Structure–Function Analysis of Mycobacterium tuberculosis Drug Target Cytochrome P450 125 (CYP125) Enzyme Family
Abstract
1. Introduction
2. Results and Discussion
2.1. CYP125A Members’ Active Site Amino Acids Are Highly Dynamic
| P450 Name | Species Name | PDB Code | Ligand(s) | Resolution (Å) | Surface Area (Å2) | Volume (Å3) | Conformation | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| CYP125A1 | Mycobacterium tuberculosis H37Rv | 2X5L | HEM | 1.48 | 1117.598 | 680.452 | Open | [38] |
| CYP125A1 | M. tuberculosis H37Rv | 2XN8 | HEM | 1.64 | 972.112 | 588.815 | Open | [38] |
| CYP125A1 | M. tuberculosis H37Rv | 3IVY | HEM | 1.35 | 1155.315 | 559.857 | Open | [20] |
| CYP125A1 | M. tuberculosis H37Rv | 3IW0 | HEM | 1.70 | 1148.149 | 632.7 | Open | [20] |
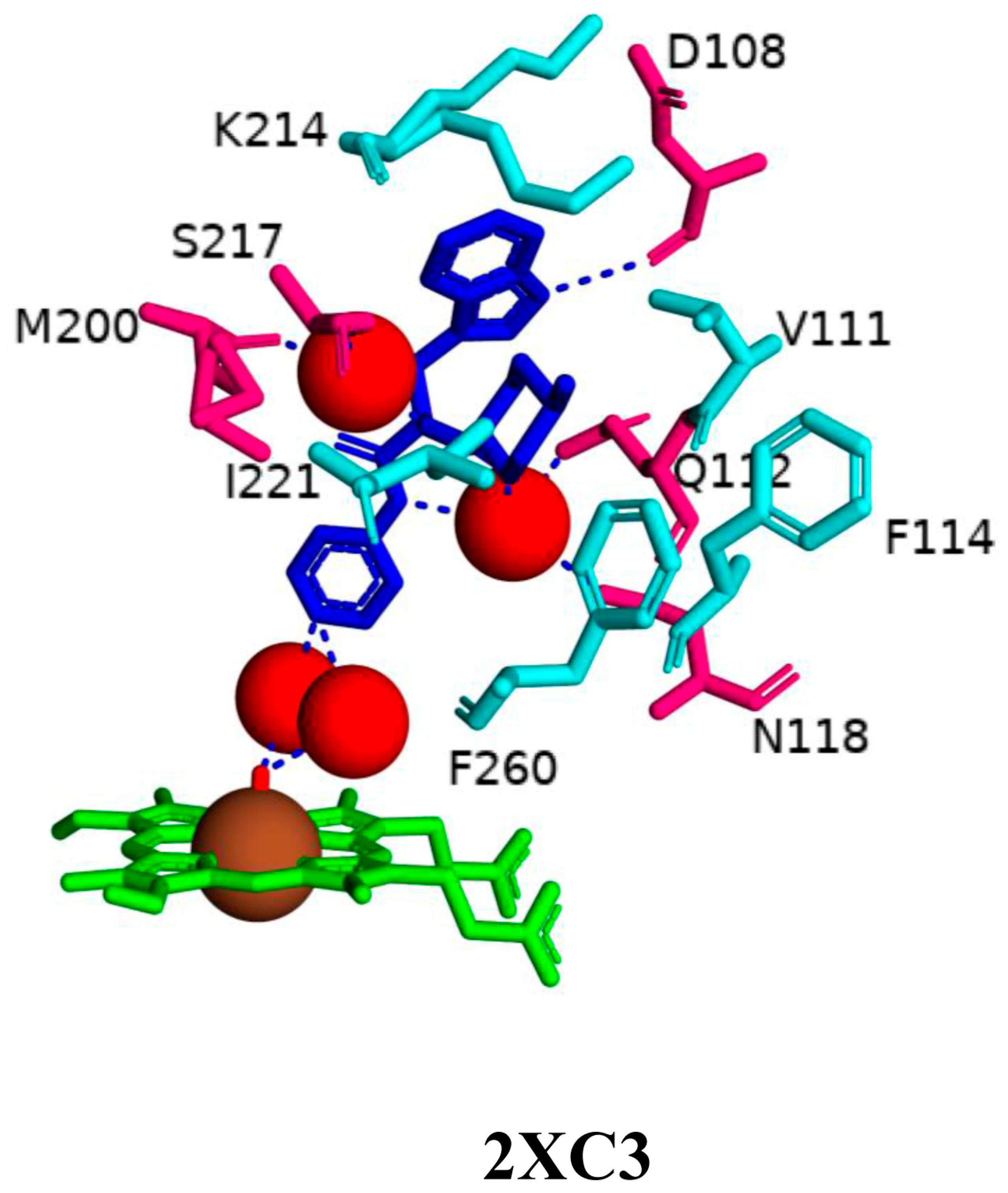
| CYP125A1 | M. tuberculosis H37Rv | 2XC3 | RT8 | 1.50 | 1098.993 | 616.083 | Closed | [38] |
| CYP125A1 | M. tuberculosis H37Rv | 7QKE | E1V | 2.30 | 938.417 | 487.117 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7QNN | E93 | 2.47 | 954.942 | 542.408 | Closed | [34] |
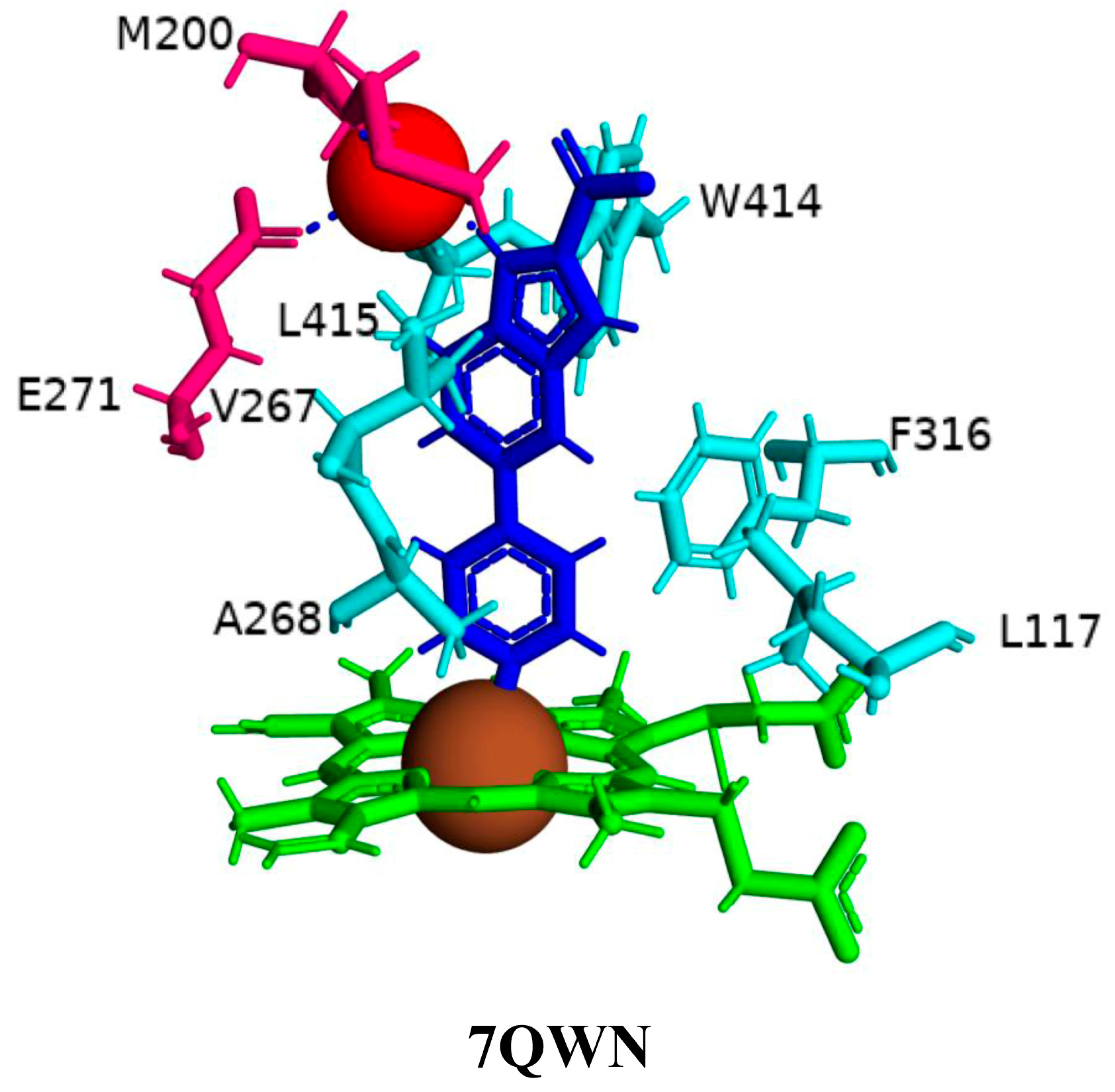
