Integrative In Silico and Experimental Characterization of Endolysin LysPALS22: Structural Diversity, Ligand Binding Affinity, and Heterologous Expression
Abstract
1. Introduction
2. Result
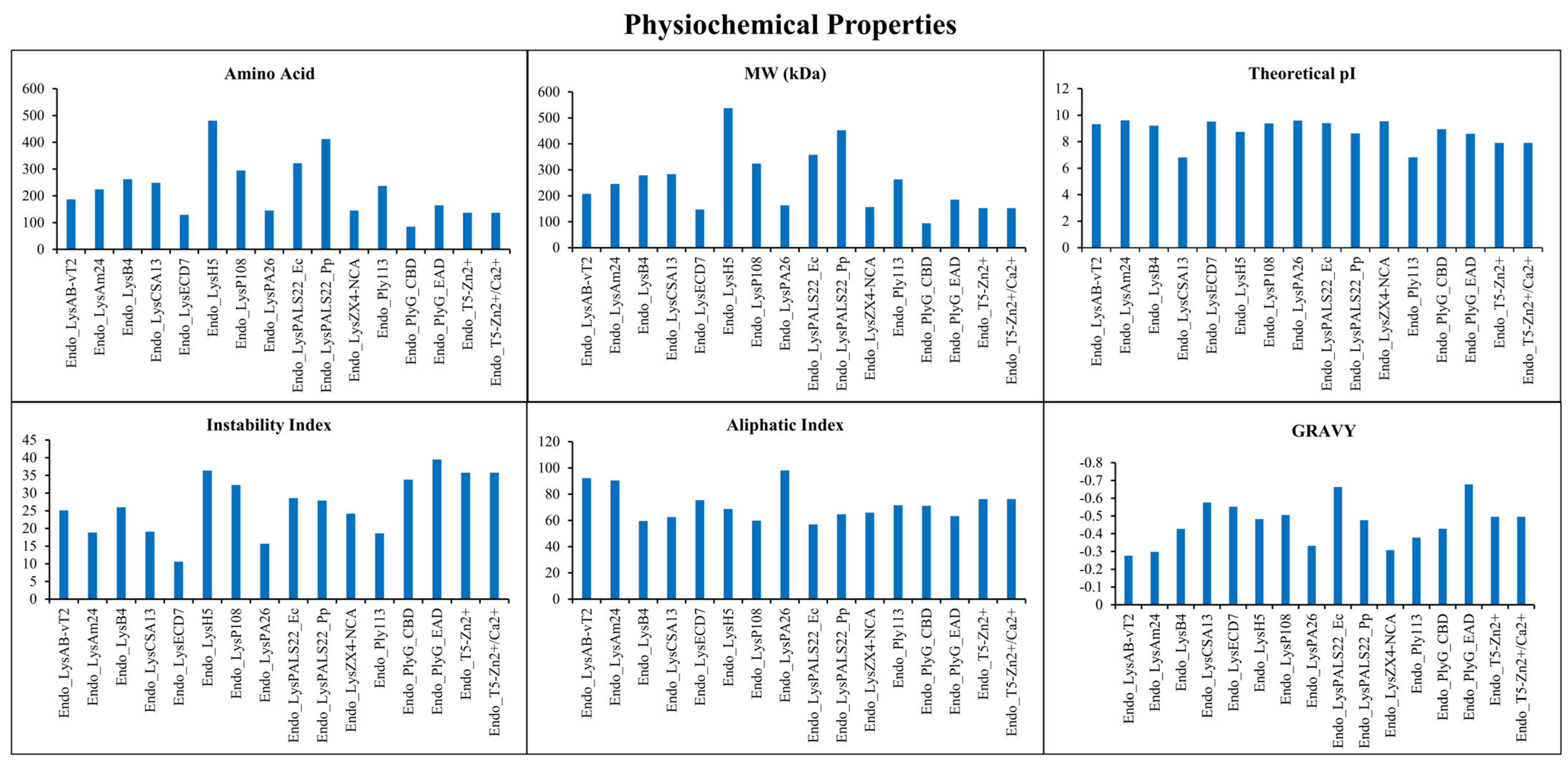
2.1. Identification and Physicochemical Properties
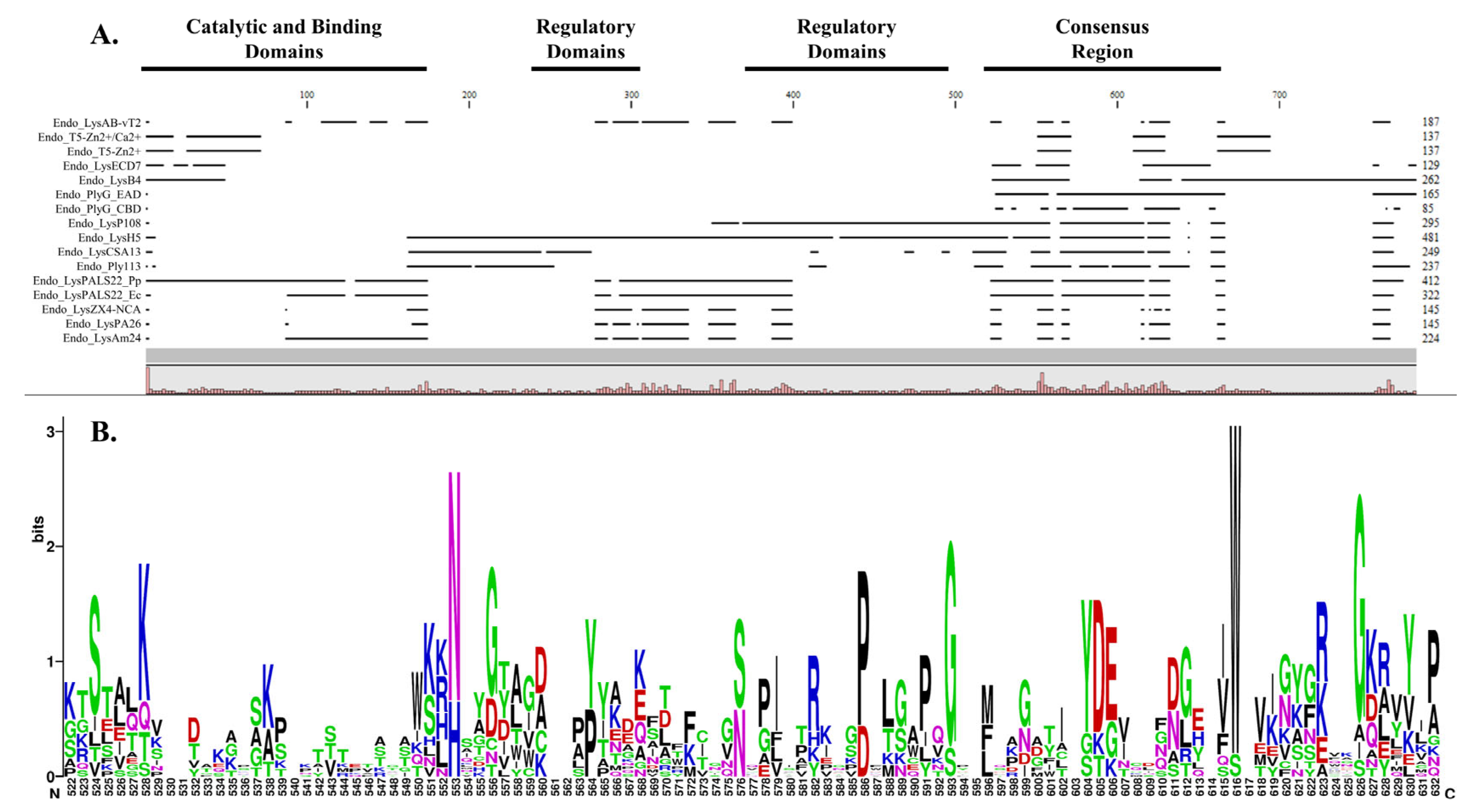
2.2. Multiple Sequence Alignment and Conserved Domains
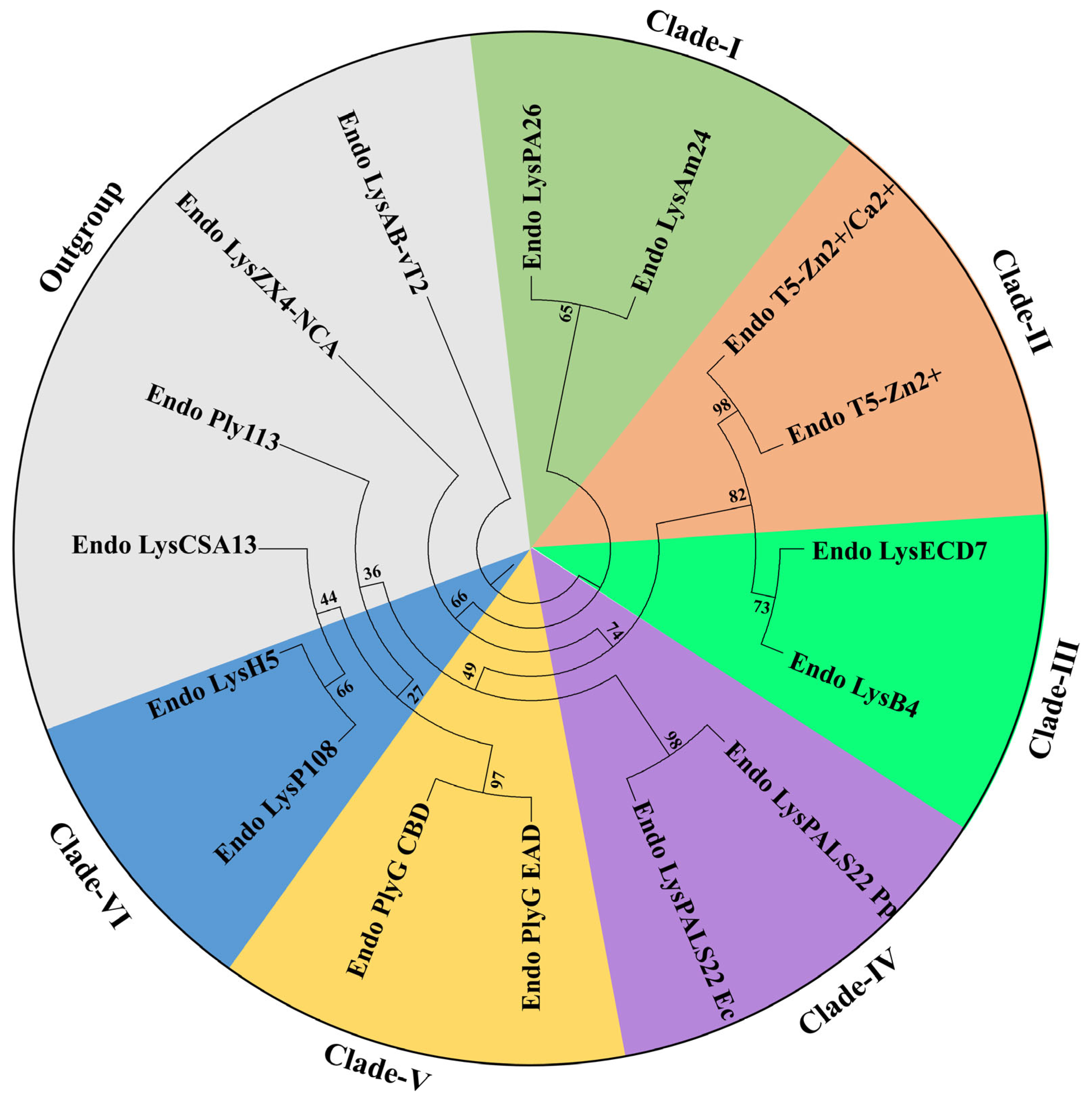
2.3. Phylogenetic Tree
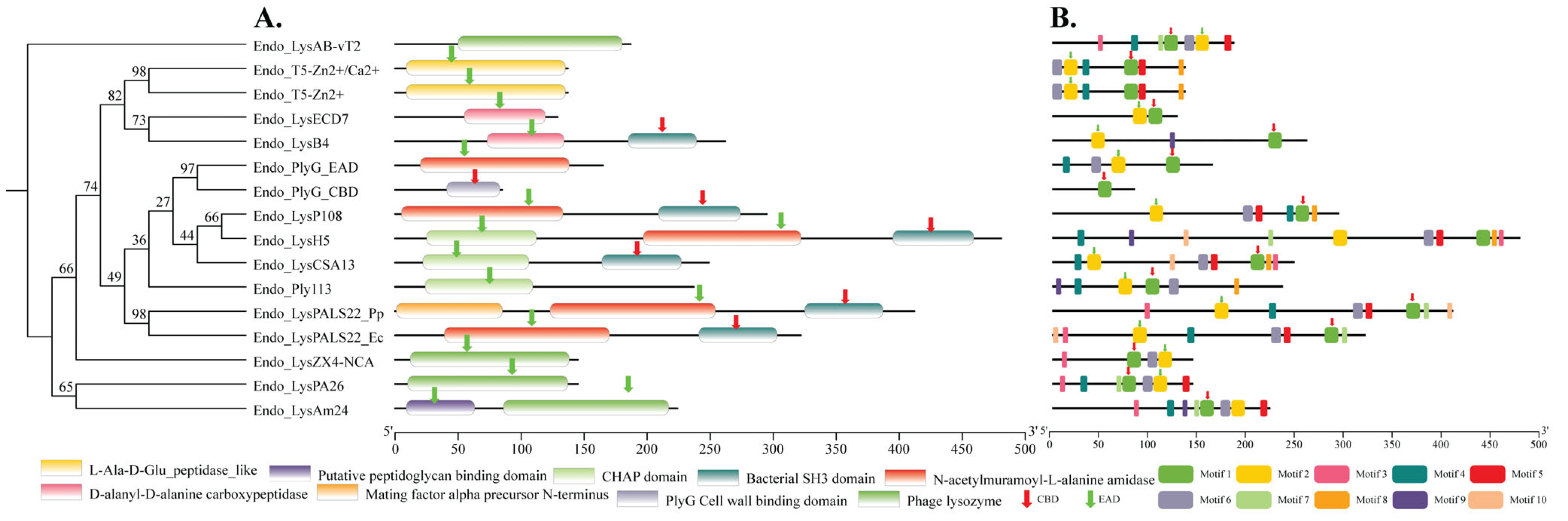
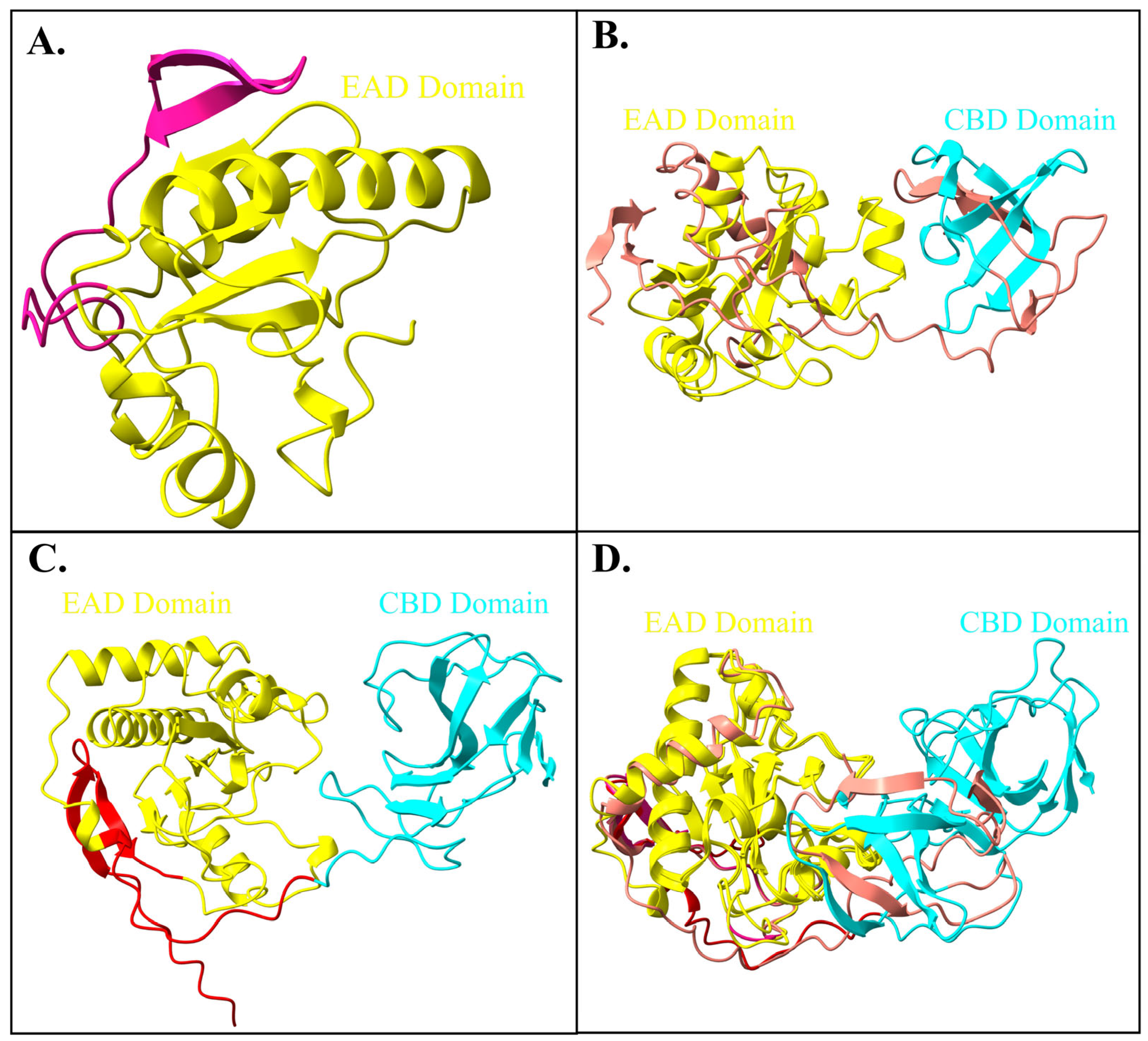
2.4. Domains and Motifs
2.5. Protein Structure Comparison
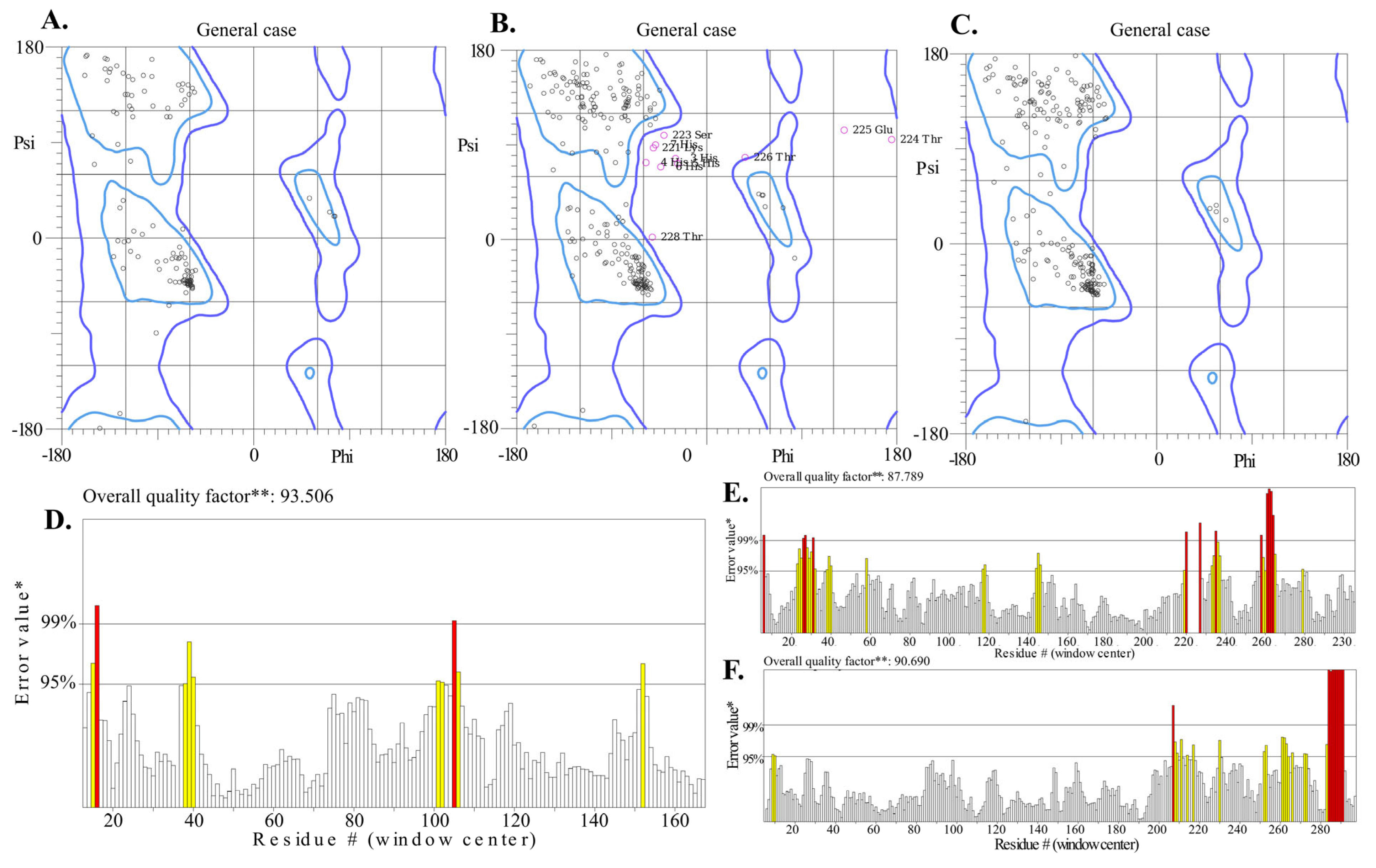
2.6. Solubility, Interaction Energy, and Charge Distribution of Endo_LysPALS22_Ec
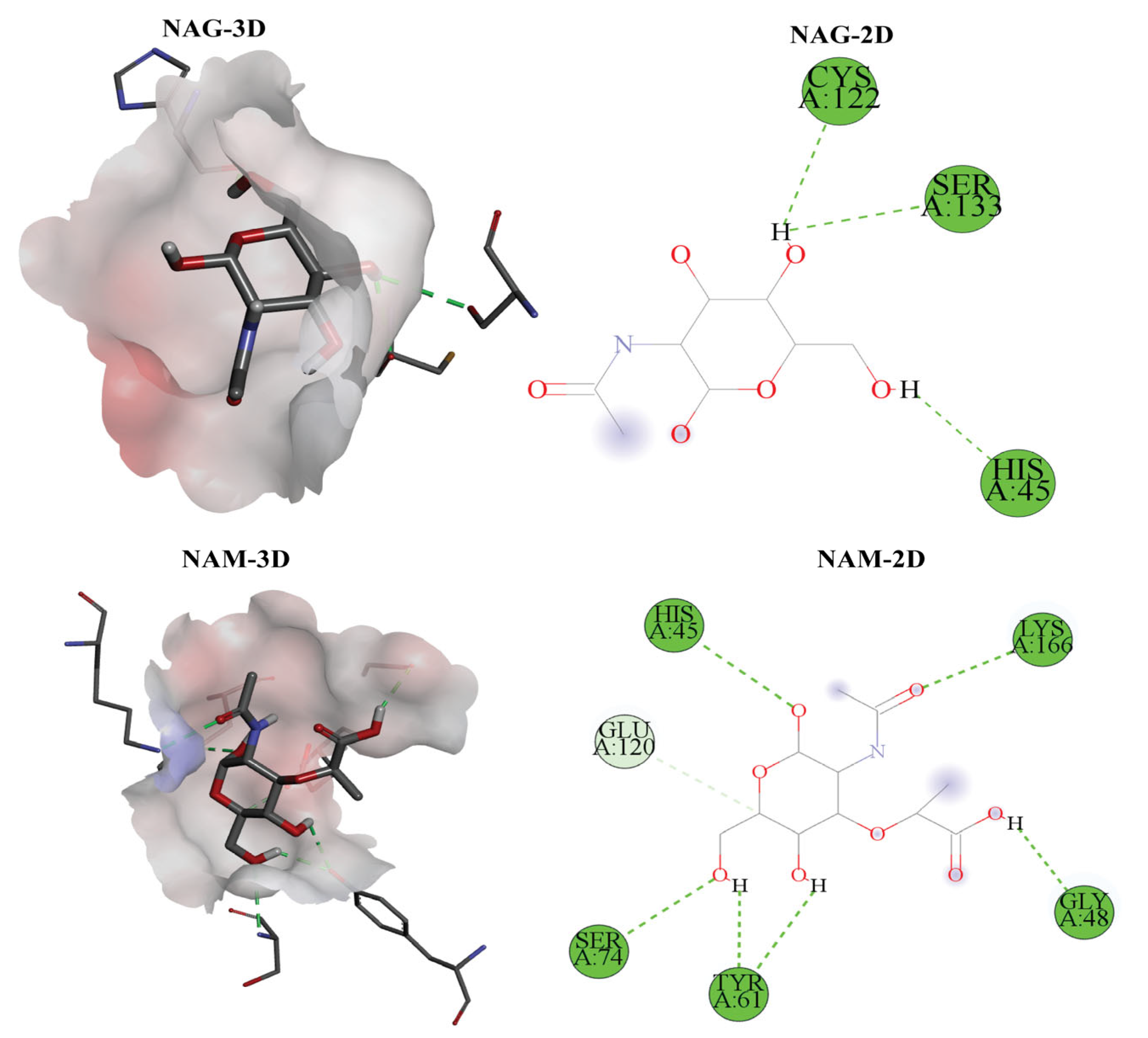
2.7. Insights into Protein–Ligand Docking
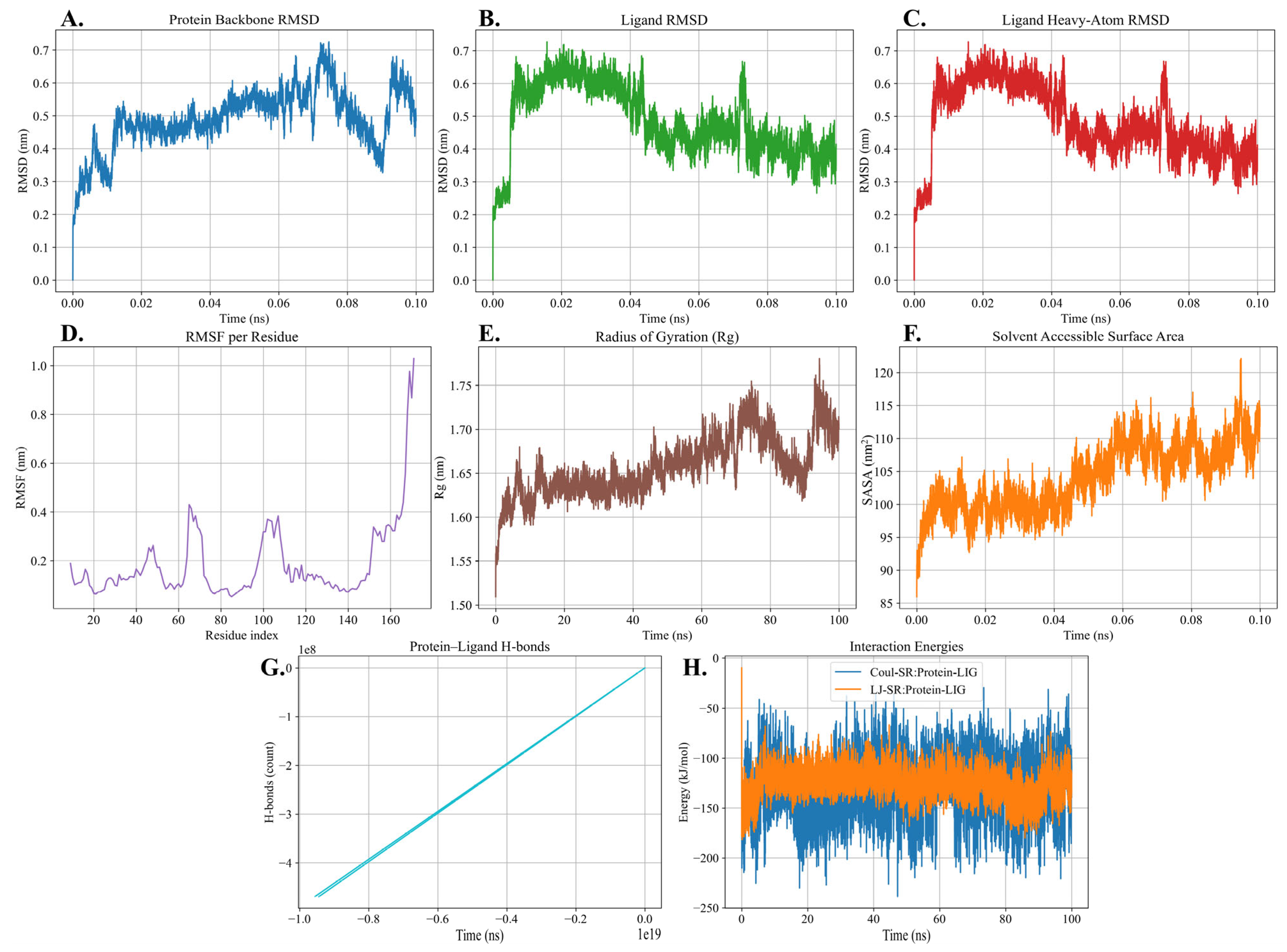
2.8. Molecular Dynamics Simulation
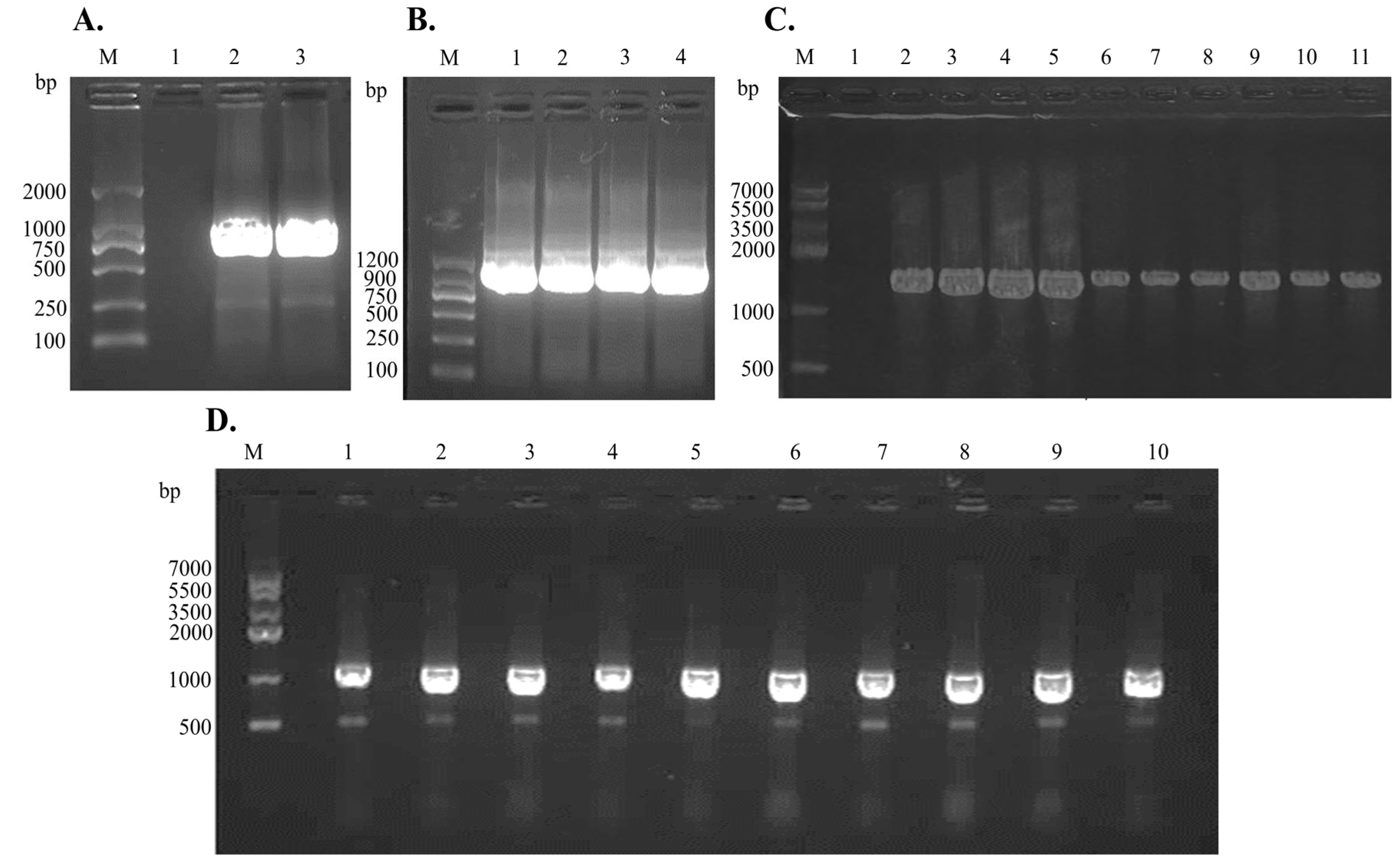
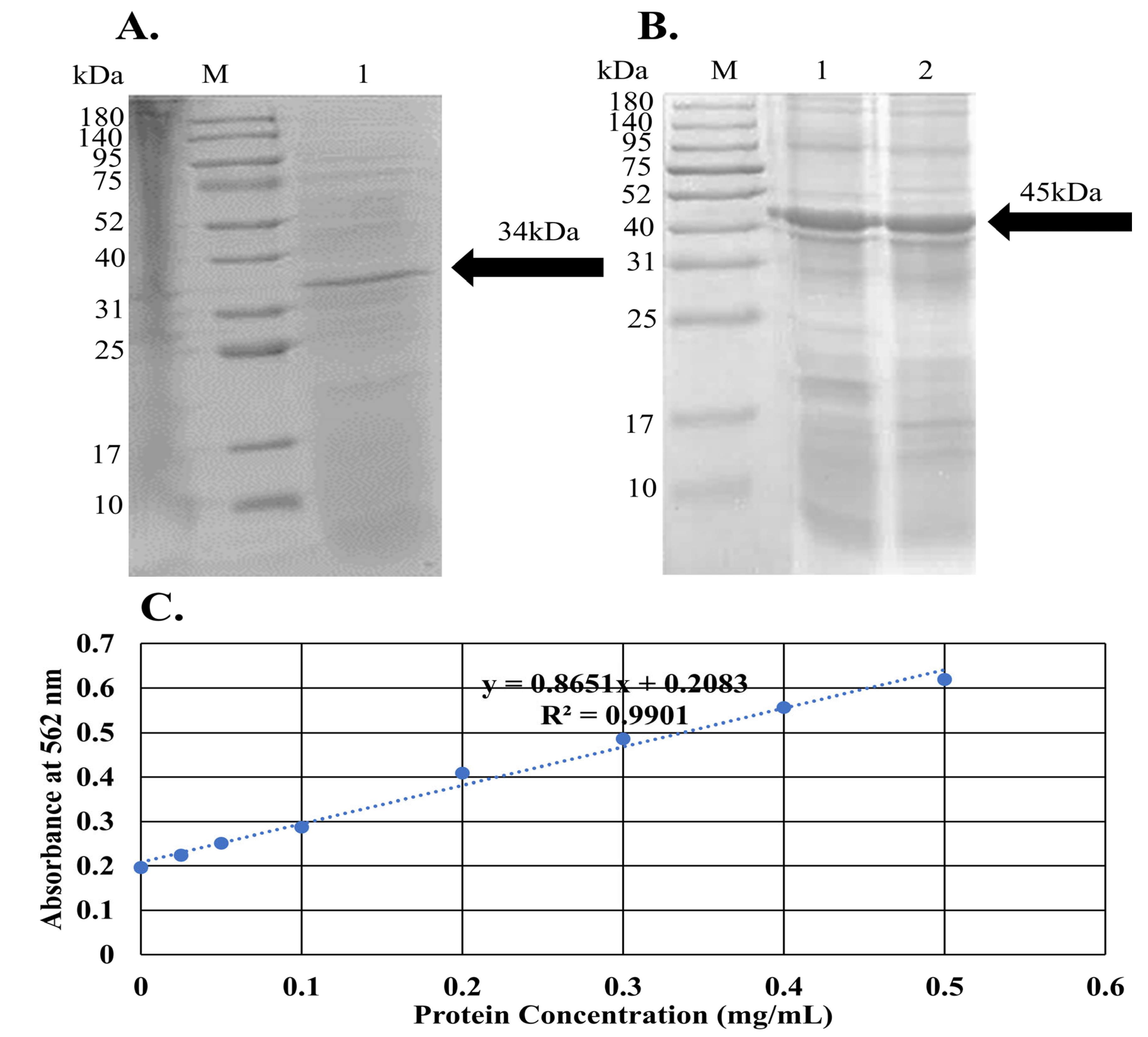
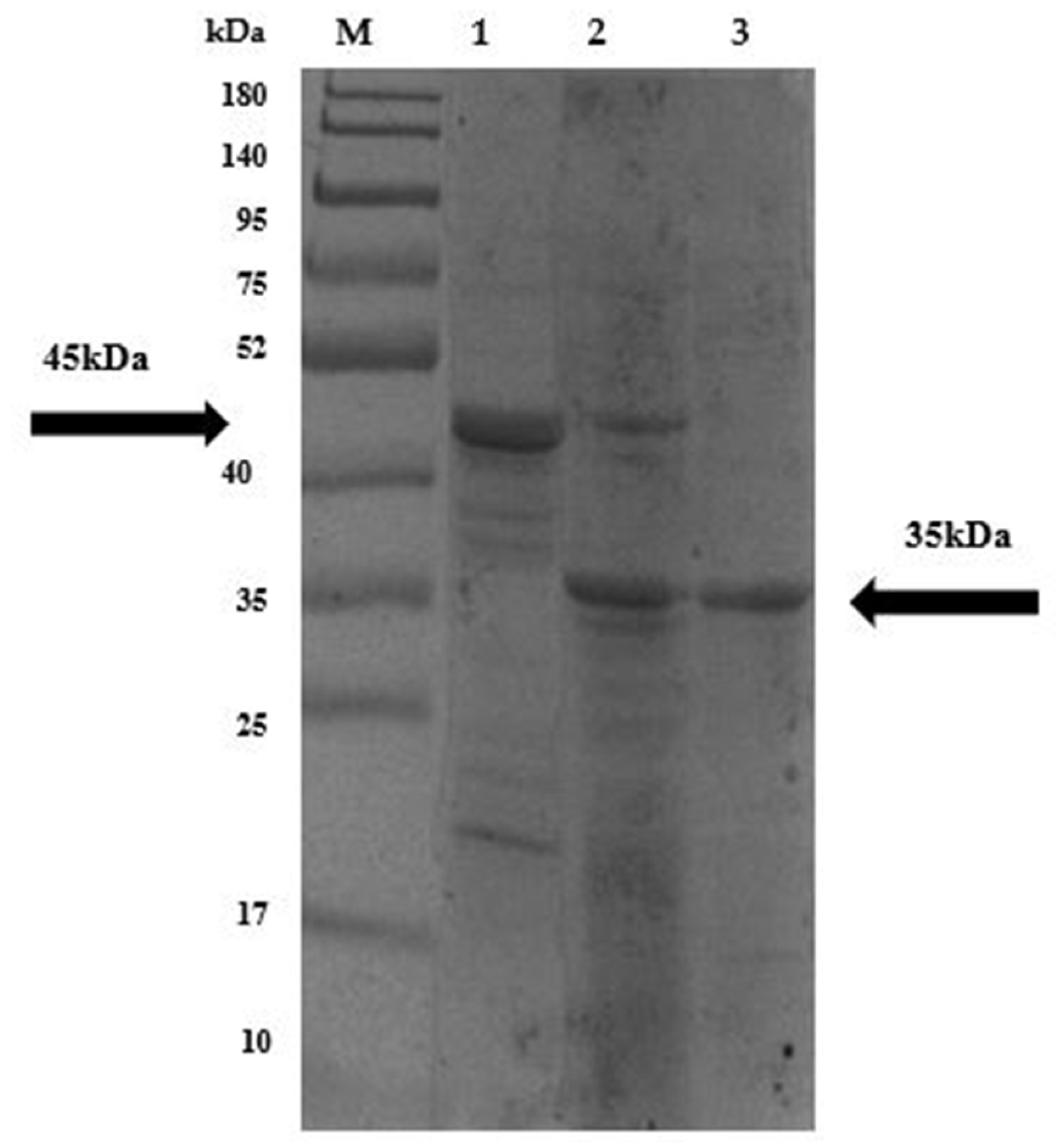
2.9. Molecular Verification, Expression, and Purification of Recombinant LysPALS22 in Heterologous Hosts
PNGase F Deglycosylation Confirms Post-Translational Modifications of the P. pastoris-Expressed LysPALS22
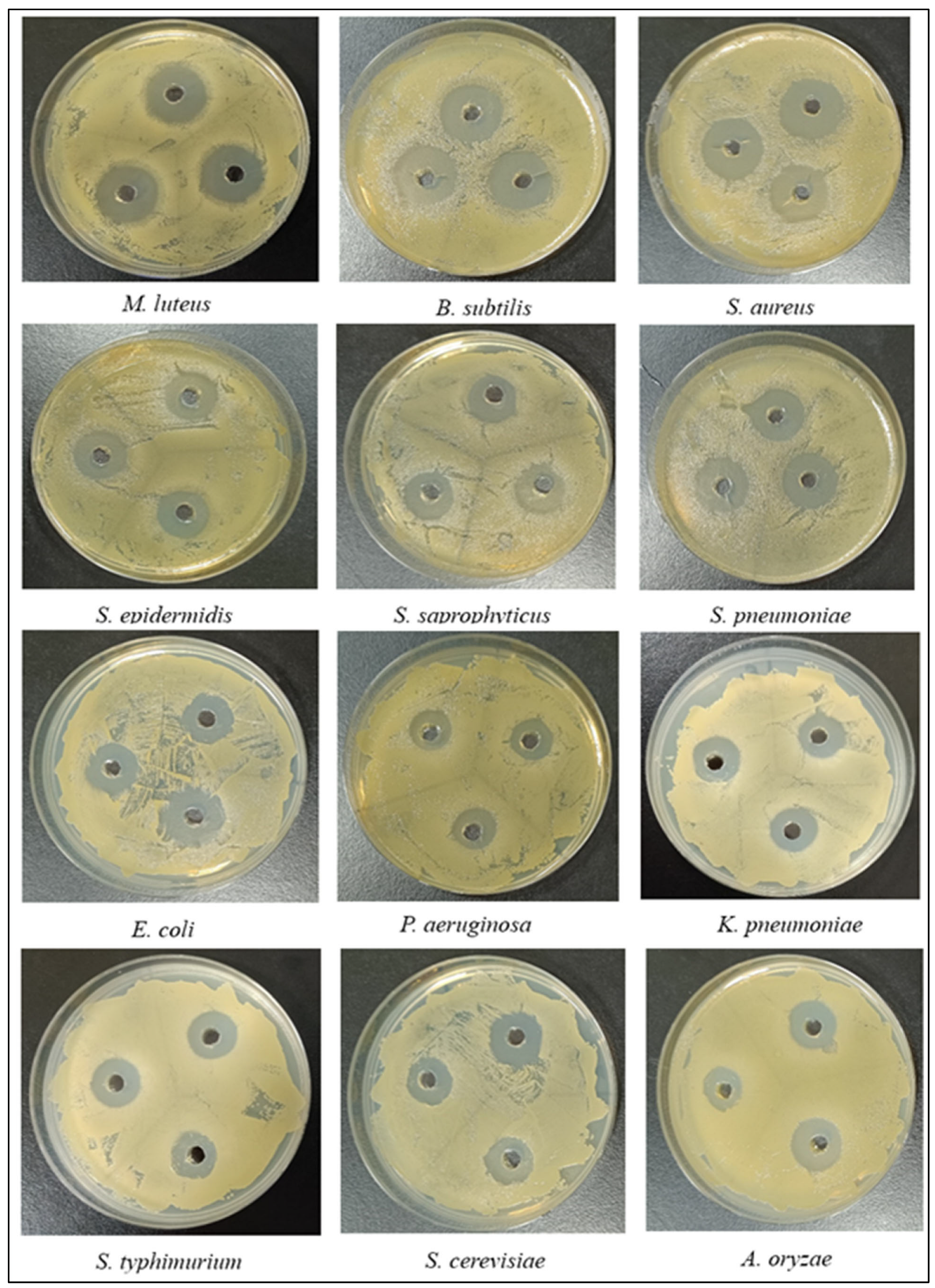
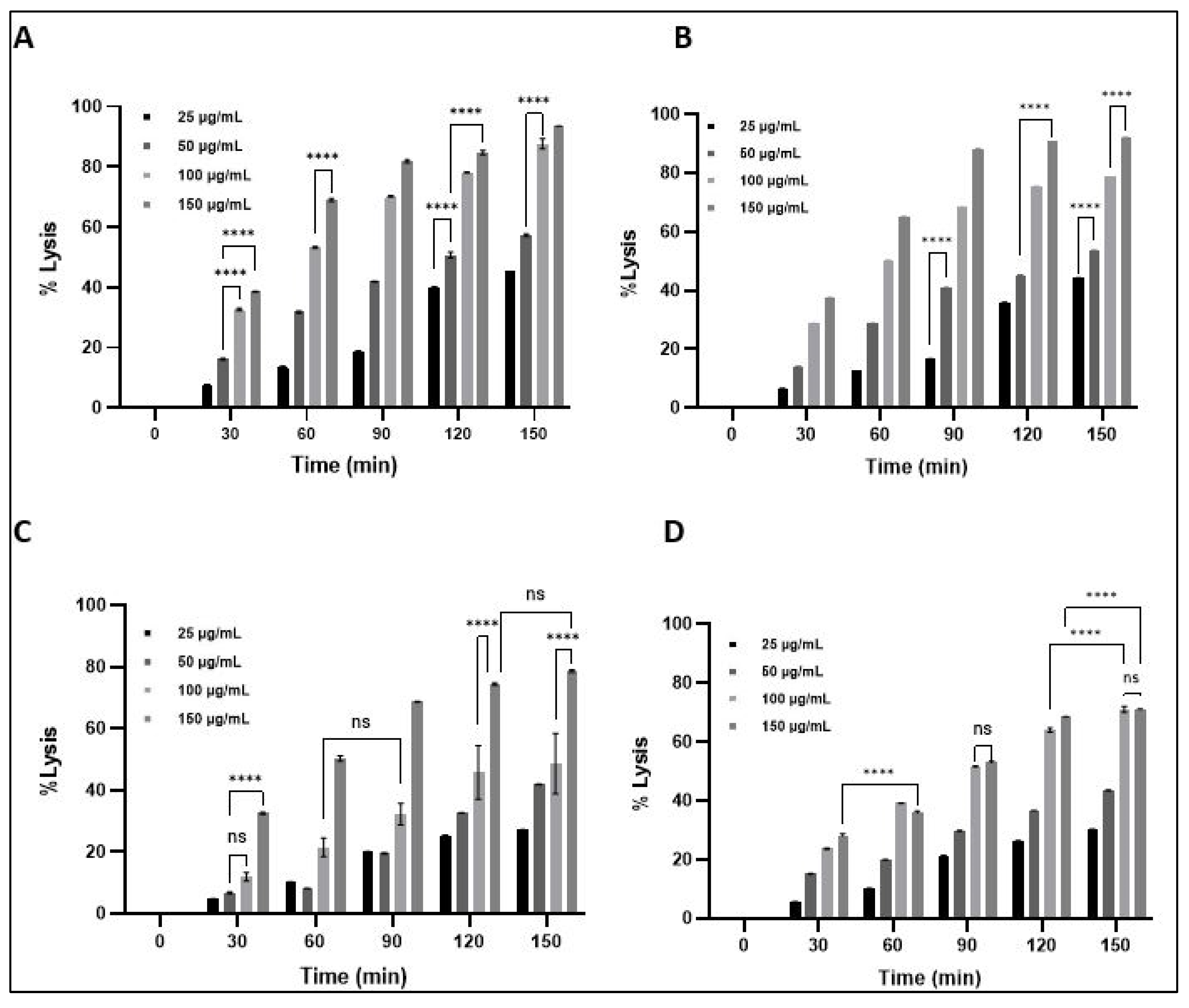
2.10. Antimicrobial Activity of LysPALS22
Turbidity Reduction and Bacterial Lysis
3. Discussion
4. Methods and Materials
4.1. Sequence Assembly and Contig Generation Using BioEdit
4.2. Protein Translation and Open Reading Frame Selection
4.3. Selection of Endolysins from Literature and Sequence Retrieval
4.4. Physicochemical Properties Analysis
4.5. Multiple Sequence Alignment and Phylogenetic Analysis of Endolysins
4.6. Domain and Motif Analyses
4.7. Protein Structure Prediction and Comparison
4.8. Protein Solubility, Interaction Energy, and Charge Analysis
4.9. Molecular Docking of Ligands with Endolysins
4.10. Molecular Dynamics Simulation Analysis
4.11. Experimental Validation
4.11.1. Bacterial Strains and Reagents
4.11.2. Cloning and Expression of the Endolysin LysPALS22 in E. coli and P. pastoris
4.11.3. Protein Purification, SDS-PAGE Analysis, and Protein Quantification
4.12. PNGase F Denaturing Assay
4.13. Antibacterial Activity Assays for LysPALS22
4.13.1. Well Diffusion Assay (ZOI)
4.13.2. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal/Fungicidal Concentration (MBC/MFC) Assays
4.13.3. Turbidity Reduction and Bacterial Lysis Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EAD | Enzymatically Active Domain |
| CBD | Cell Wall Binding Domain |
| CAP | Contig Assembly Program |
| ORF | Open Reading Frame |
| PDB | Protein Data Base |
