Regulatory Mechanisms of SPARC Overexpression in Melanoma Progression
Abstract
1. Introduction
2. Results
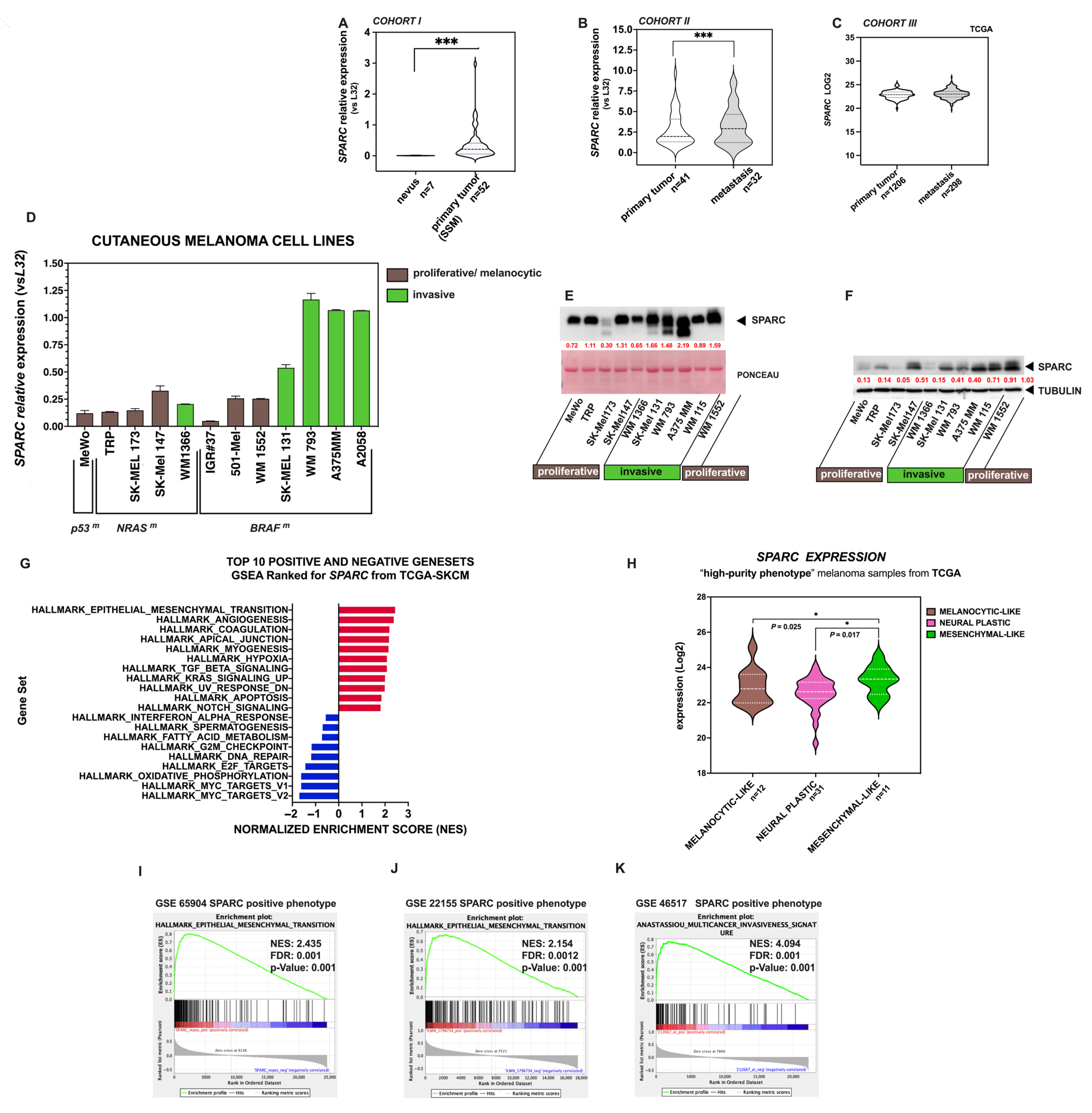
2.1. SPARC Is a Key EMT Gene in Melanoma
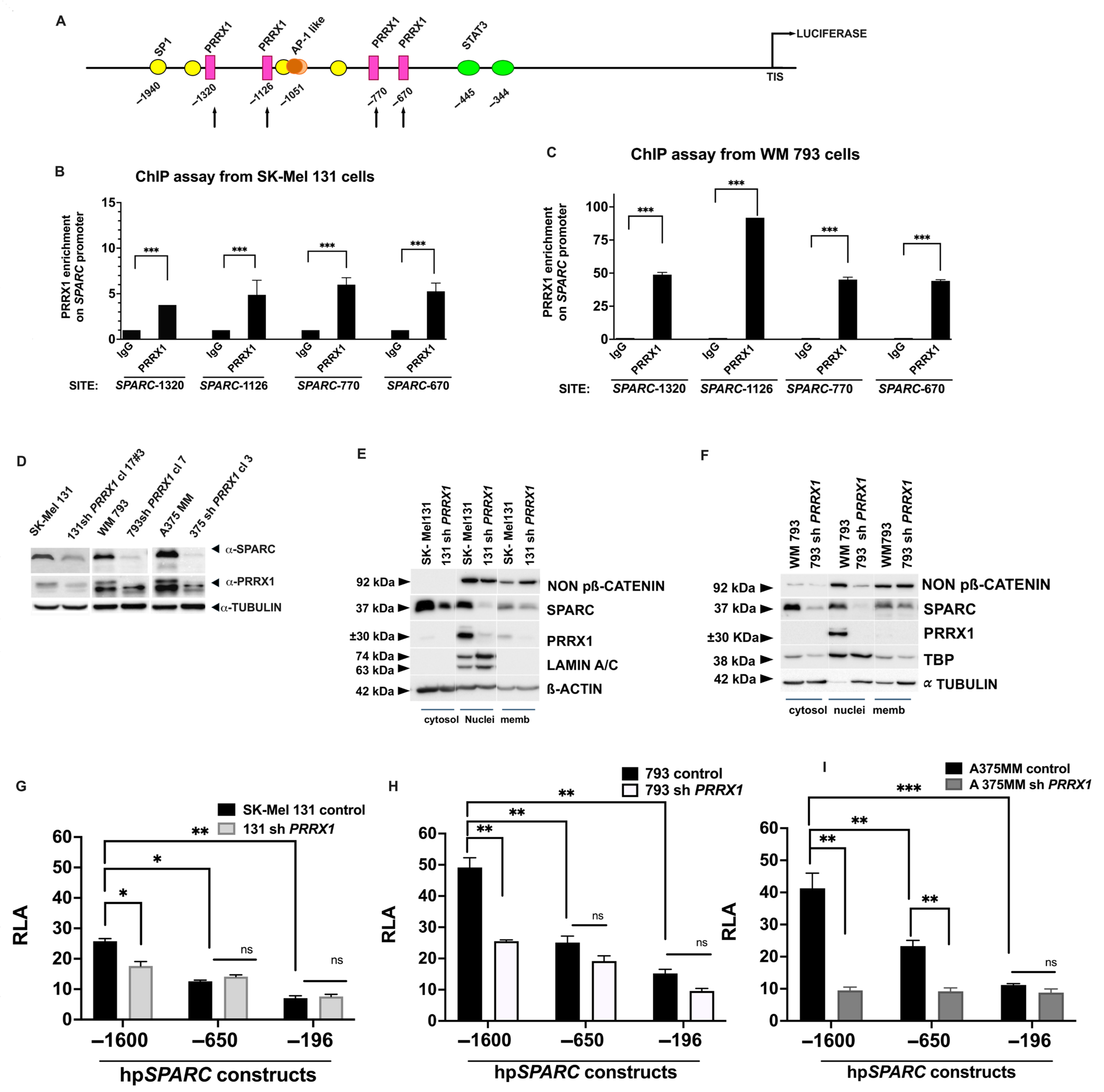
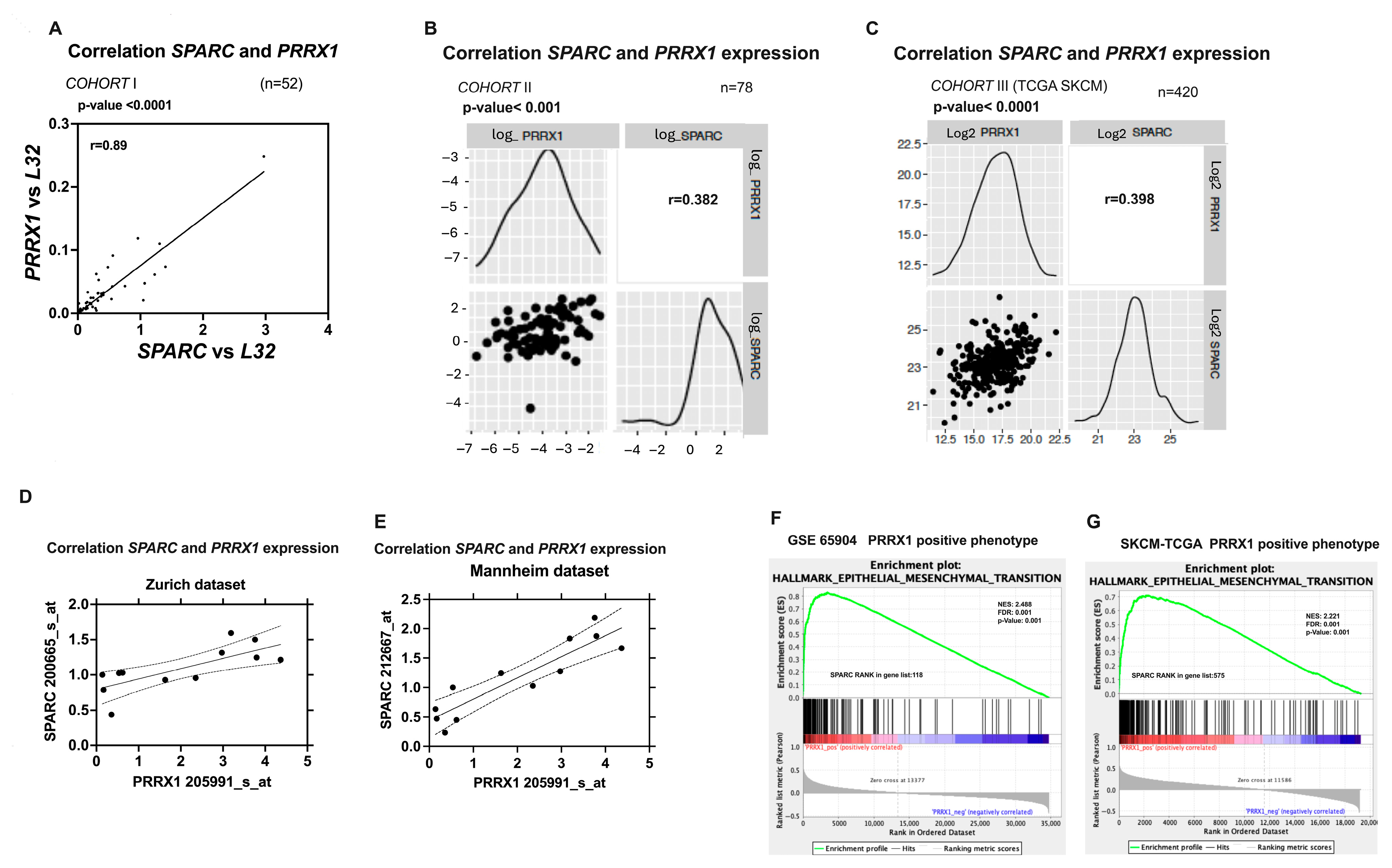
2.2. SPARC Overexpression Is Associated with PRRX1 Expression in Melanoma Cell Lines
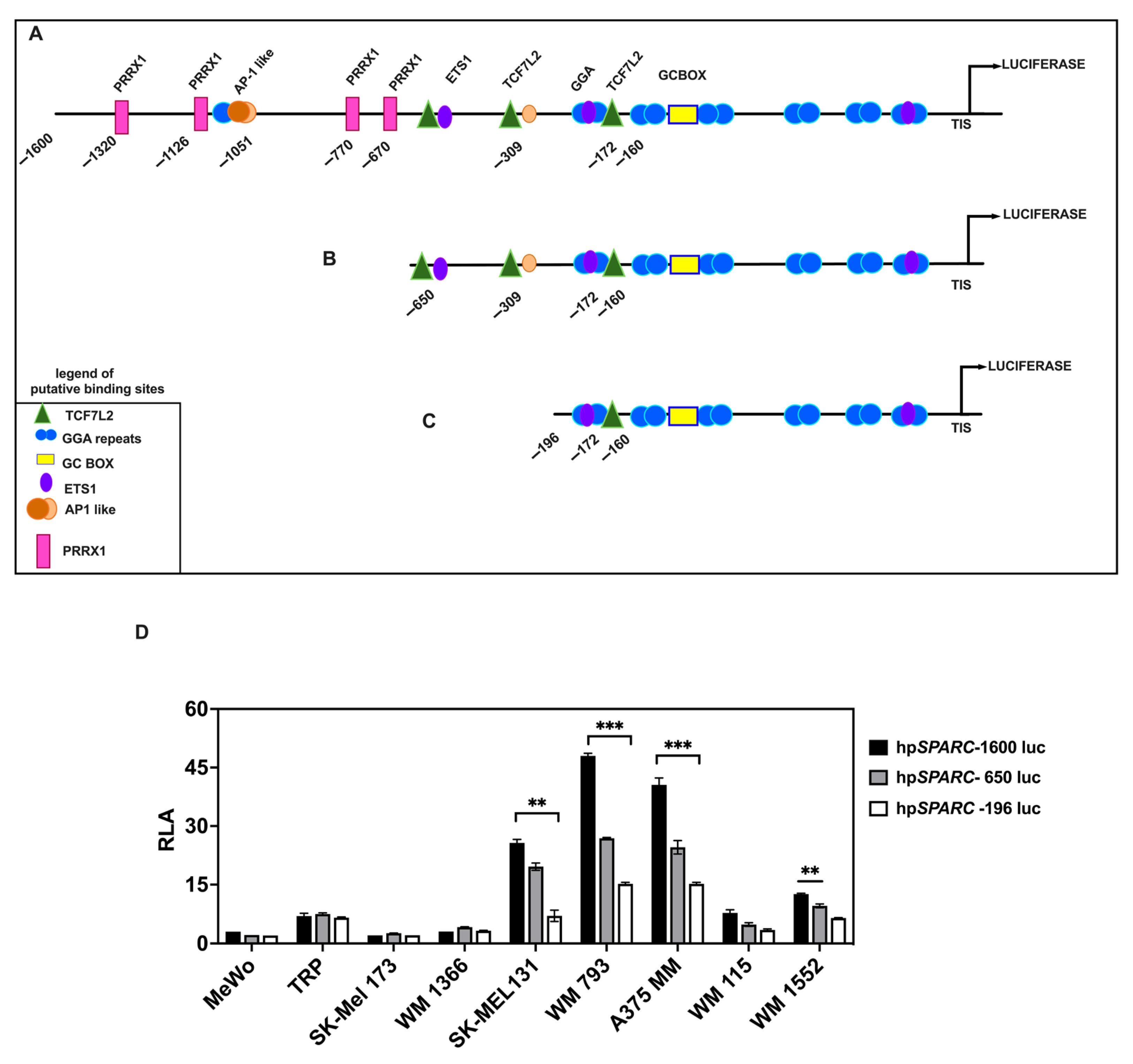
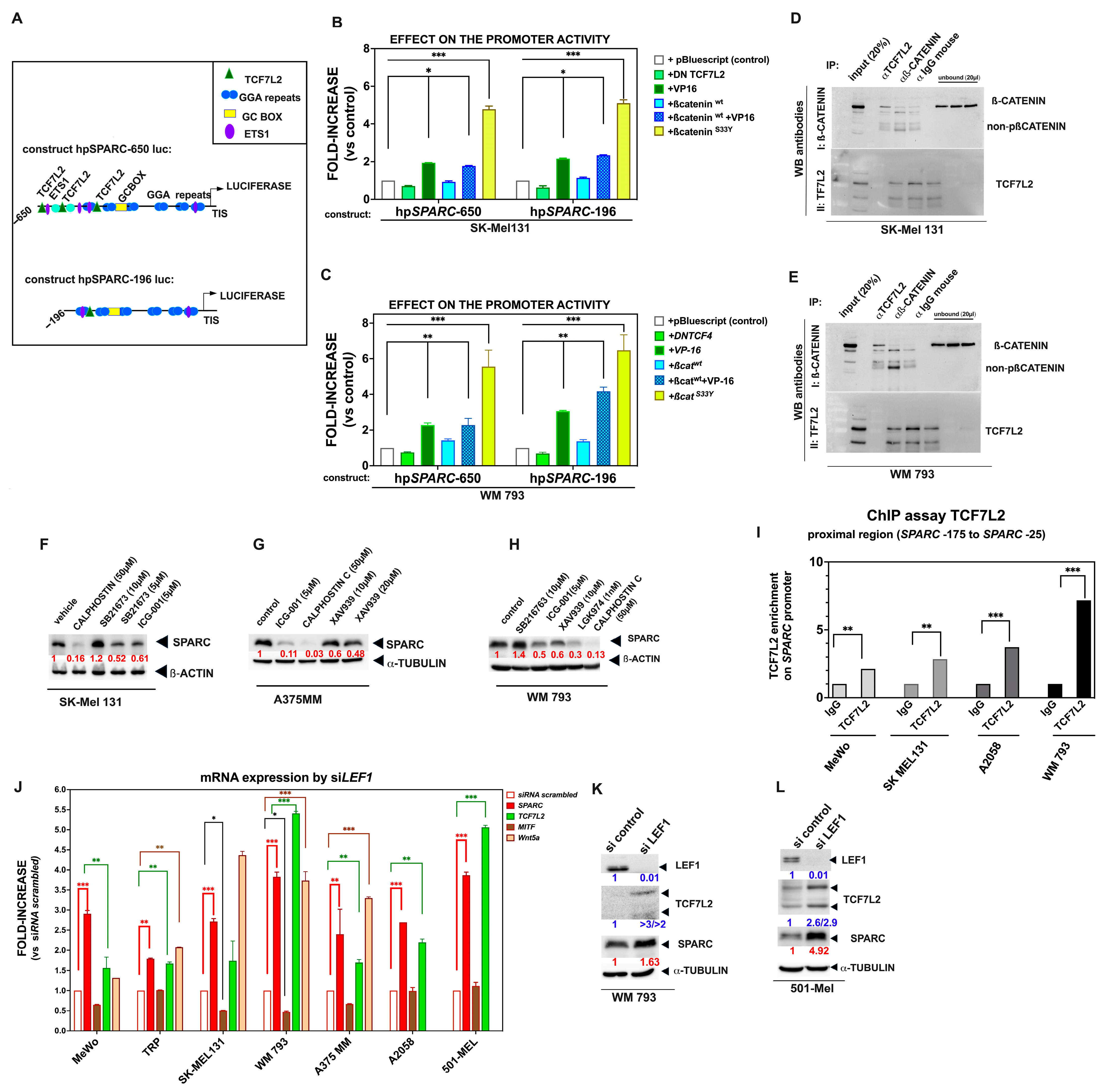
2.3. The Binding of TCF7L2 to the SPARC Promoter Results in SPARC Overexpression During Phenotypic Transitions Toward an Invasive Melanoma Cell State
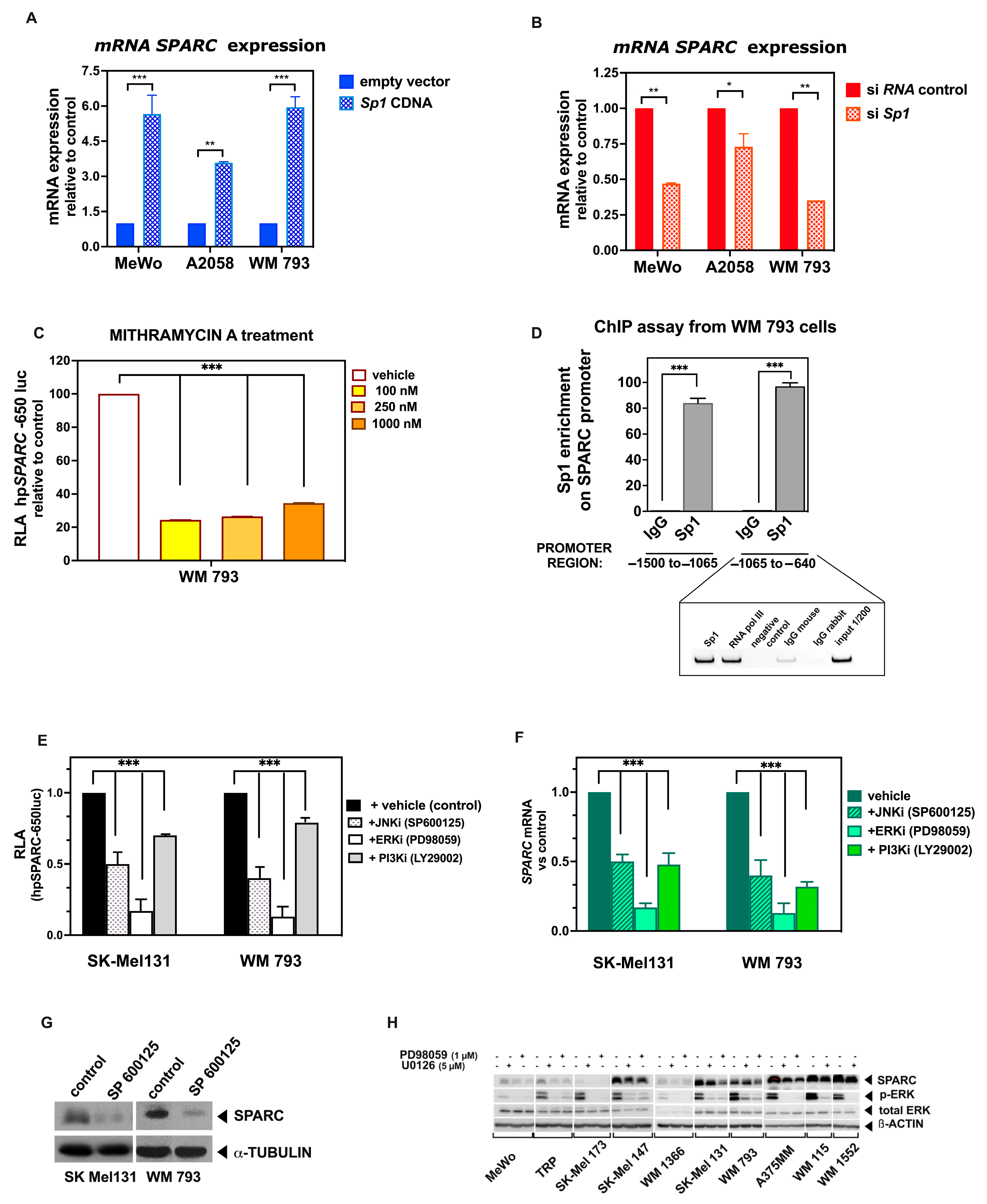
2.4. Sp1 Binds to the SPARC Proximal Promoter in Melanoma Cell Lines, Ensuring Its Basal Expression
2.5. Activated MAPK Signaling Pathways Contribute to SPARC Expression in Melanoma Cell Lines
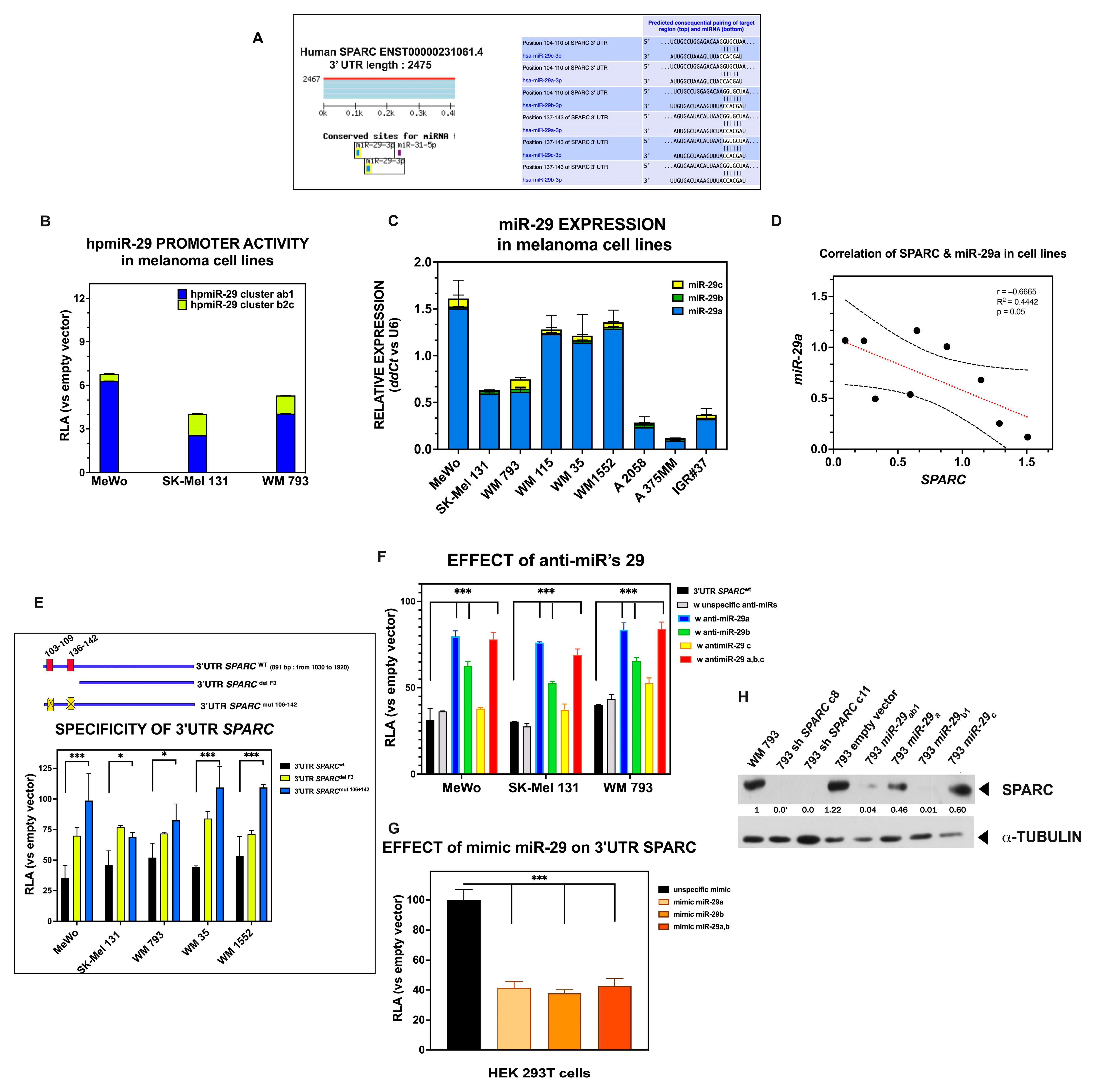
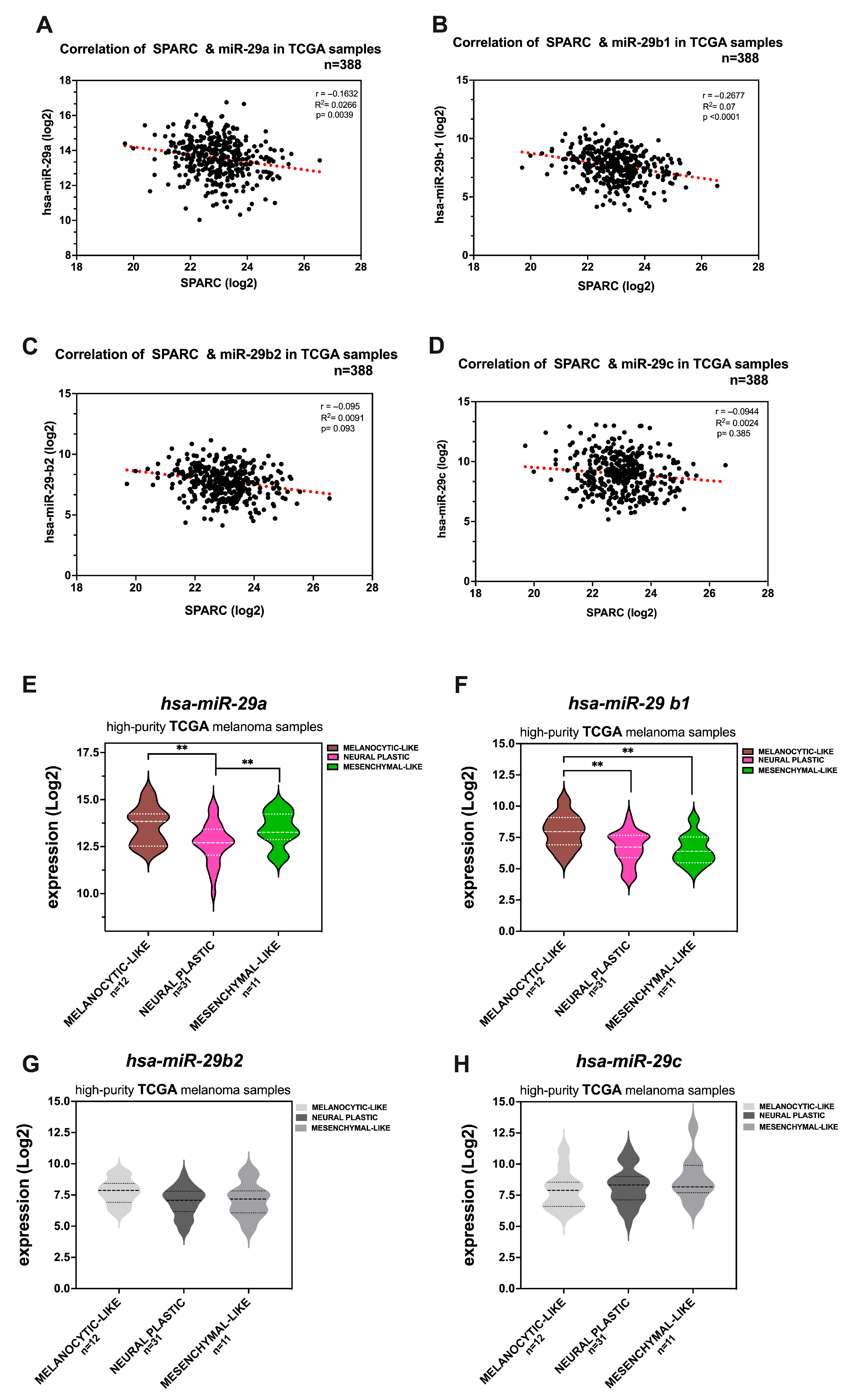
2.6. Members of the miR-29 Family of microRNAs Are Bona Fide Regulators of SPARC Expression in Melanoma
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatments
4.2. Melanoma Samples
4.3. Expression Vectors and Transfections
4.4. RNA Interference
4.5. Promoter Constructs
4.6. microRNA Activity Assay
4.7. Luciferase Reporter Assays
4.8. Generation of Stable Melanoma Cell Lines with Either PRRX1 Knockdown or miR-29
4.9. Gene Expression Analysis
4.10. Immunoprecipitation and Immunoblot Analysis
4.11. ChIP Assays
4.12. Meta-Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Youssef, K.K.; Narwade, N.; Arcas, A.; Marquez-Galera, A.; Jimenez-Castano, R.; Lopez-Blau, C.; Fazilaty, H.; Garcia-Gutierrez, D.; Cano, A.; Galceran, J.; et al. Two distinct epithelial-to-mesenchymal transition programs control invasion and inflammation in segregated tumor cell populations. Nat. Cancer 2024, 5, 1660–1680. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, C.; Di Leo, L.; De Zio, D. New Insights into the Phenotype Switching of Melanoma. Cancers 2022, 14, 6118. [Google Scholar] [CrossRef]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Dobbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef]
- Widmer, D.S.; Cheng, P.F.; Eichhoff, O.M.; Belloni, B.C.; Zipser, M.C.; Schlegel, N.C.; Javelaud, D.; Mauviel, A.; Dummer, R.; Hoek, K.S. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 2012, 25, 343–353. [Google Scholar] [CrossRef]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef]
- Pedri, D.; Karras, P.; Landeloos, E.; Marine, J.C.; Rambow, F. Epithelial-to-mesenchymal-like transition events in melanoma. FEBS J. 2022, 289, 1352–1368. [Google Scholar] [CrossRef] [PubMed]