| CYP125A1 | M. tuberculosis H37Rv | 7QWN | DQE | 1.93 | 1034.257 | 676.798 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7R1I | HIH | 2.24 | 939.011 | 506.177 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7R3U | 2QC | 1.86 | 1245.124 | 604.994 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7YXF | I6Y | 1.85 | 1136.627 | 696.791 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7ZQR | JK9 | 1.79 | 1498.941 | 859.045 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7ZLZ | JFC | 1.89 | 994.537 | 517.368 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7ZSU | JV3 | 2.20 | 910.116 | 504.892 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7ZT0 | JUR | 1.99 | 1171.918 | 691.505 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 7ZXD | KB9 | 2.09 | 1013.23 | 549.261 | Closed | [34] |
| CYP125A1 | M. tuberculosis H37Rv | 2X5W | K2B | 1.58 | 892.144 | 507.427 | Closed | [24] |
| CYP125A1 | M. tuberculosis H37Rv | 3IW1 | ASD | 2.00 | 1035.662 | 491.798 | Closed | [20] |
| CYP125A1 | M. tuberculosis H37Rv | 3IW2 | EKO | 2.19 | 1064.239 | 540.218 | Closed | [20] |
| CYP125A4 | M. smegmatis MC2 155 | 5DQN | HEM | 2.26 | 1152.161 | 785.506 | Open | [25] |
| CYP125A3 | M. smegmatis MC2 155 | 4APY | HEM | 2.00 | 1267.39 | 763.058 | Open | [26] |
| P450 Name | Change in Average Surface Area (Å2) * | Change in Average Volume (Å3) * | RMSD (Å) | Reference |
