KRAS Copy Number Gain in Cell-Free DNA Analysis-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms
Abstract
1. Introduction
2. Results
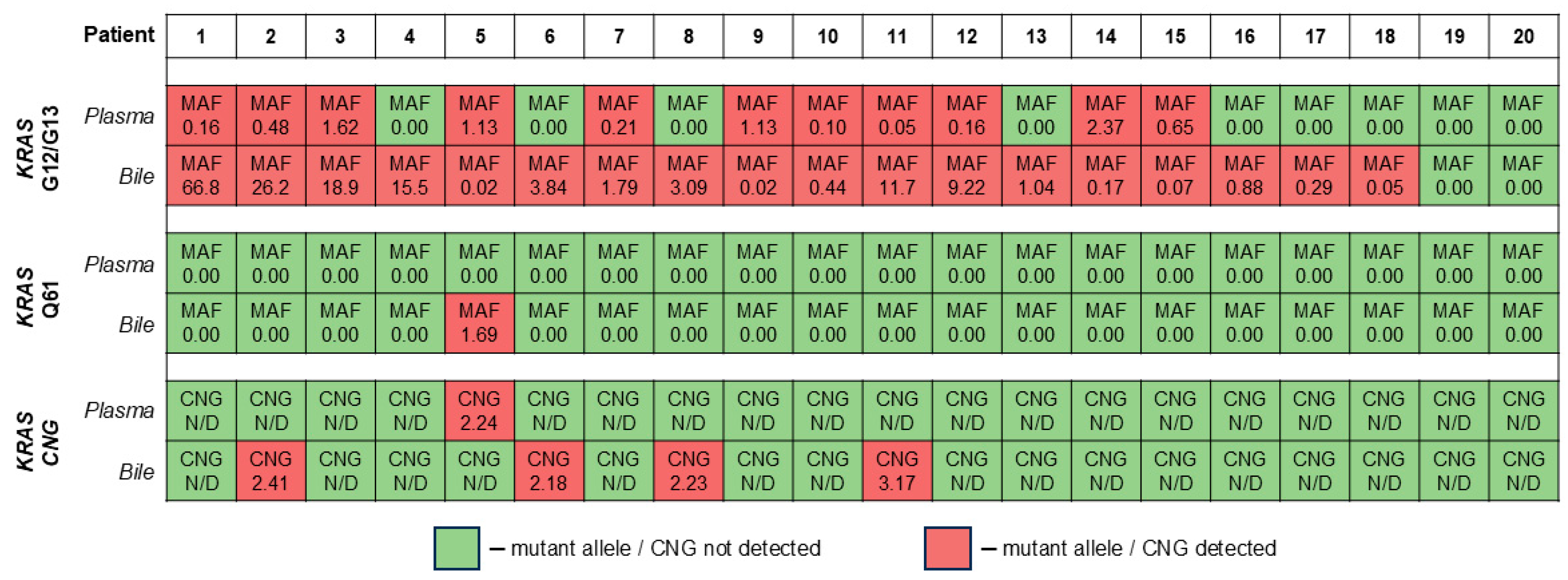
2.1. Diagnostic Performance of the KRAS CNG Analysis in cfDNA
2.2. Relation of KRAS CNG in cfDNA to Clinical and Demographic Parameters
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Biomaterial Collection and Processing
4.3. Cell-Free DNA Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area under curve |
| CA | Carbohydrate antigen |
| cfDNA | Cell-free DNA |
| cftDNA | Cell-free tumor DNA |
| CHIP | Clonal hematopoiesis of indeterminate potential |
| CI | Confidence interval |
| CN | Copy number |
| CNG | Copy number gain |
| CNV | Copy number variation |
| ddPCR | Digital droplet polymerase chain reaction |
| HR | Hazard ratio |
| IPMN | Intraductal papillary mucinous neoplasm |
| OPN | Other pancreatic neoplasms |
| PC | Pancreatic cancer |
| PCR | Polymerase chain reaction |
| PDAC | Pancreatic ductal adenocarcinoma |
| ROC | Receiver operating characteristics |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921, Erratum in Cancer Res. 2014, 74, 4006. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic Cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Ilic, M.; Ilic, I. Epidemiology of Pancreatic Cancer. World J. Gastroenterol. 2016, 22, 9694. [Google Scholar] [CrossRef]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic Adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef]
- Jin, C.; Bai, L. Pancreatic Cancer: Current Situation and Challenges. Gastroenterol. Hepatol. Lett. 2020, 2, 1–3. [Google Scholar] [CrossRef]
- Kearney, J.F.; Adsay, V.; Yeh, J.J. Pathology and Molecular Characteristics of Pancreatic Cancer. Surg. Oncol. Clin. 2021, 30, 609–619. [Google Scholar] [CrossRef]
- Han, Y.; Kwon, W.; Lee, M.; Jung, H.S.; Yun, W.G.; Cho, Y.J.; Chae, Y.S.; Fernández-Del Castillo, C.; Marchegiani, G.; Salvia, R.; et al. Optimal Surveillance Interval of Branch Duct Intraductal Papillary Mucinous Neoplasm of the Pancreas. JAMA Surg. 2024, 159, 389–396. [Google Scholar] [CrossRef]
- Zhao, B.; Zhao, B.; Chen, F. Diagnostic Value of Serum Carbohydrate Antigen 19-9 in Pancreatic Cancer: A Systematic Review and Meta-Analysis. Eur. J. Gastroenterol. Hepatol. 2022, 34, 891–904. [Google Scholar] [CrossRef]
