Genome-Wide Association Study and Transcriptome Analysis Reveal Alkaline Stress-Responsive Genes in Bread Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Results
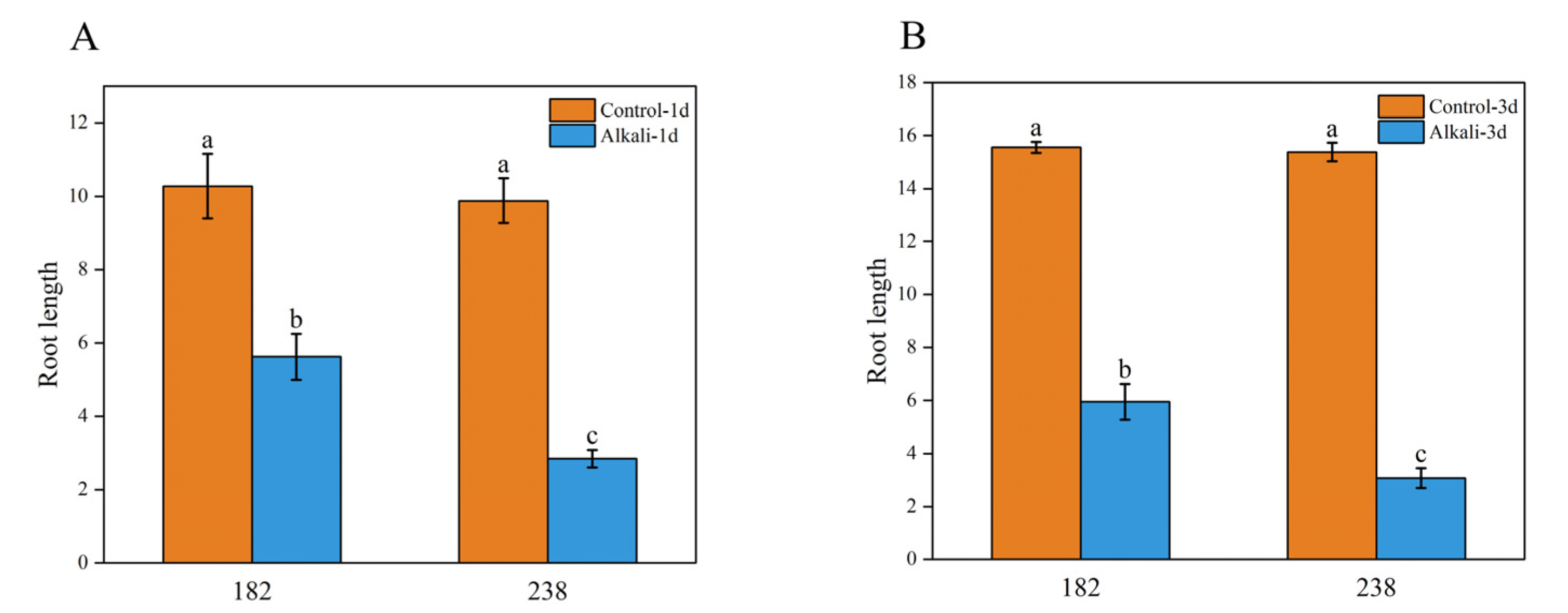
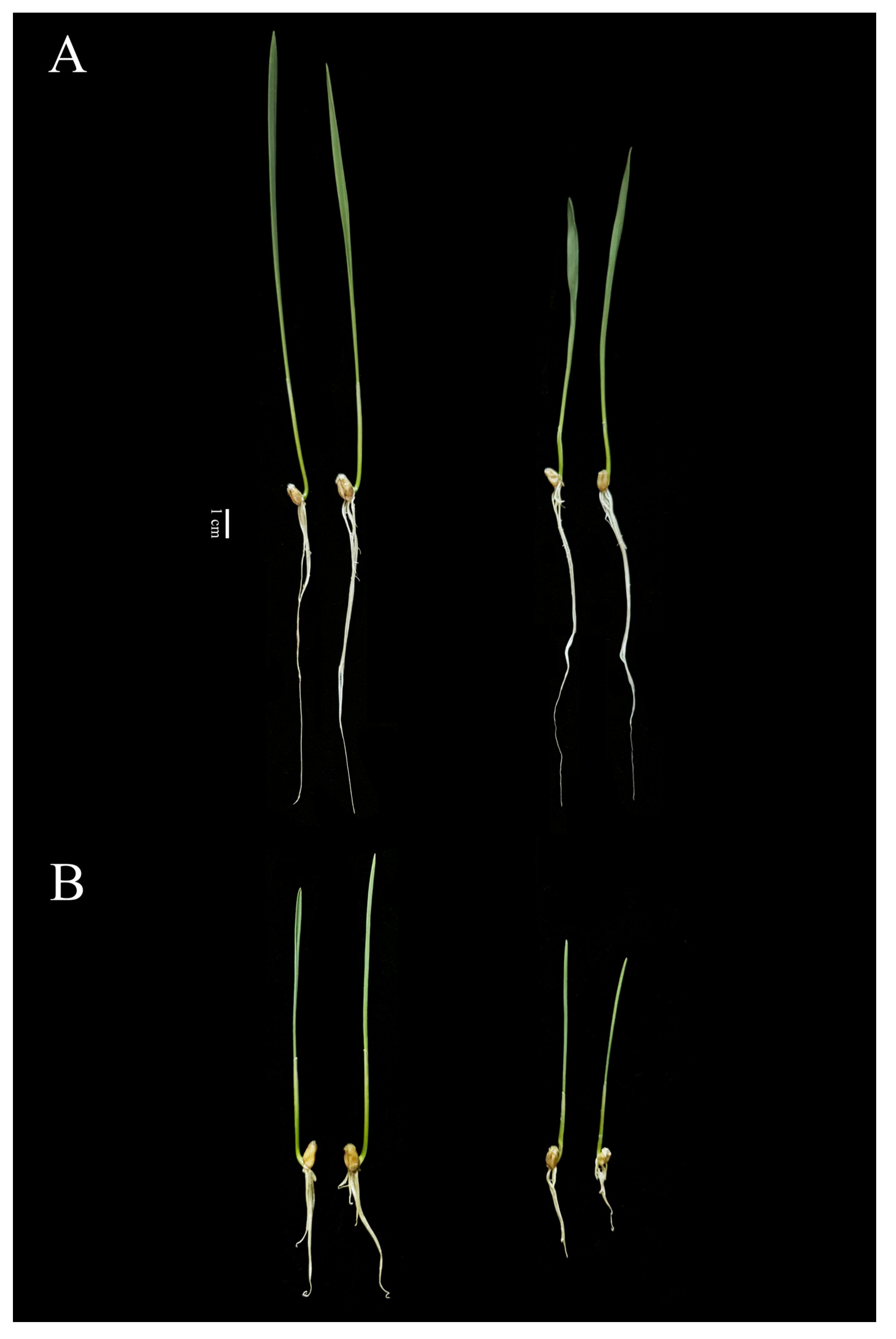
2.1. Effects of Alkaline Salt Stress on Wheat Phenotype
2.2. Population Structure Analysis
2.3. GWAS Candidate Gene Prediction
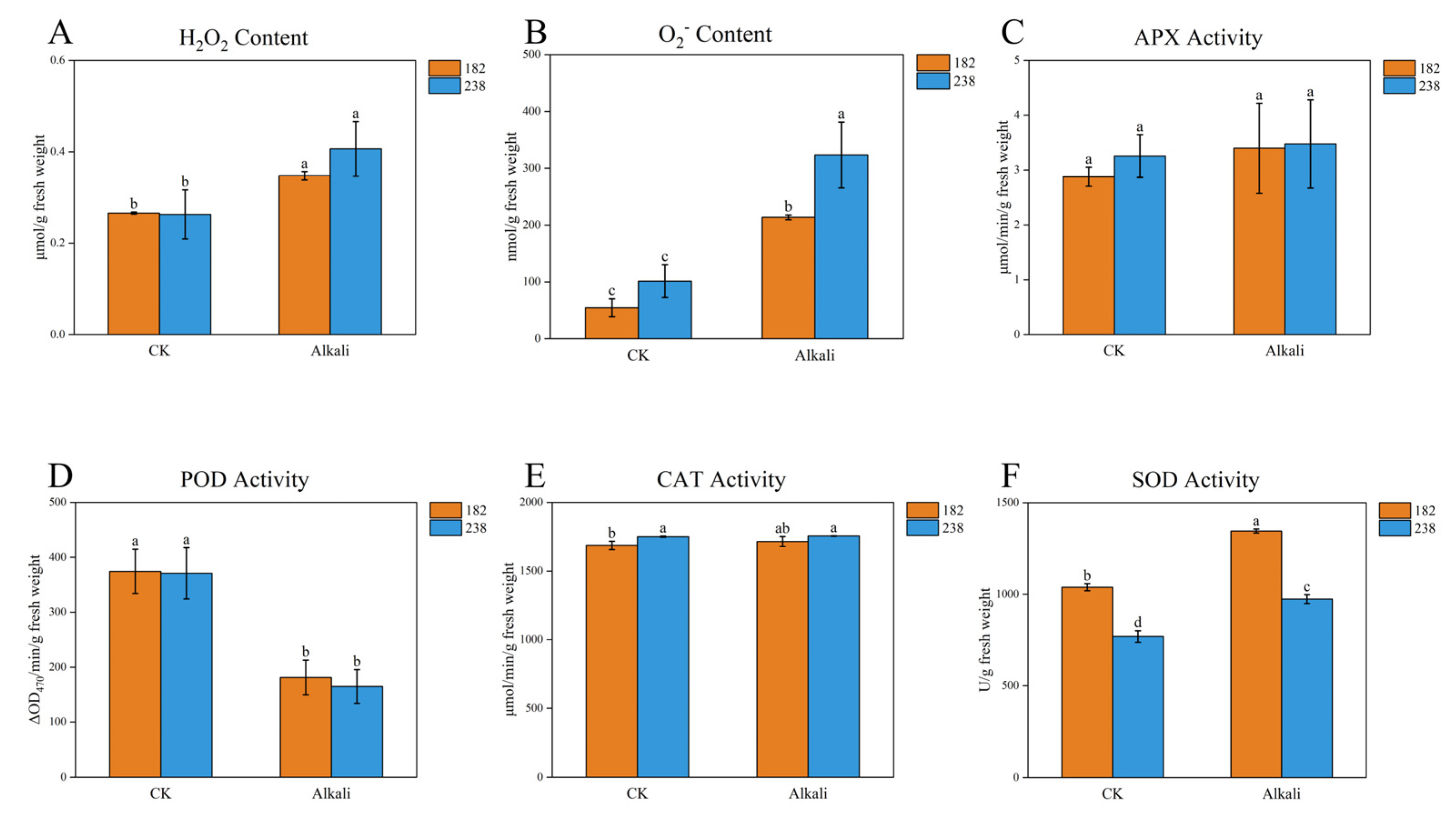
2.4. Phenotypic Analysis of Two Genotypes with Significant Differences in Alkaline Tolerance
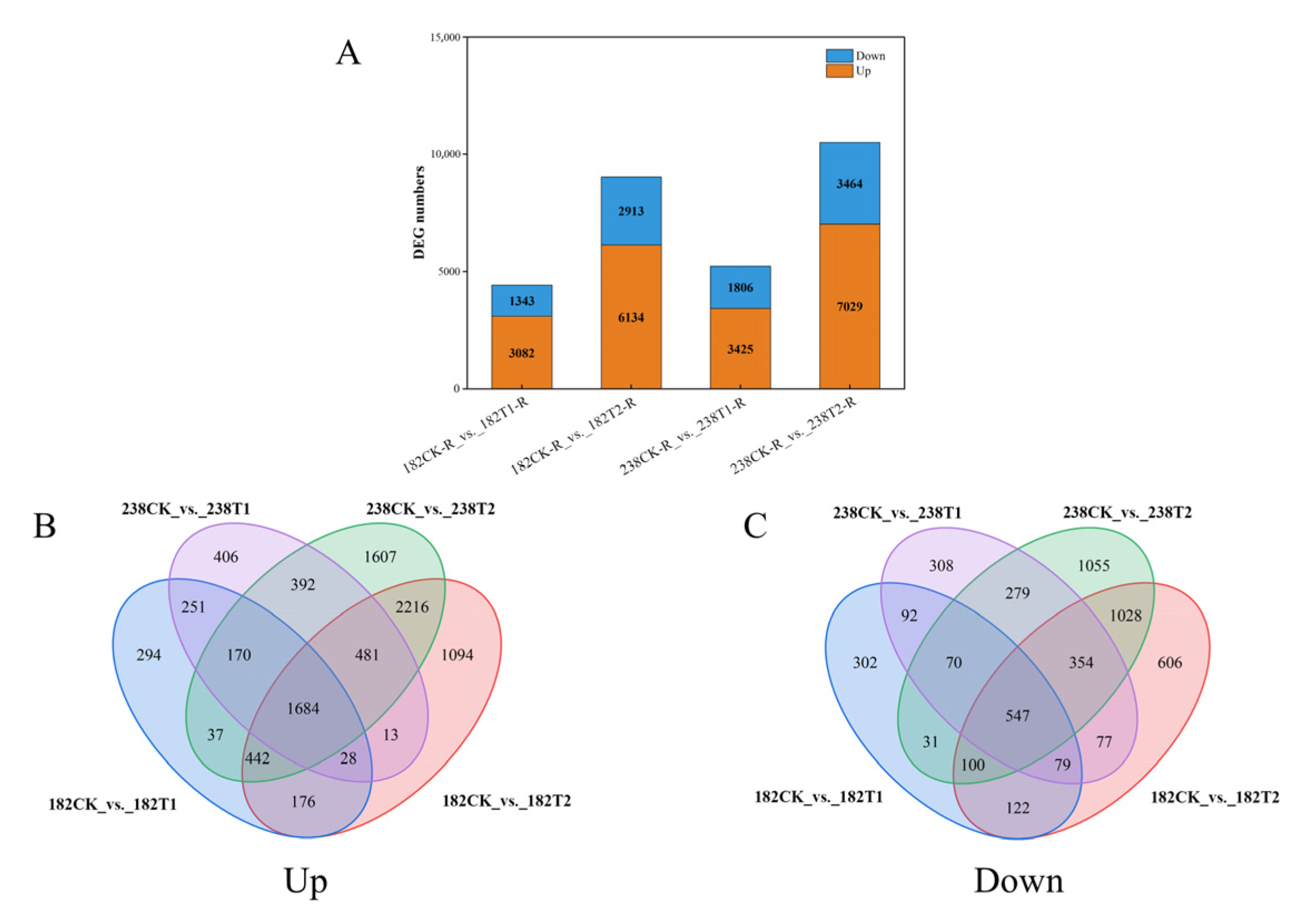
2.5. RNA-Seq Analysis Results
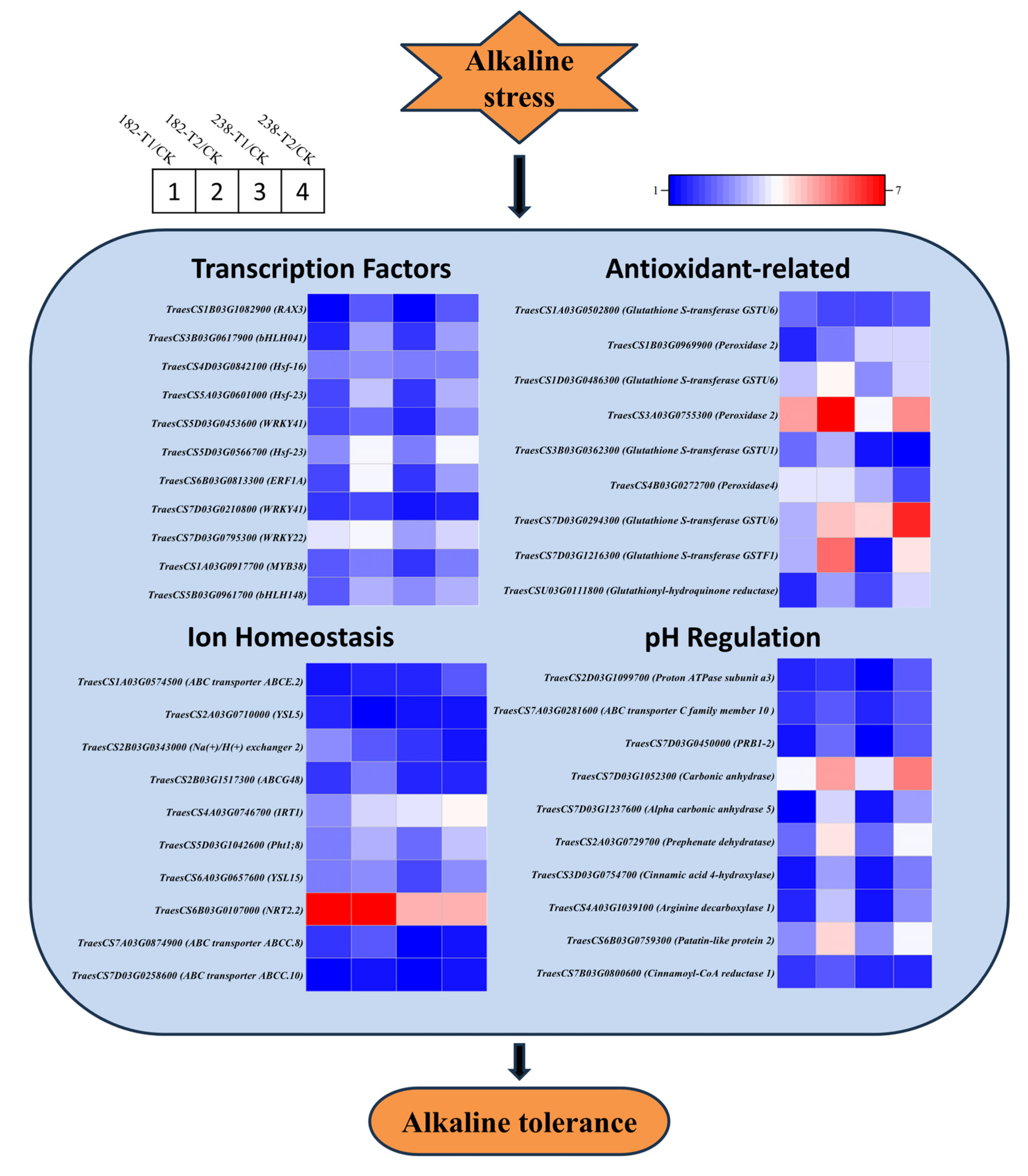
2.6. Common Genes Identified via GWAS and RNA-Seq
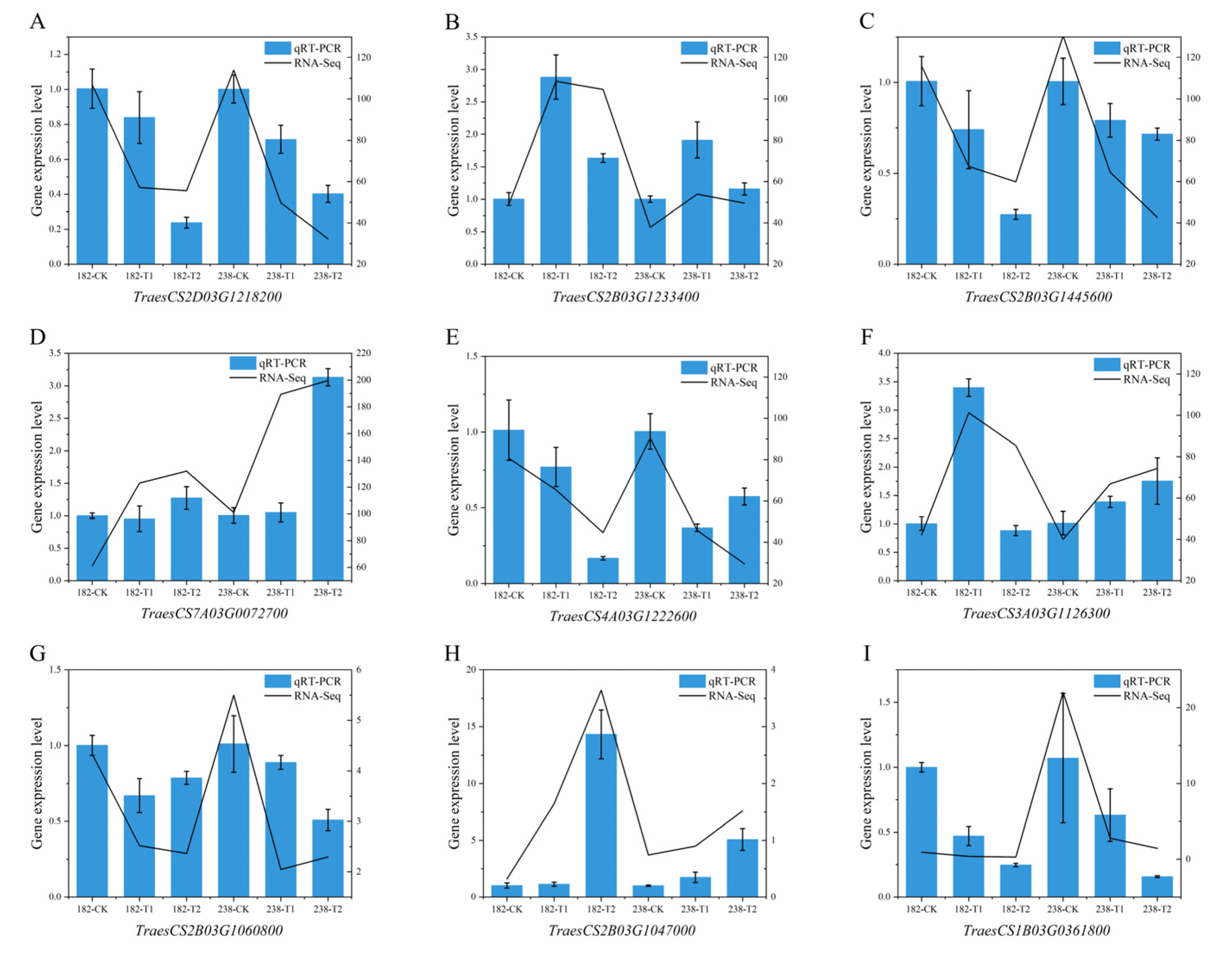
2.7. Quantitative Real-Time PCR (qPCR)
3. Discussion
3.1. Root Growth Is One of the Important Indicators for Evaluating Alkaline Tolerance
3.2. The Antioxidant Capacity Is a Key Indicator for Assessing Alkaline Stress Tolerance
3.3. The Major Loci Associated with Alkaline Stress Tolerance Identified in Wheat
3.4. Promising Candidate Genes Conferring Alkaline Tolerance Were Identified in Wheat
4. Materials and Methods
4.1. Experimental Materials
4.2. Hydroponic Experiment
4.3. Phenotypic Measurement
4.4. GWAS Analysis
4.5. RNA-Seq Analysis
4.6. Validation of RNA-Seq by Quantitative Real-Time PCR (qPCR)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du, Y.; Liu, X.; Zhang, L.; Zhou, W. Drip irrigation in agricultural saline-alkali land controls soil salinity and improves crop yield: Evidence from a global meta-analysis. Sci. Total Environ. 2023, 880, 163226. [Google Scholar] [CrossRef]
- Zhu, W.; Gu, S.; Jiang, R.; Zhang, X.; Hatano, R. Saline-alkali soil reclamation contributes to soil health improvement in China. Agriculture 2024, 14, 1210. [Google Scholar] [CrossRef]
- Liu, L.; Wang, B. Protection of halophytes and their uses for cultivation of saline-alkali soil in China. Biology 2021, 10, 353. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Jia, X.; Wang, H.; Svetla, S.; Zhu, Y.; Hu, Y.; Cheng, L.; Zhao, T.; Wang, Y. Comparative physiological responses and adaptive strategies of apple Malus halliana to salt, alkali and saline-alkali stress. Sci. Hortic. 2019, 245, 154–162. [Google Scholar] [CrossRef]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 2008, 56, 179–190. [Google Scholar] [CrossRef]