|---|---|---|---|---|
| CYP107FH5 | 276 # | 494 # | 3.0 | [32] |
| CYP121A1 | 37 | 8 | 0.2 | [31] |
| CYP102A1 | 179 # | 23 # | 4.4 | [39] |
| CYP109E1 | 151 # | 545 | 2.9 | [40] |
| CYP125A | 68 # | 82 # | 0.6 and 0.7 * | Current work |
2.2. The Shape of the Active Site Cavity Determines the Substrate Specificity
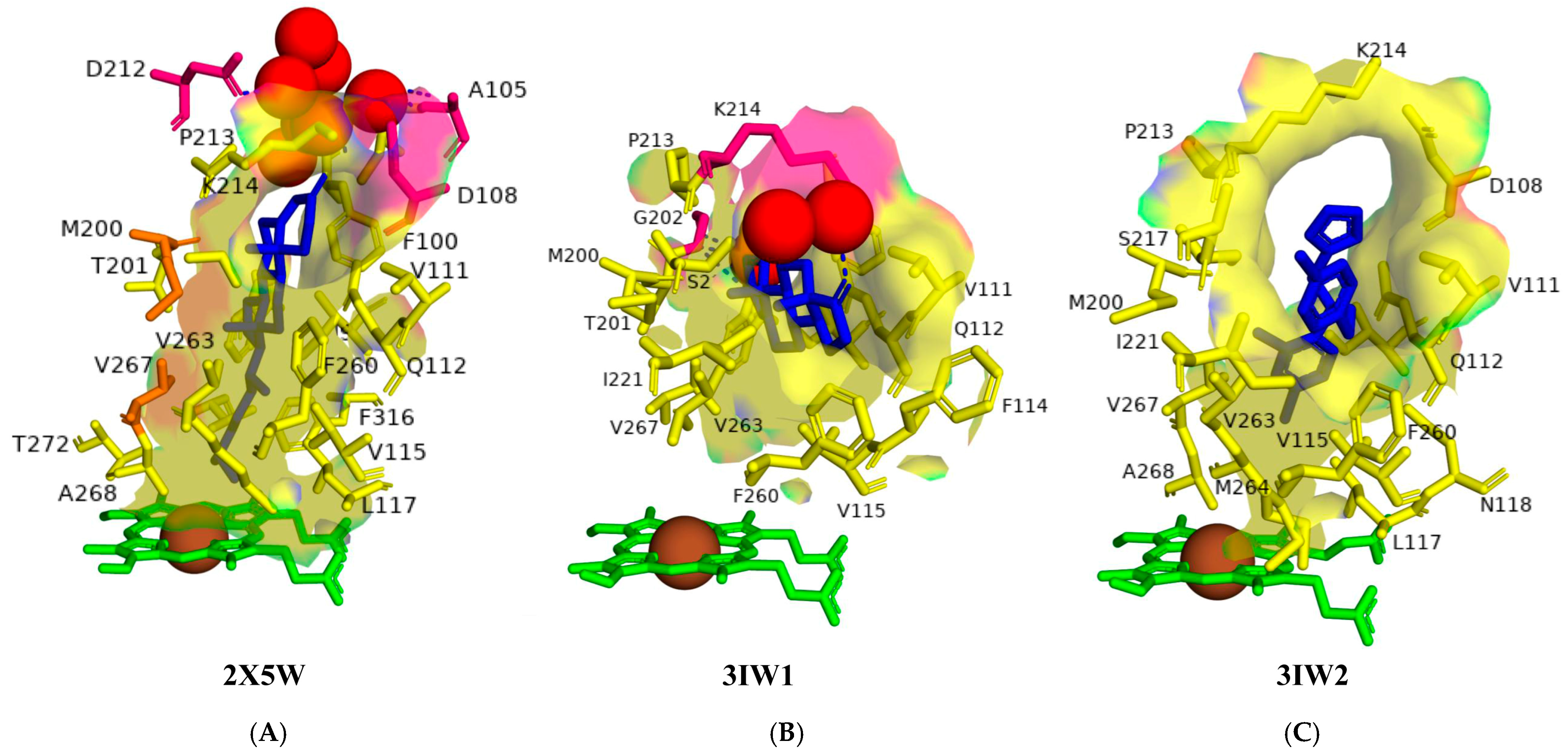
| PDB Code | Amino Acids |
|---|---|
| 2X5W | Ile97, Phe100, Ile104, Asp108, Val111, Gln112, Val115, Leu117, Met200, Thr201, Pro213, Lys214, Ser217, Phe260, Ala263, Met264, Val267, Ala268, Thr272, Val313, Phe316, Trp414 |
| 2XC3 | Ile97, Phe100, Ile104, Asp108, Ile109, Val111, Gln112, Phe114, Val115, Leu117, Asn118, Met200, Thr201, Gly202, Pro213, Lys214, Ser217, Ile221, Phe260, Val263, Met264, Val267, Ala268, Thr272, Phe316, Trp414 |
| 3IW1 | Ile97, Phe100, Val111, Gln112, Phe114, Val115, Met200, Thr201, Gly202, Pro213, Lys214, Ser217, Ile221, Phe260, Val263, Val267, Trp414 |
| 3IW2 | Ile97, Asp108, Val111, Gln112, Val115, Leu117, Asn118, Met200, Pro213, Lys214, Ser217, Ala218, Ile221, Phe260, Val263, Met264, Val267, Ala268 |
| 7QKE | Ile97, Phe100, Ile104, Asp108, Ile109, Val111, Gln112, Phe114, Val115, Leu117, Asn118, Met200, Thr201, Asn203, Pro213, Lys214, Ser217, Ile221, Phe260, Val263, Met264, Val267, Ala268, Val313, Phe316, Trp414 |