- Katsanos, K.H.; Kitsanou, M.; Christodoulou, D.K.; Tsianos, E. V High CA 19-9 Levels in Benign Biliary Tract Diseases. Report of Four Cases and Review of the Literature. Eur. J. Intern. Med. 2002, 13, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Parra-Robert, M.; Santos, V.; Canis, S.; Pla, X.; Fradera, J.; Porto, R. Relationship Between CA 19.9 and the Lewis Phenotype: Options to Improve Diagnostic Efficiency. Anticancer Res. 2018, 38, 5883–5888. [Google Scholar] [CrossRef]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid Biopsy in Cancer: Current Status, Challenges and Future Prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef]
- Boukovala, M.; Westphalen, C.B.; Probst, V. Liquid Biopsy into the Clinics: Current Evidence and Future Perspectives. J. Liq. Biopsy 2024, 4, 100146. [Google Scholar] [CrossRef]
- Atayan, D.P.; Rakhmatullin, T.I.; Jain, M.; Samokhodskaya, L.M.; Egorov, V.I. Liquid Biopsy in Pancreatic Ductal Adenocarcinoma and Precancerous Lesions. Kazan Med. J. 2025, 106, 243–257. [Google Scholar] [CrossRef]
- Zhou, L.; Baba, Y.; Kitano, Y.; Miyake, K.; Zhang, X.; Yamamura, K.; Kosumi, K.; Kaida, T.; Arima, K.; Taki, K.; et al. KRAS, BRAF, and PIK3CA Mutations, and Patient Prognosis in 126 Pancreatic Cancers: Pyrosequencing Technology and Literature Review. Med. Oncol. 2016, 33, 32. [Google Scholar] [CrossRef]
- Oketch, D.J.A.; Giulietti, M.; Piva, F. Copy Number Variations in Pancreatic Cancer: From Biological Significance to Clinical Utility. Int. J. Mol. Sci. 2024, 25, 391. [Google Scholar] [CrossRef] [PubMed]
- Chan-Seng-Yue, M.; Kim, J.C.; Wilson, G.W.; Ng, K.; Figueroa, E.F.; O’Kane, G.M.; Connor, A.A.; Denroche, R.E.; Grant, R.C.; McLeod, J.; et al. Transcription Phenotypes of Pancreatic Cancer Are Driven by Genomic Events During Tumor Evolution. Nat. Genet. 2020, 52, 231–240, Erratum in Nat. Genet. 2020, 52, 463. https://doi.org/10.1038/s41588-020-0588-3. [Google Scholar] [CrossRef] [PubMed]
- Mohan, S.; Ayub, M.; Rothwell, D.G.; Gulati, S.; Kilerci, B.; Hollebecque, A.; Sun Leong, H.; Smith, N.K.; Sahoo, S.; Descamps, T.; et al. Analysis of Circulating Cell-Free DNA Identifies KRAS Copy Number Gain and Mutation as a Novel Prognostic Marker in Pancreatic Cancer. Sci. Rep. 2019, 9, 11610. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, H.; Chen, N.; Hao, J.; Jin, H.; Ma, X. Diagnostic Value of Various Liquid Biopsy Methods for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Medicine 2020, 99, e18581. [Google Scholar] [CrossRef]