- Ennen, M.; Keime, C.; Gambi, G.; Kieny, A.; Coassolo, S.; Thibault-Carpentier, C.; Margerin-Schaller, F.; Davidson, G.; Vagne, C.; Lipsker, D.; et al. MITF-High and MITF-Low Cells and a Novel Subpopulation Expressing Genes of Both Cell States Contribute to Intra- and Intertumoral Heterogeneity of Primary Melanoma. Clin. Cancer Res. 2017, 23, 7097–7107. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Rago, L.; Kass Youssef, K.; Ocana, O.H.; Garcia-Asencio, F.; Arcas, A.; Galceran, J.; Nieto, M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019, 10, 5115. [Google Scholar] [CrossRef]
- Nieto, M.A. Context-specific roles of EMT programmes in cancer cell dissemination. Nat. Cell Biol. 2017, 19, 416–418. [Google Scholar] [CrossRef]
- Ocana, O.H.; Corcoles, R.; Fabra, A.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012, 22, 709–724. [Google Scholar] [CrossRef]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Rowling, E.J.; Miskolczi, Z.; Nagaraju, R.; Wilcock, D.J.; Wang, P.; Telfer, B.; Li, Y.; Lasheras-Otero, I.; Redondo-Munoz, M.; Sharrocks, A.D.; et al. Cooperative behaviour and phenotype plasticity evolve during melanoma progression. Pigment Cell Melanoma Res. 2020, 33, 695–708. [Google Scholar] [CrossRef]
- Andrews, M.C.; Oba, J.; Wu, C.J.; Zhu, H.; Karpinets, T.; Creasy, C.A.; Forget, M.A.; Yu, X.; Song, X.; Mao, X.; et al. Multi-modal molecular programs regulate melanoma cell state. Nat. Commun. 2022, 13, 4000. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Canciello, A.; Cervero-Varona, A.; Peserico, A.; Mauro, A.; Russo, V.; Morrione, A.; Giordano, A.; Barboni, B. "In medio stat virtus": Insights into hybrid E/M phenotype attitudes. Front. Cell Dev. Biol. 2022, 10, 1038841. [Google Scholar] [CrossRef] [PubMed]
- Sage, E.H.; Bornstein, P. Extracellular proteins that modulate cell-matrix interactions. SPARC, tenascin, and thrombospondin. J. Biol. Chem. 1991, 266, 14831–14834. [Google Scholar] [CrossRef] [PubMed]
- Brekken, R.A.; Sage, E.H. SPARC, a matricellular protein: At the crossroads of cell-matrix communication. Matrix Biol. 2001, 19, 816–827. [Google Scholar]
- Popovic, A.; Tartare-Deckert, S. Role of extracellular matrix architecture and signaling in melanoma therapeutic resistance. Front. Oncol. 2022, 12, 924553. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012, 44, 480–488. [Google Scholar] [CrossRef]
- Murphy-Ullrich, J.E. The de-adhesive activity of matricellular proteins: Is intermediate cell adhesion an adaptive state? J. Clin. Invest. 2001, 107, 785–790. [Google Scholar] [CrossRef]
- Robert, G.; Gaggioli, C.; Bailet, O.; Chavey, C.; Abbe, P.; Aberdam, E.; Sabatie, E.; Cano, A.; Garcia de Herreros, A.; Ballotti, R.; et al. SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res. 2006, 66, 7516–7523. [Google Scholar] [CrossRef] [PubMed]