| 7QNN | Ile97, Phe100, Ile104, Asp108, Val111, Gln112, Phe114, Val115, Leu117, Met200, Thr201, Pro213, Ser217, Ile221, Phe260, Val263, Met264, Val267, Ala268, Phe316, Trp414 |
| 7QWN | Ile97, Phe100, Gln112, Leu117, Met200, Thr201, Asn203, Val267, Ala268, Gly269, Asn270, Glu271, Thr272, Val313, Phe316, Cys377, Trp414, Leu415 |
| 7R1I | Ile97, Arg99, Phe100, Val111, Gln112, Val115, Leu117, Met200, Thr201, Asn203, Pro213, Ser217, Phe260, Val263, Met264, Val267, Ala268, Thr272, Val313, Phe316, Trp414 |
| 7R3U | Ile97, Gln112, Leu117, Met200, Thr201, Met264, Val267, Ala268, Gly269, Thr272, Val313, Phe316, Cys377, Trp414, Leu415 |
| 7YXF | Ile97, Phe100, Ile104, Asp108, Val111, Asn112, Leu117, Met200, Thr201, Gly202, Pro213, Lys214, Ser217, Met264, Val267, Ala268, Gly269, Asn270, Glu271, Thr272, Val313, Phe316, Cys377, Trp414, Leu415 |
| 7ZLZ | Ile97, Phe100, Ile104, Asp108, Val111, Gln112, Phe114, Val115, Leu117, Met200, Thr201, Gly202, Asn203, Pro213, Lys214, Ser217, Ile221, Phe260, Val263, Met264, Val267, Ala268, Thr272, Val313, Phe316, Trp414, Leu415 |
| 7ZQR | Ile97, Leu117, Met200, Thr201, Val267, Ala268, Thr272, Val313, Phe316, Cys377, Trp414, Leu415 |