- Jain, M.; Atayan, D.; Rakhmatullin, T.; Dakhtler, T.; Popov, P.; Kim, P.; Gontareva, J.; Samokhodskaya, L.; Egorov, V.; Kamalov, A. Tumor Cell-Free DNA Detection-Based Liquid Biopsy of Plasma and Bile in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Genes Cells 2023, 18, 41–51. [Google Scholar] [CrossRef]
- Jain, M.; Atayan, D.; Rakhmatullin, T.; Dakhtler, T.; Popov, P.; Kim, P.; Viborniy, M.; Gontareva, I.; Samokhodskaya, L.; Egorov, V. Cell-Free Tumor DNA Detection-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms. Biomedicines 2024, 12, 220. [Google Scholar] [CrossRef]
- Kruse, E.J. Palliation in Pancreatic Cancer. Surg. Clin. North Am. 2010, 90, 355–364. [Google Scholar] [CrossRef]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global Variation in Copy Number in the Human Genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef]
- Sebat, J.; Lakshmi, B.; Troge, J.; Alexander, J.; Young, J.; Lundin, P.; Månér, S.; Massa, H.; Walker, M.; Chi, M.; et al. Large-Scale Copy Number Polymorphism in the Human Genome. Science 2004, 305, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Masood, D.; Ren, L.; Nguyen, C.; Brundu, F.G.; Zheng, L.; Zhao, Y.; Jaeger, E.; Li, Y.; Cha, S.W.; Halpern, A.; et al. Evaluation of Somatic Copy Number Variation Detection by NGS Technologies and Bioinformatics Tools on a Hyper-Diploid Cancer Genome. Genome Biol. 2024, 25, 163. [Google Scholar] [CrossRef] [PubMed]
- Gilson, P.; Merlin, J.L.; Harlé, A. Deciphering Tumour Heterogeneity: From Tissue to Liquid Biopsy. Cancers 2022, 14, 1384. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Ye, Z.; Zhao, Q.; Jiang, T. Complication Incidence of EUS-Guided Pancreas Biopsy: A Systematic Review and Meta-Analysis of 11 Thousand Population from 78 Cohort Studies. Asian J. Surg. 2020, 43, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Vincent, A.; Kanda, M.; Leclerc, J.; Omura, N.; Borges, M.; Klein, A.P.; Canto, M.I.; Hruban, R.H.; Goggins, M. Genome-Wide Somatic Copy Number Alterations in Low-Grade PanINs and IPMNs from Individuals with a Family History of Pancreatic Cancer. Clin. Cancer Res. 2012, 18, 4303–4312. [Google Scholar] [CrossRef]