- Francki, A.; McClure, T.D.; Brekken, R.A.; Motamed, K.; Murri, C.; Wang, T.; Sage, E.H. SPARC regulates TGF-beta1-dependent signaling in primary glomerular mesangial cells. J. Cell. Biochem. 2004, 91, 915–925. [Google Scholar] [CrossRef]
- Fenouille, N.; Puissant, A.; Tichet, M.; Zimniak, G.; Abbe, P.; Mallavialle, A.; Rocchi, S.; Ortonne, J.P.; Deckert, M.; Ballotti, R.; et al. SPARC functions as an anti-stress factor by inactivating p53 through Akt-mediated MDM2 phosphorylation to promote melanoma cell survival. Oncogene 2011, 30, 4887–4900. [Google Scholar] [CrossRef]
- Arnold, S.A.; Brekken, R.A. SPARC: A matricellular regulator of tumorigenesis. J. Cell Commun. Signal. 2009, 3, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Framson, P.E.; Sage, E.H. SPARC and tumor growth: Where the seed meets the soil? J. Cell. Biochem. 2004, 92, 679–690. [Google Scholar] [CrossRef]
- Clark, C.J.; Sage, E.H. A prototypic matricellular protein in the tumor microenvironment--where there’s SPARC, there’s fire. J. Cell. Biochem. 2008, 104, 721–732. [Google Scholar] [CrossRef]
- Defresne, F.; Bouzin, C.; Grandjean, M.; Dieu, M.; Raes, M.; Hatzopoulos, A.K.; Kupatt, C.; Feron, O. Preconditioned endothelial progenitor cells reduce formation of melanoma metastases through SPARC-driven cell-cell interactions and endocytosis. Cancer Res. 2011, 71, 4748–4757. [Google Scholar] [CrossRef]
- Sangaletti, S.; Stoppacciaro, A.; Guiducci, C.; Torrisi, M.R.; Colombo, M.P. Leukocyte, rather than tumor-produced SPARC, determines stroma and collagen type IV deposition in mammary carcinoma. J. Exp. Med. 2003, 198, 1475–1485. [Google Scholar] [CrossRef]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Perez-Gomez, B.; Palacios, J.; Pollan, M.; Linares, J.; Serrano, S.; Saez-Castillo, A.I.; Sanchez, L.; et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef]
- Massi, D.; Franchi, A.; Borgognoni, L.; Reali, U.M.; Santucci, M. Osteonectin expression correlates with clinical outcome in thin cutaneous malignant melanomas. Hum. Pathol. 1999, 30, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Ledda, F.; Bravo, A.I.; Adris, S.; Bover, L.; Mordoh, J.; Podhajcer, O.L. The expression of the secreted protein acidic and rich in cysteine (SPARC) is associated with the neoplastic progression of human melanoma. J. Investig. Dermatol. 1997, 108, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Tai, I.T.; Tang, M.J. SPARC in cancer biology: Its role in cancer progression and potential for therapy. Drug Resist. Updat. 2008, 11, 231–246. [Google Scholar] [CrossRef] [PubMed]
- Latchana, N.; Ganju, A.; Howard, J.H.; Carson, W.E., 3rd. MicroRNA dysregulation in melanoma. Surg. Oncol. 2016, 25, 184–189. [Google Scholar] [CrossRef]
- Mione, M.; Bosserhoff, A. MicroRNAs in melanocyte and melanoma biology. Pigment Cell Melanoma Res. 2015, 28, 340–354. [Google Scholar] [CrossRef]