| 7ZSU | Ile97, Gln112, Leu115, Val115, Leu117, Asn118, Met200, Thr201, Met264, Val267, Ala268, Gly269, Glu271, Thr272, Val313, Phe316, Cys377, Trp414 |
| 7ZT0 | Ile97, Phe100, Gln112, Leu117, Met200, Thr201, Gly202, Asn203, Pro213, Val267, Ala268, Glu271, Thr272, Val313, Phe316, Cys377, Trp414, Leu415 |
| 7ZXD | Ile97, Arg99, Phe100, Val111, Gln112, Val115, Leu117, Met200, Thr201, Asn203, Glu204, Pro213, Ser217, Ile221, Phe260, Val263, Met264, Val267, Ala268, Thr272, Val313, Phe316, Trp414 |
2.3. CYP125A1 Interactions with the LP10 Molecule Revealed Structural Determinants for Developing Selective Inhibitors
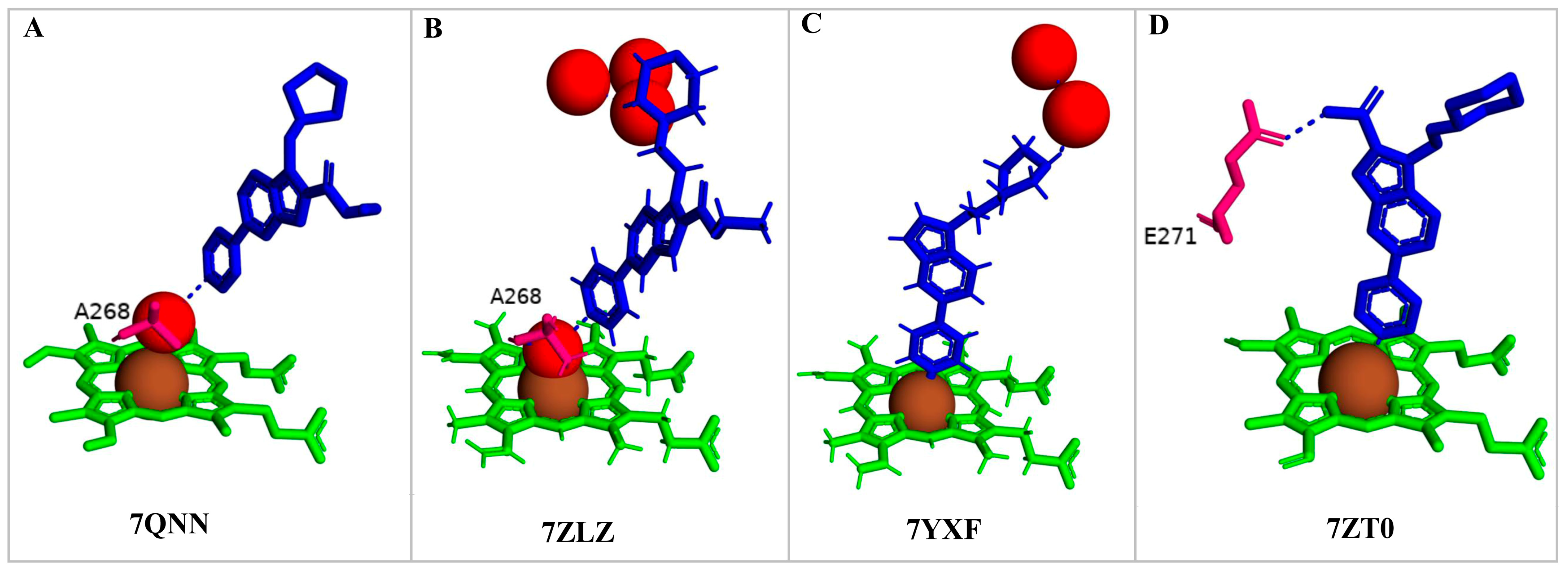
2.4. Indole-Derived Inhibitors Interact Differently Compared to the Steroids
3. Materials and Methods
3.1. Retrieving the Crystal Structures of CYP125 P450s
3.2. Assigning Crystal Structures of CYP125 into an Open and Closed Conformation
3.3. Analysis of CYP125 Active Site
3.4. Analysis of CYP125A-Ligand Interactions
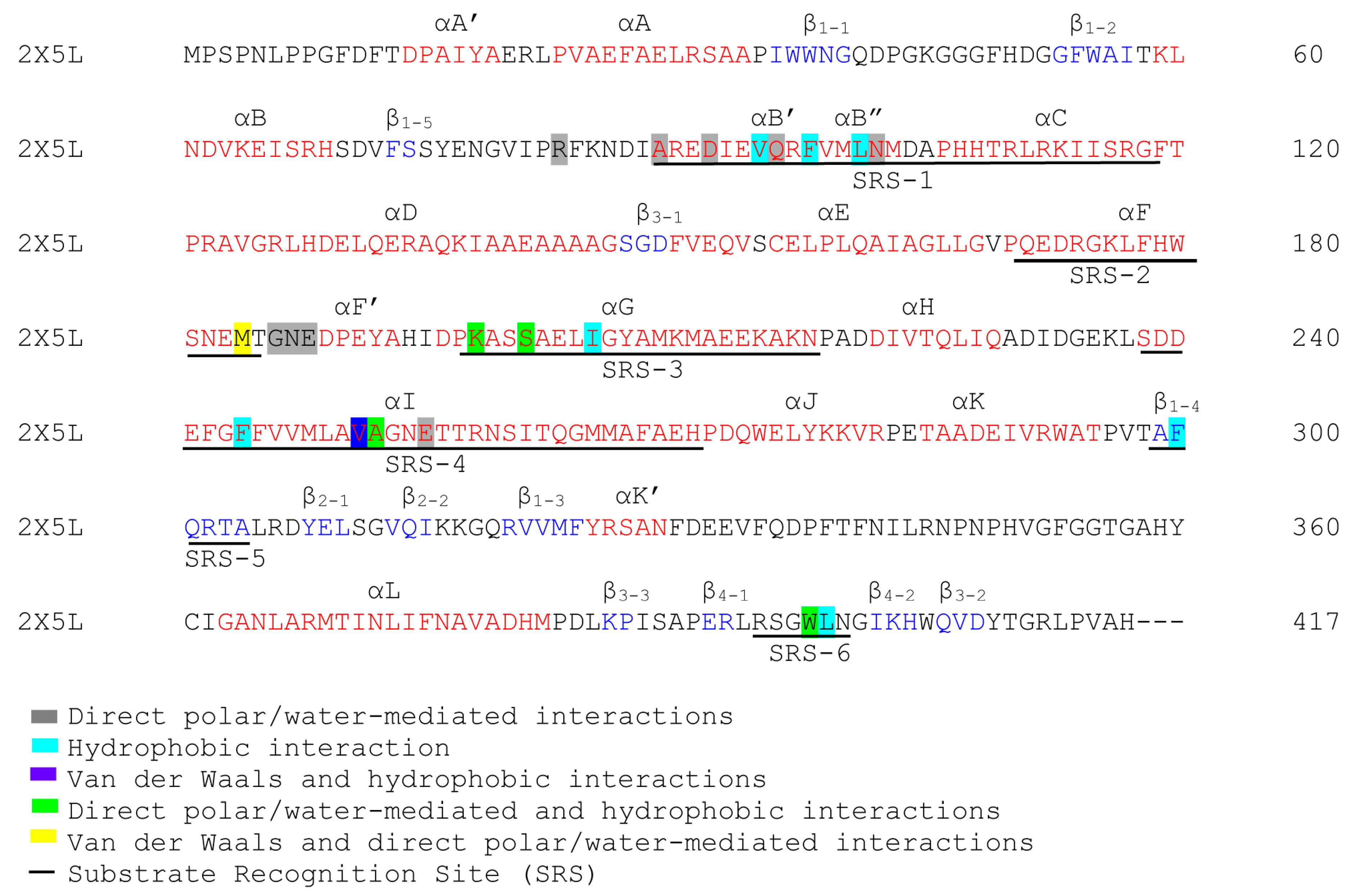
3.5. Annotation of P450 Characteristic Secondary Structures and Identification of Substrate Recognition Sites (SRSs)
