q.S.; Yu, F.; Huang, Y.J.; Wu, J.Y.; Wang, Q.N.; Xu, L.N.; Zhang, M.Q.; et al. Chromosome behavior during meiosis in pollen mother cells from Saccharum officinarum x Erianthus arundinaceus F1 hybrids. BMC Plant Biol. 2021, 21, 139. [Google Scholar] [CrossRef]
- Falistocco, E.; Passeri, V.; Marconi, G. Investigations of 5S rDNA of Vitis vinifera L.: Sequence analysis and physical mapping. Genome 2007, 50, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Liu, B.; Dong, F.; Chen, R.; Li, X.; Chen, C. Multicolor FISH analysis of rDNA and telomere on spinach. Hereditas 2007, 29, 1405–1408. [Google Scholar] [CrossRef]
- Jiang, J.; Gill, B.S. New 18S.26S ribosomal RNA gene loci: Chromosomal landmarks for the evolution of polyploid wheats. Chromosoma 1994, 103, 179–185. [Google Scholar] [CrossRef]
- Kulak, S.; Hasterok, R.; Maluszynska, J. Karyotyping of Brassica amphidiploids using 5S and 25S rDNA as chromosome markers. Hereditas 2002, 136, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Moraes, A.P.; Mirkov, T.E.; Guerra, M. Mapping the chromosomes of Poncirus trifoliata Raf. by BAC-FISH. Cytogenet. Genome Res. 2008, 121, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Morton, C.M. Phylogenetic relationships of the Aurantioideae (Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions, atpB-rbcL spacer, rps16, and trnL-trnF. Org. Divers. Evol. 2009, 9, 52–68. [Google Scholar] [CrossRef]
- Wai, C.M.; Ming, R.; Moore, P.H.; Paull, R.E.; Yu, Q. Development of Chromosome-specific Cytogenetic Markers and Merging of Linkage Fragments in Papaya. Trop. Plant Biol. 2010, 3, 171–181. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, M.; Liu, J.; Huang, Y.; Xu, L.; Deng, Z.; Zhao, X. Efficient Anchoring of Erianthus arundinaceus Chromatin Introgressed into Sugarcane by Specific Molecular Markers. Int. J. Mol. Sci. 2022, 23, 9435. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Guo, Y.; Guo, Z.; Zhang, N.; Lei, Y.; Cai, E.; Deng, Z.; Wu, J. Research Progress on 35S rDNA and 5S rDNA in Sugarcane: Challenges and Prospects. Int. J. Mol. Sci. 2025, 26, 8773. https://doi.org/10.3390/ijms26188773
Li X, Guo Y, Guo Z, Zhang N, Lei Y, Cai E, Deng Z, Wu J. Research Progress on 35S rDNA and 5S rDNA in Sugarcane: Challenges and Prospects. International Journal of Molecular Sciences. 2025; 26(18):8773. https://doi.org/10.3390/ijms26188773
Chicago/Turabian StyleLi, Xueting, Yirong Guo, Zhejun Guo, Nannan Zhang, Yawen Lei, Enping Cai, Zuhu Deng, and Jiayun Wu. 2025. "Research Progress on 35S rDNA and 5S rDNA in Sugarcane: Challenges and Prospects" International Journal of Molecular Sciences 26, no. 18: 8773. https://doi.org/10.3390/ijms26188773
APA StyleLi, X., Guo, Y., Guo, Z., Zhang, N., Lei, Y., Cai, E., Deng, Z., & Wu, J. (2025). Research Progress on 35S rDNA and 5S rDNA in Sugarcane: Challenges and Prospects. International Journal of Molecular Sciences, 26(18), 8773. https://doi.org/10.3390/ijms26188773