- Yang, C.; Xu, H.; Wang, L.; Liu, J.; Shi, D.; Wang, D. Comparative effects of salt-stress and alkali-stress on the growth, photosynthesis, solute accumulation, and ion balance of barley plants. Photosynthetica 2009, 47, 79–86. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y. How do plants maintain pH and ion homeostasis under saline-alkali stress? Front. Plant Sci. 2023, 14, 1217193. [Google Scholar] [CrossRef]
- Guo, J.; Lu, X.; Tao, Y.; Guo, H.; Min, W. Comparative ionomics and metabolic responses and adaptive strategies of cotton to salt and alkali stress. Front. Plant Sci. 2022, 13, 871387. [Google Scholar] [CrossRef]
- Li, R.; Shi, F.; Fukuda, K.; Yang, Y. Effects of salt and alkali stresses on germination, growth, photosynthesis and ion accumulation in alfalfa (Medicago sativa L.). Soil Sci. Plant Nutr. 2010, 56, 725–733. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, B.; Zhang, F.; Liu, X.; Wang, X.; Zhang, C.; Zhao, K.; Yuan, R.; Lamlom, S.F.; Abdelghany, A.M.; et al. Integration of physiological and transcriptomic approaches in investigating salt-alkali stress resilience in soybean. Plant Stress 2024, 11, 100375. [Google Scholar] [CrossRef]
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 9586. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yu, F.; Xie, P.; Sun, S.; Qiao, X.; Tang, S.; Tang, S.; Chen, C.; Yang, S.; Mei, C.; et al. A Gγ protein regulates alkaline sensitivity in crops. Science 2023, 379, eade8416. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, N.; Wang, S.; Yang, Z.; Chen, X.; Lu, X.; Huang, H.; Chen, X.; Zhao, L.; Zhang, M.; et al. GhGLDH35A gene-mediated ROS homeostasis and stomatal movement via the ascorbic acid pathway confers alkaline stress tolerance. J. Adv. Res. 2025. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Wang, K.; Li, D.; Liu, Z.; Li, M.; Wang, Z.; Li, X.; Lan, X.; Guan, Q. OsSAP6 positively regulates soda saline–alkaline stress tolerance in rice. Rice 2022, 15, 69. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, M.; Liang, X.; Li, F.; Shi, Y.; Yang, X.; Jiang, C. Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline-alkaline tolerance in maize. Nat. Commun. 2020, 11, 186. [Google Scholar] [CrossRef]
- Cui, M.; Li, Y.; Li, J.; Yin, F.; Chen, X.; Qin, L.; Wei, L.; Xia, G.; Liu, S. Ca2+-dependent TaCCD1 cooperates with TaSAUR215 to enhance plasma membrane H+-ATPase activity and alkali stress tolerance by inhibiting PP2C-mediated dephosphorylation of TaHA2 in wheat. Mol. Plant 2023, 16, 571–587. [Google Scholar] [CrossRef]
- Wei, L.; Ren, X.; Qin, L.; Zhang, R.; Cui, M.; Xia, G.; Liu, S. TaWRKY55-TaPLATZ2 module negatively regulate saline-alkali stress tolerance in wheat. J. Integr. Plant Biol. 2025, 67, 19–34. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.M.; Testerink, C. Roots withstanding their environment: Exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef]
- Wen, T.; Dong, L.; Wang, L.; Ma, F.; Zou, Y.; Li, C. Changes in root architecture and endogenous hormone levels in two Malus rootstocks under alkali stress. Sci. Hortic. 2018, 235, 198–204. [Google Scholar] [CrossRef]
- Liu, S.; An, T.; Gao, Y.; Kuang, Q.; Xu, B.; Zhang, S.; Deng, X.; Zhao, T.; Lam, H.; Shabala, S.; et al. Soybean genotypes with contrasting root system size differ in saline-alkaline tolerance. J. Agron. Crop. Sci. 2025, 211, e70040. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Zhang, R.; Yuan, H.; Wang, M.; Yang, H.; Ma, H.; Liu, D.; Jiang, C.; Liang, Z. Root damage under alkaline stress is associated with reactive oxygen species accumulation in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1580. [Google Scholar] [CrossRef]
- Meng, C.; Quan, T.; Li, Z.; Cui, K.; Yan, L.; Liang, Y.; Dai, J.; Liu, S. Transcriptome profiling reveals the genetic basis of alkalinity tolerance in wheat. BMC Genom. 2017, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef]
- Li, B. Identification of genes conferring plant salt tolerance using GWAS: Current success and perspectives. Plant Cell Physiol. 2020, 61, 1419–1426. [Google Scholar] [CrossRef]