- Sun, V.; Zhou, W.B.; Majid, S.; Kashani-Sabet, M.; Dar, A.A. MicroRNA-mediated regulation of melanoma. Br. J. Dermatol. 2014, 171, 234–241. [Google Scholar] [CrossRef]
- Thyagarajan, A.; Tsai, K.Y.; Sahu, R.P. MicroRNA heterogeneity in melanoma progression. Semin. Cancer Biol. 2019, 59, 208–220. [Google Scholar] [CrossRef]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Gebeshuber, C.A.; Zatloukal, K.; Martinez, J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009, 10, 400–405. [Google Scholar] [CrossRef]
- Grant, J.L.; Fishbein, M.C.; Hong, L.S.; Krysan, K.; Minna, J.D.; Shay, J.W.; Walser, T.C.; Dubinett, S.M. A novel molecular pathway for Snail-dependent, SPARC-mediated invasion in non-small cell lung cancer pathogenesis. Cancer Prev. Res. 2014, 7, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.C.; Cursons, J.; Hurley, D.G.; Anaka, M.; Cebon, J.S.; Behren, A.; Crampin, E.J. Systems analysis identifies miR-29b regulation of invasiveness in melanoma. Mol. Cancer 2016, 15, 72. [Google Scholar] [CrossRef]
- Gasque Schoof, C.R.; Izzotti, A.; Jasiulionis, M.G.; Vasques Ldos, R. The Roles of miR-26, miR-29, and miR-203 in the Silencing of the Epigenetic Machinery during Melanocyte Transformation. Biomed. Res. Int. 2015, 2015, 634749. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.J.; Philippidou, D.; Reinsbach, S.E.; Margue, C.; Wienecke-Baldacchino, A.; Nashan, D.; Behrmann, I.; Kreis, S. Interferon-gamma-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun. Signal. 2012, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Vera, O.; Bok, I.; Jasani, N.; Nakamura, K.; Xu, X.; Mecozzi, N.; Angarita, A.; Wang, K.; Tsai, K.Y.; Karreth, F.A. A MAPK/miR-29 Axis Suppresses Melanoma by Targeting MAFG and MYBL2. Cancers 2021, 13, 1408. [Google Scholar] [CrossRef]
- Subramanian, M.; Rao, S.R.; Thacker, P.; Chatterjee, S.; Karunagaran, D. MiR-29b downregulates canonical Wnt signaling by suppressing coactivators of beta-catenin in human colorectal cancer cells. J. Cell. Biochem. 2014, 115, 1974–1984. [Google Scholar]
- Qiu, F.; Sun, R.; Deng, N.; Guo, T.; Cao, Y.; Yu, Y.; Wang, X.; Zou, B.; Zhang, S.; Jing, T.; et al. miR-29a/b enhances cell migration and invasion in nasopharyngeal carcinoma progression by regulating SPARC and COL3A1 gene expression. PLoS ONE 2015, 10, e0120969. [Google Scholar] [CrossRef]
- Wu, L.; de Perrot, M. Omics Overview of the SPARC Gene in Mesothelioma. Biomolecules 2023, 13, 1103. [Google Scholar] [CrossRef]