| GRAVY | grand average of hydropathicity |
| MSA | multiple sequence alignment |
| NJ | Neighbor-Joining |
| VADAR | Volume, Area, Dihedral Angle Reporter |
| RMSD | Root Mean Square Deviation |
| SDS-PAGE | Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| BCA | Bicinchoninic acid |
References
- Golban, M.; Charostad, J.; Kazemian, H.; Heidari, H. Phage-Derived Endolysins Against Resistant Staphylococcus spp.: A Review of Features, Antibacterial Activities, and Recent Applications. Infect. Dis. Ther. 2025, 14, 13–57. [Google Scholar] [CrossRef]
- Kim, H.; Seo, J. A Novel Strategy to Identify Endolysins with Lytic Activity against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2023, 24, 5772. [Google Scholar] [CrossRef]
- Kocot, A.M.; Briers, Y.; Plotka, M. Phages and engineered lysins as an effective tool to combat Gram-negative foodborne pathogens. Compr. Rev. Food. Sci. Food. Saf. 2023, 22, 2235–2266. [Google Scholar] [CrossRef]
- Anastassopoulou, C.; Ferous, S.; Petsimeri, A.; Gioula, G.; Tsakris, A. Phage-Based Therapy in Combination with Antibiotics: A Promising Alternative against Multidrug-Resistant Gram-Negative Pathogens. Pathogens 2024, 13, 896. [Google Scholar] [CrossRef]
- Pattnaik, A.; Pati, S.; Samal, S.K. Bacteriophage as a potential biotherapeutics to combat present-day crisis of multi-drug resistant pathogens. Heliyon 2024, 10, e37489. [Google Scholar] [CrossRef]
- Aliakbarlu, J.; Manafi, L.; Mortazavi, N.; Lin, L.; Kaboudari, A. The antibacterial activity of endolysins against food-borne pathogenic bacteria in vitro and foods. Crit. Rev. Food. Sci. Nutr. 2025, 65, 1–15. [Google Scholar] [CrossRef]
- Hausbeck, M.K.; Bell, J.; Medina-Mora, C.; Podolsky, R.; Fulbright, D.W. Effect of bactericides on population sizes and spread of Clavibacter michiganensis subsp. michiganensis on tomatoes in the greenhouse and on disease development and crop yield in the field. Phytopathology 2000, 90, 38–44. [Google Scholar] [CrossRef]
- de Vries, J.; Harms, K.; Broer, I.; Kriete, G.; Mahn, A.; Düring, K.; Wackernagel, W. The bacteriolytic activity in transgenic potatoes expressing a chimeric T4 lysozyme gene and the effect of T4 lysozyme on soil-and phytopathogenic bacteria. Syst. Appl. Microbiol. 1999, 22, 280–286. [Google Scholar] [CrossRef]
- Liu, J.; Yue, J.; Wang, H.; Xie, L.; Zhao, Y.; Zhao, M.; Zhou, H. Strategies for Engineering Virus Resistance in Potato. Plants 2023, 12, 1736. [Google Scholar] [CrossRef]
- Behera, M.; De, S.; Ghorai, S.M. The Synergistic and Chimeric Mechanism of Bacteriophage Endolysins: Opportunities for Application in Biotherapeutics, Food, and Health Sectors. Probiotics Antimicrob. Proteins 2025, 17, 807–831. [Google Scholar] [CrossRef]