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.X.; Miao, F.Y.; Yang, J.; Zhou, W.T.; Lv, S.; Wei, F.N.; Wang, Y.; Hu, X.J.; Yin, P.; Zheng, P.Y.; et al. Global, regional, and national burden of HIV-negative tuberculosis, 1990–2021: Findings from the Global Burden of Disease Study 2021. Infect. Dis. Poverty 2024, 13, 60. [Google Scholar] [CrossRef]
- WHO. Global Tuberculosis Report 2024; WHO: Geneva, Switzerland, 2024; ISBN 978-92-4-010153-1. [Google Scholar]
- Olivier, C.; Luies, L. WHO Goals and Beyond: Managing HIV/TB Co-infection in South Africa. SN Compr. Clin. Med. 2023, 5, 251. [Google Scholar] [CrossRef]
- Perveen, S.; Kumari, D.; Singh, K.; Sharma, R. Tuberculosis drug discovery: Progression and future interventions in the wake of emerging resistance. Eur. J. Med. Chem. 2022, 229, 114066. [Google Scholar] [CrossRef]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Isin, E.M.; Guengerich, F.P. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim. Biophys. Acta 2007, 1770, 314–329. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Munro, A.W. Unusual cytochrome p450 enzymes and reactions. J. Biol. Chem. 2013, 288, 17065–17073. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef]
- Ngcobo, P.E.; Nkosi, B.V.Z.; Chen, W.; Nelson, D.R.; Syed, K. Evolution of cytochrome P450 enzymes and their redox partners in Archaea. Int. J. Mol. Sci. 2023, 24, 4161. [Google Scholar] [CrossRef]