- Alachkar, H.; Santhanam, R.; Maharry, K.; Metzeler, K.H.; Huang, X.; Kohlschmidt, J.; Mendler, J.H.; Benito, J.M.; Hickey, C.; Neviani, P.; et al. SPARC promotes leukemic cell growth and predicts acute myeloid leukemia outcome. J. Clin. Investig. 2014, 124, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.C.; Dong, Q.Z.; Zhang, X.F.; Deng, B.; Jia, H.L.; Ye, Q.H.; Qin, L.X.; Wu, X.Z. microRNA-29a suppresses cell proliferation by targeting SPARC in hepatocellular carcinoma. Int. J. Mol. Med. 2012, 30, 1321–1326. [Google Scholar] [CrossRef]
- Kapinas, K.; Kessler, C.B.; Delany, A.M. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 2009, 108, 216–224. [Google Scholar] [CrossRef]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.H.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e819. [Google Scholar] [CrossRef]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.L.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904.e895. [Google Scholar] [CrossRef]
- Verfaillie, A.; Imrichova, H.; Atak, Z.K.; Dewaele, M.; Rambow, F.; Hulselmans, G.; Christiaens, V.; Svetlichnyy, D.; Luciani, F.; Van den Mooter, L.; et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat. Commun. 2015, 6, 6683. [Google Scholar] [CrossRef]
- Cserjesi, P.; Lilly, B.; Bryson, L.; Wang, Y.; Sassoon, D.A.; Olson, E.N. MHox: A mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development 1992, 115, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Grueneberg, D.A.; Natesan, S.; Alexandre, C.; Gilman, M.Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science 1992, 257, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Ferreres, J.R.; Vinyals, A.; Campos-Martin, R.; Espin, R.; Podlipnik, S.; Ramos, R.; Bertran, E.; Carrera, C.; Marcoval, J.; Malvehy, J.; et al. PRRX1 silencing is required for metastatic outgrowth in melanoma and is an independent prognostic of reduced survival in patients. Mol. Oncol. 2024, 18, 2471–2494. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Kaochar, S.; Dong, J.; Torres, M.; Rajapakshe, K.; Nikolos, F.; Davis, C.M.; Ehli, E.A.; Coarfa, C.; Mitsiades, N.; Poulaki, V. ICG-001 Exerts Potent Anticancer Activity Against Uveal Melanoma Cells. Investig. Ophthalmol. Vis. Sci. 2018, 59, 132–143. [Google Scholar] [CrossRef]
- Chen, R.H.; Ding, W.V.; McCormick, F. Wnt signaling to beta-catenin involves two interactive components Glycogen synthase kinase-3beta inhibition activation of protein kinase, C. J. Biol. Chem. 2000, 275, 17894–17899. [Google Scholar] [CrossRef]
- Eichhoff, O.M.; Weeraratna, A.; Zipser, M.C.; Denat, L.; Widmer, D.S.; Xu, M.; Kriegl, L.; Kirchner, T.; Larue, L.; Dummer, R.; et al. Differential LEF1 and TCF4 expression is involved in melanoma cell phenotype switching. Pigment Cell Melanoma Res. 2011, 24, 631–642. [Google Scholar] [CrossRef]