- Khan, R.W.A.; Khan, R.S.A.; Awan, F.S.; Akrem, A.; Iftikhar, A.; Anwar, F.N.; Alzahrani, H.A.S.; Alsamadany, H.; Iqbal, R.K. Genome-wide association studies of seedling quantitative trait loci against salt tolerance in wheat. Front. Genet. 2022, 13, 946869. [Google Scholar] [CrossRef]
- Quan, X.; Liu, J.; Zhang, N.; Xie, C.; Li, H.; Xia, X.; He, W.; Qin, Y. Genome-wide association study uncover the genetic architecture of salt tolerance-related traits in common wheat (Triticum aestivum L.). Front. Genet. 2021, 12, 663941. [Google Scholar] [CrossRef]
- Luo, Q.; Hu, P.; Yang, G.; Li, H.; Liu, L.; Wang, Z.; Li, B.; Li, Z.; Zheng, Q. Map QTL for seedling morphological and physiological traits under normal and salt treatments in a RIL wheat population. Theor. Appl. Genet. 2021, 134, 2991–3011. [Google Scholar] [CrossRef]
- Ren, Y.; Xu, Y.; Teng, W.; Li, B.; Lin, T. QTLs for seedling traits under salinity stress in hexaploid wheat. Cienc. Rural. 2018, 48, e20170446. [Google Scholar] [CrossRef]
- Sheng, W.; Zhang, G.; Zhai, L.; Xu, J. Candidate genes for alkali tolerance identified by genome-wide association study at the seedling stage in rice (Oryza sativa L.). Sci. Rep. 2024, 14, 30063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Peng, Y.; Zhou, J.; Tan, Z.; Jin, C.; Fang, S.; Zhong, S.; Jin, C.; Wang, R.; Wen, X.; et al. Genome-wide association studies of salt-alkali tolerance at seedling and mature stages in Brassica napus. Front. Plant Sci. 2022, 13, 857149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Peng, Y.; Zhou, J.; Tan, Z.; Jin, C.; Fang, S.; Zhong, S.; Jin, C.; Wang, R.; Wen, X.; et al. Fine mapping of major QTLs for alkaline tolerance at the seedling stage in maize (Zea mays L.) through genetic linkage analysis combined with high-throughput DNA sequencing. Euphytica 2018, 214, 120. [Google Scholar] [CrossRef]
- Wang, W.; Pang, J.; Zhang, F.; Sun, L.; Yang, L.; Zhao, Y.; Yang, Y.; Wang, Y.; Siddique, K.H.M. Integrated transcriptomics and metabolomics analysis to characterize alkali stress responses in canola (Brassica napus L.). Plant Physiol. Biochem. 2021, 166, 605–620. [Google Scholar] [CrossRef]