- Koposova, O.N.; Kazantseva, O.A.; Shadrin, A.M. Diversity of Endolysin Domain Architectures in Bacteriophages Infecting Bacilli. Biomolecules 2024, 14, 1586. [Google Scholar] [CrossRef]
- Vázquez, R.; Gutiérrez, D.; Grimon, D.; Fernández, L.; García, P.; Rodríguez, A.; Briers, Y. The New SH3b_T Domain Increases the Structural and Functional Variability Among SH3b-Like CBDs from Staphylococcal Phage Endolysins. Probiotics Antimicrob. Proteins 2024, 16, 1–14. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-encoded endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Tham, H.Y.; Chong, L.C.; Krishnan, M.; Khan, A.M.; Choi, S.B.; Tamura, T.; Yusoff, K.; Tan, G.H.; Song, A.A.L. Characterization of the host specificity of the SH3 cell wall binding domain of the staphylococcal phage 88 endolysin. Arch. Microbiol. 2025, 207, 47. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Arís, A.; Garcia-Fruitos, E.; Ferrer-Miralles, N. Design strategies for positively charged endolysins: Insights into Artilysin development. Biotechnol. Adv. 2023, 69, 108250. [Google Scholar] [CrossRef]
- Guo, L.; Stoffels, K.; Broos, J.; Kuipers, O.P. Altering Specificity and Enhancing Stability of the Antimicrobial Peptides Nisin and Rombocin through Dehydrated Amino Acid Residue Engineering. Peptides 2024, 174, 171152. [Google Scholar] [CrossRef]
- Arakelian, A.G.; Chuev, G.N.; Mamedov, T.V. Molecular Docking of Endolysins for Studying Peptidoglycan Binding Mechanism. Molecules 2024, 29, 5386. [Google Scholar] [CrossRef] [PubMed]
- Maneekul, J.; Chiaha, A.; Hughes, R.; Labry, F.; Saito, J.; Almendares, M.; Banda, B.N.; Lopez, L.; McGaskey, N.; Miranda, M.; et al. Investigating novel Streptomyces bacteriophage endolysins as potential antimicrobial agents. Microbiol. Spectr. 2025, 13, e0117024. [Google Scholar] [CrossRef]
- Sabur, A.; Khan, A.; Borphukan, B.; Razzak, A.; Salimullah, M.; Khatun, M. The Unique Capability of Endolysin to Tackle Antibiotic Resistance: Cracking the Barrier. J. Xenobiot. 2025, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Chalkha, M.; Chebbac, K.; Nour, H.; Nakkabi, A.; El Moussaoui, A.; Tüzün, B.; Bourhia, M.; Chtita, S.; Bakhouch, M.; Laaroussi, H.; et al. In vitro and in silico evaluation of the antimicrobial and antioxidant activities of spiropyrazoline oxindole congeners. Arab. J. Chem. 2024, 17, 105465. [Google Scholar] [CrossRef]
- Khan, F.M.; Rasheed, F.; Yang, Y.; Liu, B.; Zhang, R. Endolysins: A new antimicrobial agent against antimicrobial resistance. Strategies and opportunities in overcoming the challenges of endolysins against Gram-negative bacteria. Front. Pharmacol. 2024, 15, 1385261. [Google Scholar] [CrossRef]