- Gront, D.; Syed, K.; Nelson, D.R. Exploring P450 superfamily diversity with P450Atlas-Online tool for automated subfamily assignment. Protein Sci. 2025, 34, e70057. [Google Scholar] [CrossRef]
- Zondo, N.M.; Padayachee, T.; Nelson, D.R.; Syed, K. Saprophytic to pathogenic mycobacteria: Loss of cytochrome P450s vis a vis their prominent involvement in natural metabolite biosynthesis. Int. J. Mol. Sci. 2023, 24, 149. [Google Scholar] [CrossRef]
- van Wyk, R.; van Wyk, M.; Mashele, S.S.; Nelson, D.R.; Syed, K. Comprehensive comparative analysis of cholesterol catabolic genes/proteins in mycobacterial species. Int. J. Mol. Sci. 2019, 20, 1032. [Google Scholar] [CrossRef]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Ortiz de Montellano, P.R. Potential drug targets in the Mycobacterium tuberculosis cytochrome P450 system. J. Inorg. Biochem. 2018, 180, 235–245. [Google Scholar] [CrossRef]
- Chang, J.C.; Harik, N.S.; Liao, R.P.; Sherman, D.R. Identification of Mycobacterial genes that alter growth and pathology in macrophages and in mice. J. Infect. Dis. 2007, 196, 788–795. [Google Scholar] [CrossRef]