- Li, M.; Guo, P.; Nan, N.; Ma, A.; Liu, W.; Wang, T.; Yun, D.; Xu, Z. Plasma membrane-localized H+-ATPase OsAHA3 functions in saline-alkaline stress tolerance in rice. Plant Cell Rep. 2024, 43, 11. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, L.H.; Guo, Z.H.; Shi, D.F.; Zhai, H.L.; Yao, Y.; Yang, T.X.; Xin, S.Q.; Cui, H.Y.; Li, J.Q.; et al. Benefits of biological nitrification inhibition of Leymus chinensis under alkaline stress: The regulatory function of ammonium-N exceeds its nutritional function. Front. Plant Sci. 2023, 14, 16. [Google Scholar] [CrossRef]
- Shen, T.; Yan, R.; Xu, F.; Wang, Q.; Chen, D.; Li, K.; Ni, L.; Jiang, M. The NADPH oxidase OsRbohD and OsRbohH negatively regulate saline-alkaline tolerance in rice. Environ. Exp. Bot. 2023, 213, 105445. [Google Scholar] [CrossRef]
- Song, C.; Guo, Y.; Qiu, Q.; Lambert, G.; Galbraith, D.W.; Jagendorf, A.; Zhu, J. A probable Na+(K+)/H+ exchanger on the chloroplast envelope functions in pH homeostasis and chloroplast development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 10211–10216. [Google Scholar] [CrossRef]
- Kamran, M.; Ramesh, S.A.; Gilliham, M.; Tyerman, S.D.; Bose, J. Role of TaALMT1 malate-GABA transporter in alkaline pH tolerance of wheat. Plant Cell Environ. 2020, 43, 2443–2459. [Google Scholar] [CrossRef]
- Casaretto, J.; El-Kereamy, A.; Zeng, B.; Stiegelmeyer, S.; Chen, X.; Bi, Y.; Rothstein, S. Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genom. 2016, 17, 312. [Google Scholar] [CrossRef]
- El-Kereamy, A.; Bi Ranathunge, K.; Beatty, P.; Good, A.; Rothstein, S. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef]
- Kumar, K.; Jaiswal, A.; Koppolu, U.M.K.; Kumar, K.R.R. Alkaline stress disrupts growth, biochemistry, and ion homeostasis of chickpea (Cicer arietinum L.) roots. Front. Agron. 2024, 6, 1497054. [Google Scholar] [CrossRef]