- Palanikumar, P.; Nathan, B.; Muthusamy, K.; Natesan, S.; Sampathrajan, V. Unravelling the Antibiotic Resistance: Molecular Insights and Combating Therapies. Appl. Biochem. Biotechnol. 2025, 197, 3549–3580. [Google Scholar] [CrossRef]
- Peterková, D.; Mancoš, M.; Godány, A. Searching and in Sillico Characterization of Streptomyces Phage Endolysins and Their Catalytic Domains. J. Microbiol. Biotechnol. Food Sci. 2020, 10, 221–229. [Google Scholar] [CrossRef]
- Love, M.J.; Abeysekera, G.S.; Muscroft-Taylor, A.C.; Billington, C.; Dobson, R.C. On the catalytic mechanism of bacteriophage endolysins: Opportunities for engineering. BBA Proteins Proteomics 2020, 1868, 140302. [Google Scholar] [CrossRef]
- Linden, S.B.; Zhang, H.; Heselpoth, R.D.; Shen, Y.; Schmelcher, M.; Eichenseher, F.; Nelson, D.C. Biochemical and biophysical characterization of PlyGRCS, a bacteriophage endolysin active against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 741–752. [Google Scholar] [CrossRef]
- Balaban, C.L.; Suárez, C.A.; Boncompain, C.A.; Peressutti-Bacci, N.; Ceccarelli, E.A.; Morbidoni, H.R. Evaluation of factors influencing expression and extraction of recombinant bacteriophage endolysins in Escherichia coli. Microb. Cell Factories. 2022, 21, 40. [Google Scholar] [CrossRef]
- Hebditch, M.; Warwicker, J. Web-based display of protein surface and pH-dependent properties for assessing the developability of biotherapeutics. Sci. Rep. 2019, 9, 1969. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.A.; Aziz, D.M.; Abdullah, M.N.; Bhat, A.R.; Dongre, R.S.; Hadda, T.B.; Almalki, F.A.; Kawsar, S.M.; Rahiman, A.K.; Ahmed, S.; et al. In vitro and in vivo evaluation of the antimicrobial, antioxidant, cytotoxic, hemolytic activities and in silico POM/DFT/DNA-binding and pharmacokinetic analyses of new sulfonamide bearing thiazolidin-4-ones. J. Biomol. Struct. Dyn. 2024, 42, 3747–3763. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, K.; Gutiérrez, D.; Tamés-Caunedo, H.; Ruas-Madiedo, P.; Safaan, A.; Khairalla, A.S.; Gaber, Y.; Dishisha, T.; Briers, Y. Engineering a lysin with intrinsic antibacterial activity (LysMK34) by cecropin a fusion enhances its antibacterial properties against Acinetobacter baumannii. Appl. Environ. Microbiol. 2022, 88, e01515-21. [Google Scholar] [CrossRef] [PubMed]
- Kawsar, S.M.; Hosen, M.A.; El Bakri, Y.; Ahmad, S.; Affi, S.T.; Goumri-Said, S. In silico approach for potential antimicrobial agents through antiviral, molecular docking, molecular dynamics, pharmacokinetic and bioactivity predictions of galactopyranoside derivatives. Arab J. Basic Appl. Sci. 2022, 29, 99–112. [Google Scholar] [CrossRef]