- Hafner, M.; Zimmermann, K.; Pottgiesser, J.; Krieg, T.; Nischt, R. A purine-rich sequence in the human BM-40 gene promoter region is a prerequisite for maximum transcription. Matrix Biol. 1995, 14, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.; Chamboredon, S.; Castellazzi, M.; Kerry, J.A.; Bos, T.J. Transcriptional upregulation of SPARC, in response to c-Jun overexpression, contributes to increased motility and invasion of MCF7 breast cancer cells. Oncogene 2002, 21, 7077–7091. [Google Scholar] [CrossRef]
- McVey, J.H.; Nomura, S.; Kelly, P.; Mason, I.J.; Hogan, B.L. Characterization of the mouse SPARC/osteonectin gene. Intron/exon organization and an unusual promoter region. J. Biol. Chem. 1988, 263, 11111–11116. [Google Scholar] [CrossRef] [PubMed]
- Vial, E.; Perez, S.; Castellazzi, M. Transcriptional control of SPARC by v-Jun and other members of the AP1 family of transcription factors. Oncogene 2000, 19, 5020–5029. [Google Scholar] [CrossRef]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef]
- Jorda, M.; Olmeda, D.; Vinyals, A.; Valero, E.; Cubillo, E.; Llorens, A.; Cano, A.; Fabra, A. Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J. Cell. Sci. 2005, 118, 3371–3385. [Google Scholar] [CrossRef]
- Portugal, J. Mithramycin and its analogs: Molecular features and antitumor action. Pharmacol. Ther. 2024, 260, 108672. [Google Scholar] [CrossRef]
- Sachrajda, I.; Ratajewski, M. Mithramycin A suppresses expression of the human melanoma-associated gene ABCB8. Mol. Genet. Genomics. 2011, 285, 57–65. [Google Scholar] [CrossRef]
- Lopez-Bergami, P. The role of mitogen- and stress-activated protein kinase pathways in melanoma. Pigment Cell Melanoma Res. 2011, 24, 902–921. [Google Scholar] [CrossRef] [PubMed]
- Kappelmann, M.; Bosserhoff, A.; Kuphal, S. AP-1/c-Jun transcription factors: Regulation and function in malignant melanoma. Eur. J. Cell. Biol. 2014, 93, 76–81. [Google Scholar] [CrossRef]
- Huang, Z.; Li, Y.; Qian, Y.; Zhai, E.; Zhao, Z.; Zhang, T.; Liu, Y.; Ye, L.; Wei, R.; Zhao, R.; et al. Tumor-secreted LCN2 impairs gastric cancer progression via autocrine inhibition of the 24p3R/JNK/c-Jun/SPARC axis. Cell Death Dis. 2024, 15, 756. [Google Scholar] [CrossRef]
- Kapinas, K.; Kessler, C.; Ricks, T.; Gronowicz, G.; Delany, A.M. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 2010, 285, 25221–25231. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics. 2019, 12, 173–194. [Google Scholar] [CrossRef]
- Botti, G.; Scognamiglio, G.; Marra, L.; Collina, F.; Di Bonito, M.; Cerrone, M.; Grilli, B.; Anniciello, A.; Franco, R.; Fulciniti, F.; et al. SPARC/osteonectin is involved in metastatic process to the lung during melanoma progression. Virchows Arch. 2014, 465, 331–338. [Google Scholar] [CrossRef]