- Thangthong, N.; Jogloy, S.; Punjansing, T.; Kvien, C.; Kesmala, T.; Vorasoot, N. Changes in root anatomy of peanut (Arachis hypogaea L.) under different durations of early season drought. Agronomy 2019, 9, 215. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J.; Kay, T.; Weir, B. How to estimate kinship. Mol. Ecol. 2018, 27, 4121–4135. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Wu, G.; Sun, X.; Sun, Q.; Kang, X.; Wang, J.; He, X.; Liu, W.; Xu, D.; Dai, X.; Ma, W.; et al. Genetic variation in wheat root transcriptome responses to salinity: A comparative study of tolerant and sensitive genotypes. Int. J. Mol. Sci. 2025, 26, 331. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ (–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinf. Biomath. 2013, 3, 71. [Google Scholar]




| Trait | Min (cm) | Max (cm) | Mean (cm) | SD | CV (%) |
|---|---|---|---|---|---|
| RL_CK_1d | 5.60 | 11.10 | 8.76 | 1.06 | 12.14 |
| RL_T_1d | 2.00 | 5.63 | 3.74 | 0.71 | 19.05 |
| RL_CK_3d | 9.33 | 17.55 | 14.09 | 1.40 | 9.93 |
| RL_T_3d | 1.95 | 5.95 | 3.80 | 0.74 | 19.58 |
| Traits | SNP | Chr | Pos | −log10(P) | R2% | Effect | Model |
|---|---|---|---|---|---|---|---|
| RL-CK-1d | 2A_471531993 | 2A | 471531993 | 4.18 | 5.28 | −0.54 | GLM |
| RL-CK-3d | 1B_115442787 | 1B | 115442787 | 4.96 | 7.31 | −0.67 | GLM |
| RL-CK-3d | 1B_115442787 | 1B | 115442787 | 4.18 | 7.13 | −0.66 | MLM |
| RL-CK-3d | 1B_119602852 | 1B | 119602852 | 5.20 | 7.83 | −0.69 | GLM |
| RL-CK-3d | 1B_119602852 | 1B | 119602852 | 4.47 | 7.86 | −0.69 | MLM |
| RL-CK-3d | 1B_129994772 | 1B | 129994772 | 4.25 | 6.23 | −0.61 | GLM |
| RL-CK-3d | 1B_134605692 | 1B | 134605692 | 5.03 | 7.68 | −0.67 | GLM |
| RL-CK-3d | 1B_134605692 | 1B | 134605692 | 4.25 | 7.47 | −0.66 | MLM |
| RL-CK-3d | 1B_167777201 | 1B | 167777201 | 4.67 | 7.16 | −0.65 | GLM |
| RL-CK-3d | 1B_167777201 | 1B | 167777201 | 4.05 | 7.23 | −0.65 | MLM |
| RL-CK-3d | 1B_219513011 | 1B | 219513011 | 4.60 | 6.85 | 0.63 | GLM |
| RL-CK-3d | 2A_313405793 | 2A | 313405793 | 4.49 | 15.71 | −0.80 | GLM |
| RL-CK-3d | 2A_313405793 | 2A | 313405793 | 4.12 | 16.45 | −0.82 | MLM |
| RL-CK-3d | 3B_3239089 | 3B | 3239089 | 4.52 | 6.67 | −0.64 | GLM |
| RL-CK-3d | 4B_817608 | 4B | 817608 | 4.12 | 3.89 | −0.63 | GLM |