- Alamri, M.A.; Alawam, A.S.; Alshahrani, M.M.; Kawsar, S.M.; Prinsa and Saha, S. Establishing the Role of Iridoids as Potential Kirsten Rat Sarcoma Viral Oncogene Homolog G12C Inhibitors Using Molecular Docking; Molecular Docking Simulation; Molecular Mechanics Poisson–Boltzmann Surface Area; Frontier Molecular Orbital Theory; Molecular Electrostatic Potential, and Absorption, Distribution, Metabolism, Excretion, and Toxicity Analysis. Molecules 2023, 28, 5050. [Google Scholar] [PubMed]
- Shen, K.S.; Shu, M.; Tang, M.X.; Yang, W.Y.; Wang, S.C.; Zhong, C.; Wu, G.P. Molecular cloning, expression and characterization of a bacteriophage JN01 endolysin and its antibacterial activity against E. coli O157: H7. LWT 2022, 165, 113705. [Google Scholar] [CrossRef]
- Zhang, S.; Chang, Y.; Zhang, Q.; Yuan, Y.; Qi, Q.; Lu, X. Characterization of Salmonella endolysin XFII produced by recombinant Escherichia coli and its application combined with chitosan in lysing Gram-negative bacteria. Microb. Cell Fact. 2022, 21, 171. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Sitthisak, S.; Manrueang, S.; Khongfak, S.; Leungtongkam, U.; Thummeepak, R.; Thanwisai, A.; Burton, N.; Dhanoa, G.K.; Tsapras, P.; Sagona, A.P. Antibacterial activity of vB_AbaM_PhT2 phage hydrophobic amino acid fusion endolysin, combined with colistin against Acinetobacter baumannii. Sci. Rep. 2023, 13, 7470. [Google Scholar] [CrossRef]
- Antonova, N.P.; Vasina, D.V.; Lendel, A.M.; Usachev, E.V.; Makarov, V.V.; Gintsburg, A.L.; Tkachuk, A.P.; Gushchin, V.A. Broad bactericidal activity of the Myoviridae bacteriophage lysins LysAm24, LysECD7, and LysSi3 against Gram-negative ESKAPE pathogens. Viruses 2019, 11, 284. [Google Scholar] [CrossRef]
- Antonova, N.P.; Vasina, D.V.; Rubalsky, E.O.; Fursov, M.V.; Savinova, A.S.; Grigoriev, I.V.; Usachev, E.V.; Shevlyagina, N.V.; Zhukhovitsky, V.G.; Balabanyan, V.U.; et al. Modulation of endolysin LysECD7 bactericidal activity by different peptide tag fusion. Biomolecules 2020, 10, 440. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Y.; Wang, J.; Zhao, Y.; Zhong, Q.; Li, G.; Fu, Z.; Lu, S. Phage endolysin LysP108 showed promising antibacterial potential against methicillin-resistant Staphylococcus aureus. Front. Cell. Infect. 2021, 11, 668430. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shen, M.; Ma, W.; Zhou, X.; Shen, J. LysZX4-NCA, a new endolysin with broad-spectrum antibacterial activity for topical treatment. Virus Res. 2024, 340, 199296. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, S.; Lu, X.; Xu, Q.; Zhu, Y.; Yu, S.; Zhang, W.; Liu, S.; Xie, F. Bacteriophage endolysin Ply113 as a potent antibacterial agent against polymicrobial biofilms formed by enterococci and Staphylococcus aureus. Front. Microbiol. 2023, 14, 1304932. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 49, D10–D17. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- QCG (QIAGEN CLC Genomics). QIAGEN Digital Insights. Available online: https://digitalinsights.qiagen.com/ (accessed on 25 June 2025.).
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Waterhouse, A.M.; Bienert, S.; Studer, G.; Alexander, L.T.; Tauriello, G.; Schwede, T.; Pereira, J. The SWISS-Model Repository of 3D Protein Structures and Models. In Open Access Databases and Datasets for Drug Discovery; Wiley: New York, NY, USA, 2024; pp. 175–199. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.; Levy Karin, E.; Kim, H.; Moriwaki, Y.; Ovchinnikov, S.; Steinegger, M.; Mirdita, M. Easy and accurate protein structure prediction using ColabFold. Nat. Protoc. 2024, 20, 620–642. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Ying, B.W.; Saito, K.; Jin, W.; Takada, S.; Ueda, T.; Taguchi, H. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire Escherichia coli proteome. Proc. Natl. Acad. Sci. USA 2009, 106, 4201–4206. [Google Scholar] [CrossRef] [PubMed]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology: Methods and Protocols; Springer: Cham, Switzerland, 2014; pp. 243–250. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
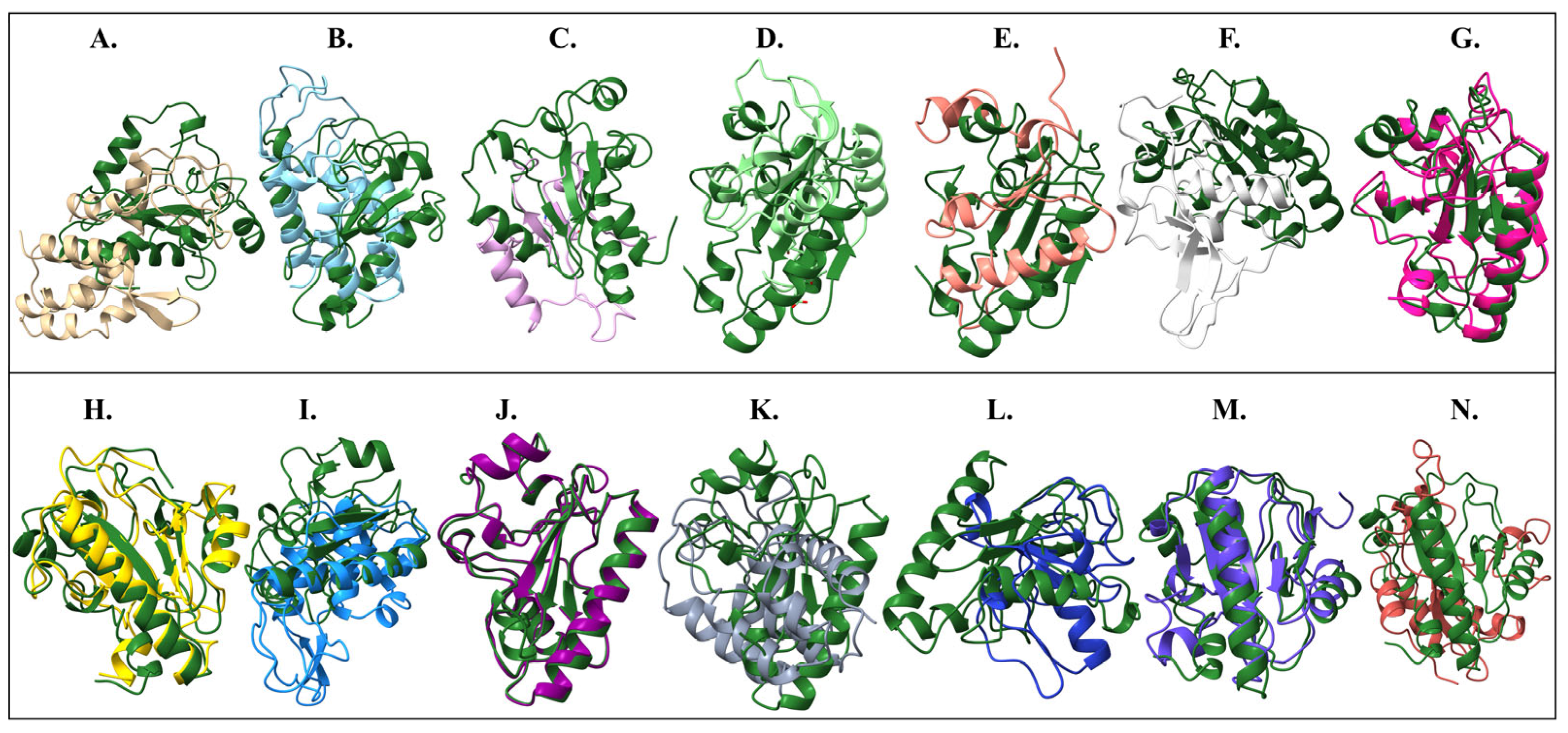
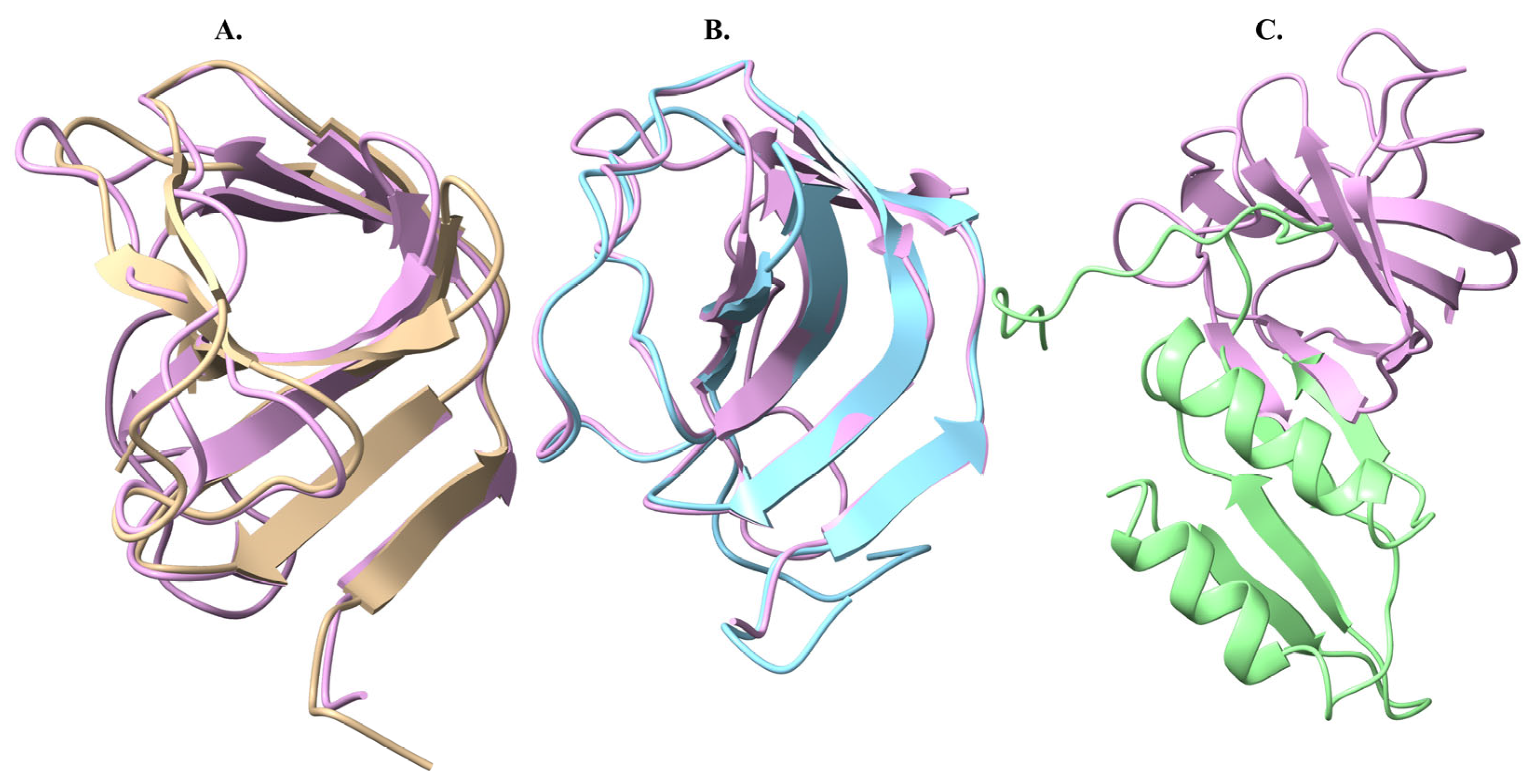

| Reference | Target Protein | Chain | Sequence Alignment Score | Pruned Atom Pairs | RMSD Pruned (Å) | RMSD All Pairs (Å) |
|---|---|---|---|---|---|---|
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysAB-vT2_EAD | D | 23.2 | 4 | 1.112 | 16.823 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysAm24_EAD | A | 20.9 | 6 | 1.549 | 20.871 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysCSA13_EAD | A | 31.7 | 4 | 0.677 | 15.02 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysECD7_EAD | A | 34.6 | 8 | 1.215 | 11.834 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysH5_EAD1 | A | 14.3 | 4 | 0.731 | 14.053 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysH5_EAD2 | A | 177.7 | 69 | 0.973 | 4.165 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysPA26_EAD | A | 24.4 | 23 | 0.965 | 20.343 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysPALS22_Pp_EAD | A | 748.5 | 138 | 0.509 | 0.63 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysZX4-NCA_EAD | A | 24.9 | 7 | 1.473 | 16.224 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_Ply113_EAD | C | 14.6 | 5 | 0.987 | 12.155 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_T5-Zn2+_EAD | A | 32.8 | 5 | 1.188 | 14.828 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysB4_EAD | A | 17.6 | 7 | 0.983 | 14.942 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_LysP108_EAD | A | 149.8 | 65 | 1.121 | 4.796 |
| Endo_PALS22_Ec_AlphaFold_EAD | Endo_PlyG_EAD_EAD | A | 227.5 | 97 | 1.116 | 2.746 |
| Microorganism | CIP | Amp | Lys-PALS22 | MIC | MBC/ MFC | AMI (CIP) | PAI (CIP) | AMI (Amp) | PAI (Amp) |
|---|---|---|---|---|---|---|---|---|---|
| ZOI (mm) | ZOI (mm) | ZOI (mm) | (μg/mL) | (μg/mL) | |||||
| M. luteus ATCC 4698 | 20 | 21 | 22 | 30 | 30 | 1.1 | 110 | 1.04 | 104 |
| B. subtilis ATCC 6051 | 18 | 20 | 19 | 44 | 44 | 1.05 | 105 | 0.95 | 95 |
| S. aureus ATCC 25923 | 19 | 21 | 21 | 50 | 50 | 1.1 | 110 | 1.0 | 100 |
| S. epidermidis ATCC 12228 | 18.5 | 22 | 19 | 55 | 55 | 1.03 | 103 | 0.86 | 86 |
| S. saprophyticus ATCC 15305 | 19 | 22.5 | 18 | 63 | 63 | 0.95 | 95 | 0.80 | 80 |
| S. pneumoniae ATCC 6303 | 17.5 | 22 | 17 | 69 | 69 | 0.97 | 97 | 0.77 | 77 |
| E. coli ATCC 25922 | 17 | 19 | 16 | 84 | 84 | 0.94 | 94 | 0.84 | 84 |
| P. aeruginosa ATCC 9027 | 16.5 | 18 | 15 | 101 | 101 | 0.91 | 91 | 0.83 | 83 |
| K. pneumoniae ATCC 13883 | 16 | 17 | 14 | 107 | 107 | 0.88 | 88 | 0.82 | 82 |
| S. typhimurium ATCC 14028 | 14 | 15 | 13 | 111 | 111 | 0.93 | 93 | 0.86 | 86 |
| S. cerevisiae | 18 | 19 | 15 | 140 | 140 | 0.83 | 83 | 0.78 | 78 |
| A. oryzae | 15 | 16 | 14 | 105 | 105 | 0.93 | 93 | 0.87 | 87 |
| Protein Name | GB* Accession | Bacteriophage Origin |
|---|---|---|
| Endo_LysAB-vT2 | QHJ75684.1 | Acinetobacter phage vB_AbaM_PhT2 |
| Endo_LysAm24 | APD20282.1 | Acinetobacter phage AM24 |
| Endo_LysB4 | AFF27501.1 | Bacillus phage B4 |
| Endo_LysCSA13 | AWD93110.1 | Staphylococcus phage CSA13 |
| Endo_LysECD7 | ASJ80195.1 | Escherichia phage ECD7 |
| Endo_LysH5 | ACE77796.1 | Staphylococcus phage phiH5 |
| Endo_LysP108 | AIK69635.1 | Staphylococcus phage P108 |
| Endo_LysPA26 | ARB16052.1 | Bacteriophage sp. isolate |
| Endo_LysZX4-NCA | QTH79953.1 | Klebsiella phage vB_KpnS_ZX4 |
| Endo_Ply113 | QVW54600.1 | Enterococcus phage 113 |
| Endo_PlyG_CBD | PDB: 2L48_A | Bacillus phage Gamma |
| Endo_PlyG_EAD | PDB: 2L47_A | Bacillus phage Gamma |
| Endo_T5-Zn2+ | AAS19387.1 (PDB: 2MXZ) | Enterobacteria phage T5 |
| Endo_T5-Zn2+/Ca2+ | AAX11973.1 (PDB: 8P3A) | Enterobacteria phage T5 |
| Microorganism | ATCC No. | Resistance/Susceptibility Pattern |
|---|---|---|