- Basturk, O.; Berger, M.F.; Yamaguchi, H.; Adsay, V.; Askan, G.; Bhanot, U.K.; Zehir, A.; Carneiro, F.; Hong, S.M.; Zamboni, G.; et al. Pancreatic Intraductal Tubulopapillary Neoplasm Is Genetically Distinct from Intraductal Papillary Mucinous Neoplasm and Ductal Adenocarcinoma. Mod. Pathol. 2017, 30, 1760–1772. [Google Scholar] [CrossRef]
- Driescher, C.; Fuchs, K.; Haeberle, L.; Goering, W.; Frohn, L.; Opitz, F.V.; Haeussinger, D.; Knoefel, W.T.; Keitel, V.; Esposito, I. Bile-Based Cell-Free DNA Analysis Is a Reliable Diagnostic Tool in Pancreatobiliary Cancer. Cancers 2020, 13, 39. [Google Scholar] [CrossRef]
- Gou, Q.; Zhang, C.Z.; Sun, Z.H.; Wu, L.G.; Chen, Y.; Mo, Z.Q.; Mai, Q.C.; He, J.; Zhou, Z.X.; Shi, F.; et al. Cell-Free DNA from Bile Outperformed Plasma as a Potential Alternative to Tissue Biopsy in Biliary Tract Cancer. ESMO Open 2021, 6, 100275. [Google Scholar] [CrossRef]
- Grunvald, M.W.; Jacobson, R.A.; Kuzel, T.M.; Pappas, S.G.; Masood, A. Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 7651. [Google Scholar] [CrossRef]
- Toledano-Fonseca, M.; Cano, M.T.; Inga, E.; Rodríguez-Alonso, R.; Gómez-España, M.A.; Guil-Luna, S.; Mena-Osuna, R.; de la Haba-Rodríguez, J.R.; Rodríguez-Ariza, A.; Aranda, E. Circulating Cell-Free DNA-Based Liquid Biopsy Markers for the Non-Invasive Prognosis and Monitoring of Metastatic Pancreatic Cancer. Cancers 2020, 12, 1754. [Google Scholar] [CrossRef]
- Strijker, M.; Soer, E.C.; de Pastena, M.; Creemers, A.; Balduzzi, A.; Beagan, J.J.; Busch, O.R.; van Delden, O.M.; Halfwerk, H.; van Hooft, J.E.; et al. Circulating Tumor DNA Quantity Is Related to Tumor Volume and Both Predict Survival in Metastatic Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2019, 146, 1445. [Google Scholar] [CrossRef]
- Conces, M.; Ni, Y.; Bazeley, P.; Patel, B.; Funchain, P.; Carraway, H.E. Clonal Hematopoiesis of Indeterminate Potential (CHIP) Mutations in Solid Tumor Malignancies. J. Clin. Oncol. 2019, 37, 1507. [Google Scholar] [CrossRef]