| RL-CK-3d | 4B_417309850 | 4B | 417309850 | 4.03 | 4.01 | 0.41 | GLM |
| RL-CK-3d | 5B_6485240 | 5B | 6485240 | 4.04 | 3.89 | −0.42 | MLM |
| RL-CK-3d | 7B_14912720 | 7B | 14912720 | 4.80 | 8.22 | −0.70 | GLM |
| RL-CK-3d | 7B_14912720 | 7B | 14912720 | 4.92 | 8.70 | −0.72 | MLM |
| RL-T-1d | 3B_322640 | 3B | 322640 | 4.45 | 6.34 | 0.33 | GLM |
| RL-T-1d | 3B_704997 | 3B | 704997 | 4.56 | 6.54 | 0.34 | GLM |
| RL-T-1d | 3D_41688704 | 3D | 41688704 | 4.43 | 4.46 | −0.25 | GLM |
| RL-T-1d | 3D_63239049 | 3D | 63239049 | 4.11 | 4.38 | 0.22 | GLM |
| RL-T-1d | 5B_491948869 | 5B | 491948869 | 4.01 | 3.42 | 0.34 | GLM |
| RL-T-1d | 5B_557502580 | 5B | 557502580 | 4.35 | 5.03 | −0.26 | GLM |
| RL-T-3d | 3B_322640 | 3B | 322640 | 4.06 | 5.74 | 0.33 | GLM |
| RL-T-3d | 3B_704997 | 3B | 704997 | 4.14 | 5.90 | 0.33 | GLM |
| RL-T/CK-1d | 1A_551399390 | 1A | 551399390 | 4.13 | 4.22 | −0.02 | GLM |
| RL-T/CK-1d | 4A_481510017 | 4A | 481510017 | 4.22 | 4.88 | −0.05 | MLM |
| RL-T/CK-1d | 4A_481510017 | 4A | 481510017 | 4.33 | 4.42 | −0.05 | GLM |
| RL-T/CK-1d | 5B_557502580 | 5B | 557502580 | 4.67 | 6.14 | −0.03 | MLM |
| RL-T/CK-1d | 5B_561705149 | 5B | 561705149 | 4.03 | 4.24 | −0.02 | GLM |
| RL-T/CK-1d | 5B_557502580 | 5B | 557502580 | 4.83 | 5.73 | −0.03 | GLM |
| RL-T/CK-3d | 5B_557502580 | 5B | 557502580 | 4.32 | 5.03 | −0.02 | GLM |
| RL-T/CK-3d | 5B_557502580 | 5B | 557502580 | 4.22 | 5.06 | −0.02 | MLM |
| Gene ID | Associated SNP | Chr | Position (bp) | Description | Genotype 182 (Tolerant) | Genotype 238 (Sensitive) | ||
|---|---|---|---|---|---|---|---|---|
| T1/CK | T2/CK | T1/CK | T2/CK | |||||
| TraesCS1A03G0917700 | 1A_551399390 | 1A | 551,591,836–551,593,360 | Transcription factor RAX2; regulates axillary meristem development; contains Myb-like DNA-binding domain | 4.06 | 6.04 | 3.16 | 5.56 |
| TraesCS1A03G0913800 | 1A_551399390 | 1A | 550,967,927–550,969,339 | Beta-1,3-galactosyltransferase (PvGal synthesis protein 3); involved in glycosylation and stress response | 2.10 | 4.17 | 2.05 | 3.42 |
| TraesCS3D03G0225500 | 3D_63239049 | 3D | 63,007,082–63,008,233 | Zinc finger CCCH domain-containing protein 2 (OsDOS); regulates senescence and stress responses | 2.35 | 2.25 | 2.98 | 3.71 |