- Ruiz, E.M.; Alhassan, S.A.; Errami, Y.; Abd Elmageed, Z.Y.; Fang, J.S.; Wang, G.; Brooks, M.A.; Abi-Rached, J.A.; Kandil, E.; Zerfaoui, M. A Predictive Model of Adaptive Resistance to BRAF/MEK Inhibitors in Melanoma. Int. J. Mol. Sci. 2023, 24, 8407. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Li, Y.; Peng, C.; Yang, S.; Seto, E. HDAC10 inhibition represses melanoma cell growth and BRAF inhibitor resistance via upregulating SPARC expression. NAR Cancer 2024, 6, zcae018. [Google Scholar] [CrossRef]
- Garcia-Bellido, A. Genetic control of wing disc development in Drosophila. In Ciba Foundation Symposium 29—Cell Patterning; John Wiley & Sons, Ltd.: Chichester, UK, 1975; pp. 161–182. [Google Scholar]
- Guo, J.; Fu, Z.; Wei, J.; Lu, W.; Feng, J.; Zhang, S. PRRX1 promotes epithelial-mesenchymal transition through the Wnt/beta-catenin pathway in gastric cancer. Med. Oncol. 2015, 32, 393. [Google Scholar] [CrossRef]
- Lopez-Moncada, F.; Torres, M.J.; Castellon, E.A.; Contreras, H.R. Secreted protein acidic and rich in cysteine (SPARC) induces epithelial-mesenchymal transition, enhancing migration and invasion, and is associated with high Gleason score in prostate cancer. Asian J. Androl. 2019, 21, 557–564. [Google Scholar] [CrossRef]
- Girotti, M.R.; Fernandez, M.; Lopez, J.A.; Camafeita, E.; Fernandez, E.A.; Albar, J.P.; Benedetti, L.G.; Valacco, M.P.; Brekken, R.A.; Podhajcer, O.L.; et al. SPARC promotes cathepsin B-mediated melanoma invasiveness through a collagen I/alpha2beta1 integrin axis. J. Investig. Dermatol. 2011, 131, 2438–2447. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Z.; Heravi, M.; Thuraisingam, T.; Di Marco, S.; Muanza, T.; Radzioch, D. Brg-1 mediates the constitutive and fenretinide-induced expression of SPARC in mammary carcinoma cells via its interaction with transcription factor Sp1. Mol. Cancer 2010, 9, 210. [Google Scholar] [CrossRef]
- Benasciutti, E.; Pages, G.; Kenzior, O.; Folk, W.; Blasi, F.; Crippa, M.P. MAPK and JNK transduction pathways can phosphorylate Sp1 to activate the uPA minimal promoter element and endogenous gene transcription. Blood 2004, 104, 256–262. [Google Scholar] [CrossRef]
- Delany, A.M.; Canalis, E. Basic fibroblast growth factor destabilizes osteonectin mRNA in osteoblasts. Am. J. Physiol. 1998, 274, C734–C740. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, J.; Wang, G.K.; Zhang, J.T.; Huang, S.; Qin, Y.W.; Jing, Q. Uracils at nucleotide position 9–11 are required for the rapid turnover of miR-29 family. Nucleic Acids Res. 2011, 39, 4387–4395. [Google Scholar] [CrossRef]
- Wang, C.; Bian, Z.; Wei, D.; Zhang, J.G. miR-29b regulates migration of human breast cancer cells. Mol. Cell. Biochem. 2011, 352, 197–207. [Google Scholar] [CrossRef]
- Yan, B.; Guo, Q.; Fu, F.J.; Wang, Z.; Yin, Z.; Wei, Y.B.; Yang, J.R. The role of miR-29b in cancer: Regulation, function, and signaling. Onco Targets Ther. 2015, 8, 539–548. [Google Scholar] [PubMed]