- Roma, C.; Sacco, A.; Forgione, L.; Esposito Abate, R.; Lambiase, M.; Dotolo, S.; Maiello, M.R.; Frezzetti, D.; Nasti, G.; Morabito, A.; et al. Low Impact of Clonal Hematopoiesis on the Determination of RAS Mutations by Cell-Free DNA Testing in Routine Clinical Diagnostics. Diagnostics 2022, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Truty, R.; Rojahn, S.; Ouyang, K.; Kautzer, C.; Kennemer, M.; Pineda-Alvarez, D.; Johnson, B.; Stafford, A.; Basel-Salmon, L.; Saitta, S.; et al. Patterns of Mosaicism for Sequence and Copy-Number Variants Discovered Through Clinical Deep Sequencing of Disease-Related Genes in One Million Individuals. Am. J. Hum. Genet. 2023, 110, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards Standardisation of Cell-Free DNA Measurement in Plasma: Controls for Extraction Efficiency, Fragment Size Bias and Quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512. [Google Scholar] [CrossRef]


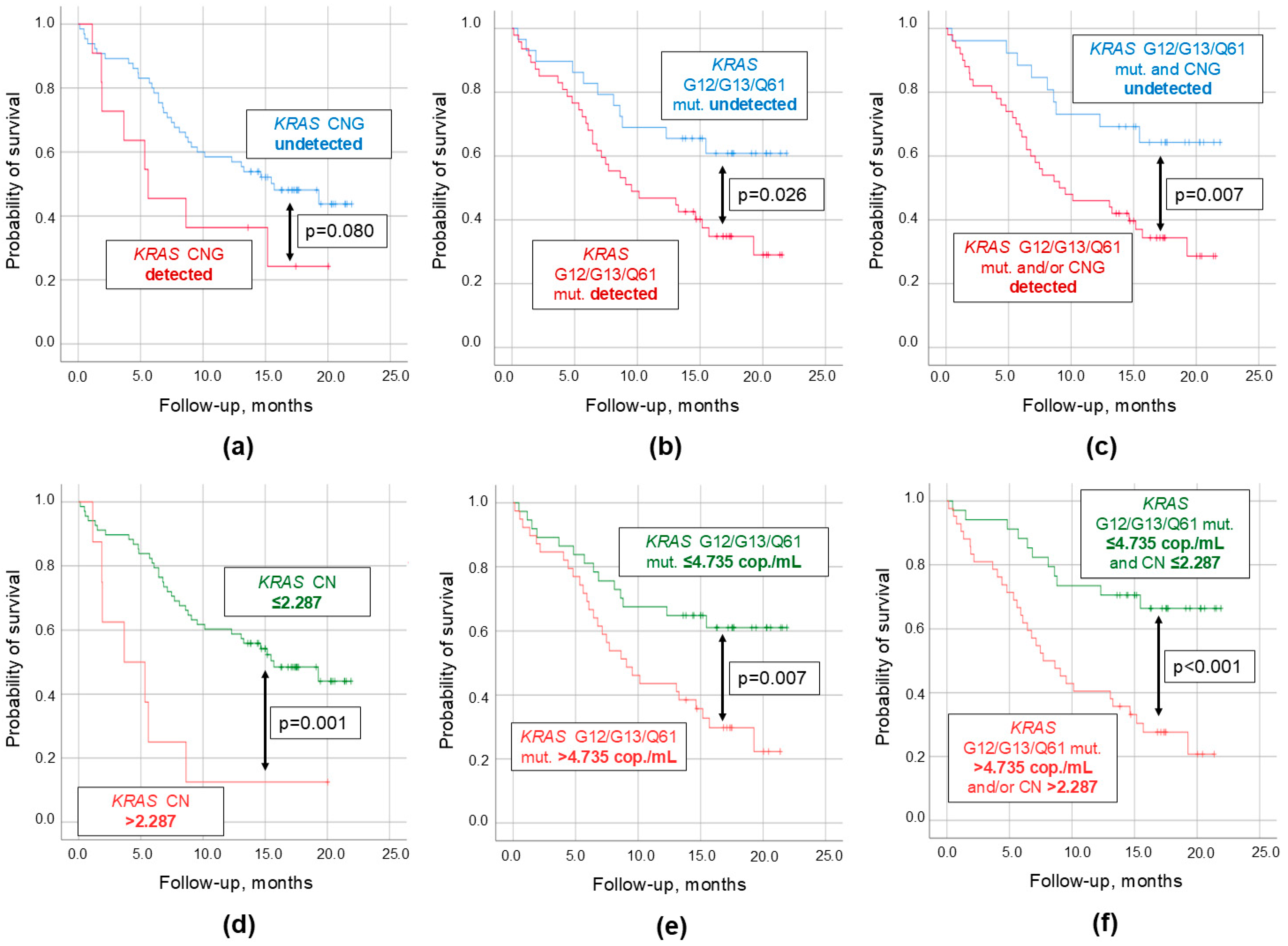
| Parameters | Survival, Months | HR (95% CI) | p-Values |
|---|---|---|---|
| Detectable KRAS CNG Undetectable KRAS CNG | 9.3 ± 2.2 14.1 ± 1.0 | 1.92 (0.91–4.27) | 0.080 |
| Detectable KRAS point mutations Undetectable KRAS point mutations | 11.8 ± 1.1 16.1 ± 1.5 | 2.15 (1.08–4.29) | 0.026 |
| Detectable KRAS CNG and/or point mutations Undetectable KRAS CNG and point mutations | 11.6 ± 1.1 17.0 ± 1.4 | 2.58 (1.23–5.41) | 0.009 |
| KRAS CNG > 2.287 KRAS CNG ≤ 2.287 | 6.0 ± 2.0 14.3 ± 1.0 | 3.54 (1.55–8.12) | 0.001 |
| KRAS point mutations > 4.735 copies/mL KRAS point mutations ≤ 4.735 copies/mL | 11.6 ± 1.1 17.0 ± 1.4 | 2.35 (1.23–4.48) | 0.007 |
| KRAS CNG and/or point mutations > thresholds 1 KRAS CNG and point mutations ≤ thresholds 1 | 10.5 ± 1.2 17.1 ± 1.3 | 3.23 (1.62–6.44) | <0.001 |
| Parameters | PDAC Group (n = 94) | OPN Group (n = 17) | Control Group (n = 69) |
|---|---|---|---|
| Age, years 1 | 65 (41–88) | 58 (43–66) | 40 (19–71) |
| Sex, n (%): | |||
| - Male | 46/94 | 4/17 | 33/69 |
| - Female | 48/94 | 13/17 | 36/69 |
| OPN types: | |||
| - mdIPMN, n | N/A | 5/17 | N/A |
| - multIPMN, n | N/A | 1/17 | N/A |
| - bdIPMN, n | N/A | 7/17 | N/A |
| - adenoma, n | N/A | 1/17 | N/A |
| - serous cystadenoma, n | N/A | 1/17 | N/A |
| - SPPN, n | N/A | 1/17 | N/A |
| - NET, n | N/A | 1/17 | N/A |
| Tumor localization in pancreas: | |||
| - head, n | 44/87 2 | N/A | N/A |
| - body, n | 6/87 2 | N/A | N/A |
| - tail, n | 11/87 2 | N/A | N/A |
| - head + body, n | 10/87 2 | N/A | N/A |
| - body + tail, n | 15/87 2 | N/A | N/A |
| - head + body + tail, n | 1/87 2 | N/A | N/A |
| Tumor size: | |||