| TraesCS3D03G0226000 | 3D_63239049 | 3D | 63,405,847–63,409,862 | ATP-dependent 6-phosphofructokinase 3 (ATP-PFK 3); key enzyme in glycolysis | 2.39 | 2.84 | 2.11 | 2.87 |
| TraesCS5B03G0953100 | 5B_557502580 | 5B | 558,417,547–558,422,277 | Organic cation/carnitine transporter 7 (AtOCT7); member of sugar and other transporter family | 2.21 | 1.90 | 2.52 | 2.37 |
| TraesCS5B03G0961700 | 5B_561705149 | 5B | 563,149,203–563,153,131 | Transcription factor bHLH148 (OsbHLH148); regulates stress-responsive genes | 4.51 | 8.10 | 6.57 | 8.62 |
| TraesCS3D03G0227400 | 3D_63239049 | 3D | 64,080,875–64,083,929 | Profilin; involved in cytoskeleton organization; contains Profilin domain | 0.36 | 0.34 | 0.40 | 0.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Kang, X.; Wang, J.; He, X.; Liu, W.; Xu, D.; Dai, X.; Ma, W.; Zeng, J. Genome-Wide Association Study and Transcriptome Analysis Reveal Alkaline Stress-Responsive Genes in Bread Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2025, 26, 8659. https://doi.org/10.3390/ijms26178659
Sun X, Kang X, Wang J, He X, Liu W, Xu D, Dai X, Ma W, Zeng J. Genome-Wide Association Study and Transcriptome Analysis Reveal Alkaline Stress-Responsive Genes in Bread Wheat (Triticum aestivum L.). International Journal of Molecular Sciences. 2025; 26(17):8659. https://doi.org/10.3390/ijms26178659
Chicago/Turabian StyleSun, Xuelian, Xin Kang, Jiayan Wang, Xiaoyan He, Wenxing Liu, Dengan Xu, Xuehuan Dai, Wujun Ma, and Jianbin Zeng. 2025. "Genome-Wide Association Study and Transcriptome Analysis Reveal Alkaline Stress-Responsive Genes in Bread Wheat (Triticum aestivum L.)" International Journal of Molecular Sciences 26, no. 17: 8659. https://doi.org/10.3390/ijms26178659
APA StyleSun, X., Kang, X., Wang, J., He, X., Liu, W., Xu, D., Dai, X., Ma, W., & Zeng, J. (2025). Genome-Wide Association Study and Transcriptome Analysis Reveal Alkaline Stress-Responsive Genes in Bread Wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 26(17), 8659. https://doi.org/10.3390/ijms26178659