| Micrococcus luteus | 4698 | Antibiotic-susceptible reference strain |
| Bacillus subtilis | 6051 | Antibiotic-susceptible reference strain |
| Staphylococcus aureus | 25923 | CLSI quality control strain; antibiotic-susceptible |
| Staphylococcus epidermidis | 12228 | Antibiotic-susceptible laboratory strain |
| Staphylococcus saprophyticus | 15305 | Antibiotic-susceptible reference strain |
| Streptococcus pneumoniae | 6303 | Antibiotic-susceptible reference strain |
| Escherichia coli | 25922 | CLSI quality control strain; antibiotic-susceptible |
| Pseudomonas aeruginosa | 9027 | Non-multidrug-resistant reference strain |
| Klebsiella pneumoniae | 13883 | Antibiotic-susceptible reference strain |
| Salmonella Typhimurium | 14028 | Antibiotic-susceptible reference strain |
| Saccharomyces cerevisiae | – | Laboratory yeast strain; not antibiotic-resistant |
| Aspergillus oryzae | – | Laboratory mold strain; not antibiotic-resistant |
| Primer | Sequences | Restriction Enzymes | Plasmids |
|---|---|---|---|
| Sense-Eco | CATGCCATGGATGCATCATCATCATCATCACGCGAAAAAAC | NcoI | pET28a—LysPALS22 |
| Antisense-Eco | GGCCTCGAGTTAAGAGAAGGTACCCCA | XhoI | |
| Primer1-F | CCGGAATTCGCTAAGAAGCATATTGG | EcoRI | pPICZαA—LysPALS22 |
| Primer1-R | ATTTGCGGCCGCTTAATGATGATGATGATGATGGGAGAAAGTACCCC | NotI |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, N.; Nawaz, S.; Hussain, A.; Anayat, M.; Wen, S.; Wang, F. Integrative In Silico and Experimental Characterization of Endolysin LysPALS22: Structural Diversity, Ligand Binding Affinity, and Heterologous Expression. Int. J. Mol. Sci. 2025, 26, 8579. https://doi.org/10.3390/ijms26178579
Nawaz N, Nawaz S, Hussain A, Anayat M, Wen S, Wang F. Integrative In Silico and Experimental Characterization of Endolysin LysPALS22: Structural Diversity, Ligand Binding Affinity, and Heterologous Expression. International Journal of Molecular Sciences. 2025; 26(17):8579. https://doi.org/10.3390/ijms26178579
Chicago/Turabian StyleNawaz, Nida, Shiza Nawaz, Athar Hussain, Maryam Anayat, Sai Wen, and Fenghuan Wang. 2025. "Integrative In Silico and Experimental Characterization of Endolysin LysPALS22: Structural Diversity, Ligand Binding Affinity, and Heterologous Expression" International Journal of Molecular Sciences 26, no. 17: 8579. https://doi.org/10.3390/ijms26178579
APA StyleNawaz, N., Nawaz, S., Hussain, A., Anayat, M., Wen, S., & Wang, F. (2025). Integrative In Silico and Experimental Characterization of Endolysin LysPALS22: Structural Diversity, Ligand Binding Affinity, and Heterologous Expression. International Journal of Molecular Sciences, 26(17), 8579. https://doi.org/10.3390/ijms26178579