| - >4 cm, n | 39/87 2 | N/A | N/A |
| - 2–4 cm, n | 41/87 2 | N/A | N/A |
| - <2 cm, n | 7/87 2 | N/A | N/A |
| Contact with arteries/veins, n | 78/87 2 | N/A | N/A |
| Bile ducts invasion, n | 39/87 2 | N/A | N/A |
| Distant metastases, n | 43/87 2 | N/A | N/A |
| CA 19-9, U/mL 3 | 36.5 [0.9; 883.3] | 3.9 [0.8; 7.9] | N/A |
| Gene. | Amplicon Size | Oligonucleotide | Sequence | Concentration 1 |
|---|---|---|---|---|
| KRAS | 82 bp | Forward primer | GTA ATT TAC TGG GAA AGC | 0.9 μM |
| Reverse primer | CAG TCT GAT GTC TGT TTA | 0.9 μM | ||
| Probe | FAM-AGC TCA TAA TCT CAA ACT TCT TGC ACA-BHQ1 | 0.25 μM | ||
| EIF2C1 | 81 bp | Forward primer | GTT CGG CTT TCA CCA GTC T | 0.9 μM |
| Reverse primer | CTC CAT AGC TCT CCC CAC TC | 0.9 μM | ||
| Probe | HEX-CGC CCT GCC ATG TGG AAG AT-BHQ1 | 0.25 μM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, M.; Atayan, D.; Rakhmatullin, T.; Dakhtler, T.; Inokenteva, V.; Popov, P.; Farmanov, A.; Viborniy, M.; Gontareva, I.; Samokhodskaya, L.; et al. KRAS Copy Number Gain in Cell-Free DNA Analysis-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms. Int. J. Mol. Sci. 2025, 26, 8763. https://doi.org/10.3390/ijms26188763
Jain M, Atayan D, Rakhmatullin T, Dakhtler T, Inokenteva V, Popov P, Farmanov A, Viborniy M, Gontareva I, Samokhodskaya L, et al. KRAS Copy Number Gain in Cell-Free DNA Analysis-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms. International Journal of Molecular Sciences. 2025; 26(18):8763. https://doi.org/10.3390/ijms26188763
Chicago/Turabian StyleJain, Mark, David Atayan, Tagir Rakhmatullin, Tatiana Dakhtler, Victoria Inokenteva, Pavel Popov, Aleksandr Farmanov, Mikhail Viborniy, Iuliia Gontareva, Larisa Samokhodskaya, and et al. 2025. "KRAS Copy Number Gain in Cell-Free DNA Analysis-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms" International Journal of Molecular Sciences 26, no. 18: 8763. https://doi.org/10.3390/ijms26188763
APA StyleJain, M., Atayan, D., Rakhmatullin, T., Dakhtler, T., Inokenteva, V., Popov, P., Farmanov, A., Viborniy, M., Gontareva, I., Samokhodskaya, L., & Egorov, V. (2025). KRAS Copy Number Gain in Cell-Free DNA Analysis-Based Liquid Biopsy of Plasma and Bile in Patients with Various Pancreatic Neoplasms. International Journal of Molecular Sciences, 26(18), 8763. https://doi.org/10.3390/ijms26188763