- Puig-Butille, J.A.; Vinyals, A.; Ferreres, J.R.; Aguilera, P.; Cabre, E.; Tell-Marti, G.; Marcoval, J.; Mateo, F.; Palomero, L.; Badenas, C.; et al. AURKA Overexpression Is Driven by FOXM1 and MAPK/ERK Activation in Melanoma Cells Harboring BRAF or NRAS Mutations: Impact on Melanoma Prognosis and Therapy. J. Investig. Dermatol. 2017, 137, 1297–1310. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Ugalde, A.P.; Ramsay, A.J.; De La Rosa, J.; Varela, I.; Marino, G.; Cadinanos, J.; Lu, J.; Freije, J.M.; López-Otín, C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011, 30, 2219–2232. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Cirenajwis, H.; Ekedahl, H.; Lauss, M.; Harbst, K.; Carneiro, A.; Enoksson, J.; Rosengren, F.; Werner-Hartman, L.; Törngren, T.; Kvist, A.; et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget 2015, 6, 12297–12309. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, G.; Busch, C.; Knappskog, S.; Geisler, J.; Miletic, H.; Ringnér, M.; Lillehaug, J.R.; Borg, Å.; Lønning, P.E. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin. Cancer Res. 2010, 16, 3356–3367. [Google Scholar] [CrossRef]
- Kabbarah, O.; Nogueira, C.; Feng, B.; Nazarian, R.M.; Bosenberg, M.; Wu, M.; Scott, K.L.; Kwong, L.N.; Xiao, Y.; Cordon-Cardo, C.; et al. Integrative genome comparison of primary and metastatic melanomas. PLoS ONE 2010, 5, e10770. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vinyals, A.; Ferreres, J.R.; Campos-Martín, R.; Torres, O.J.C.; Mainez, J.; Puig-Butillé, J.A.; Marcoval, J.; Puig, S.; Fabregat, I.; Fabra, À. Regulatory Mechanisms of SPARC Overexpression in Melanoma Progression. Int. J. Mol. Sci. 2025, 26, 8743. https://doi.org/10.3390/ijms26178743
Vinyals A, Ferreres JR, Campos-Martín R, Torres OJC, Mainez J, Puig-Butillé JA, Marcoval J, Puig S, Fabregat I, Fabra À. Regulatory Mechanisms of SPARC Overexpression in Melanoma Progression. International Journal of Molecular Sciences. 2025; 26(17):8743. https://doi.org/10.3390/ijms26178743
Chicago/Turabian StyleVinyals, Antònia, Josep R. Ferreres, Rafael Campos-Martín, Olga J. C. Torres, Jessica Mainez, Joan A. Puig-Butillé, Joaquim Marcoval, Susana Puig, Isabel Fabregat, and Àngels Fabra. 2025. "Regulatory Mechanisms of SPARC Overexpression in Melanoma Progression" International Journal of Molecular Sciences 26, no. 17: 8743. https://doi.org/10.3390/ijms26178743
APA StyleVinyals, A., Ferreres, J. R., Campos-Martín, R., Torres, O. J. C., Mainez, J., Puig-Butillé, J. A., Marcoval, J., Puig, S., Fabregat, I., & Fabra, À. (2025). Regulatory Mechanisms of SPARC Overexpression in Melanoma Progression. International Journal of Molecular Sciences, 26(17), 8743. https://doi.org/10.3390/ijms26178743