- Chang, J.C.; Miner, M.D.; Pandey, A.K.; Gill, W.P.; Harik, N.S.; Sassetti, C.M.; Sherman, D.R. igr Genes and Mycobacterium tuberculosis cholesterol metabolism. J. Bacteriol. 2009, 191, 5232–5239. [Google Scholar] [CrossRef]
- Frank, D.J.; Zhao, Y.; Wong, S.H.; Basudhar, D.; De Voss, J.J.; Ortiz de Montellano, P.R. Cholesterol Analogs with Degradation-resistant Alkyl Side Chains Are Effective Mycobacterium tuberculosis Growth Inhibitors. J. Biol. Chem. 2016, 291, 7325–7333. [Google Scholar] [CrossRef]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- McLean, K.J.; Lafite, P.; Levy, C.; Cheesman, M.R.; Mast, N.; Pikuleva, I.A.; Leys, D.; Munro, A.W. The Structure of Mycobacterium tuberculosis CYP125: Molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 2009, 284, 35524–35533. [Google Scholar] [CrossRef]
- Rosłoniec, K.Z.; Wilbrink, M.H.; Capyk, J.K.; Mohn, W.W.; Ostendorf, M.; van der Geize, R.; Dijkhuizen, L.; Eltis, L.D. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 2009, 74, 1031–1043. [Google Scholar] [CrossRef]
- Capyk, J.K.; Kalscheuer, R.; Stewart, G.R.; Liu, J.; Kwon, H.; Zhao, R.; Okamoto, S.; Jacobs, W.R., Jr.; Eltis, L.D.; Mohn, W.W. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. J. Biol. Chem. 2009, 284, 35534–35542. [Google Scholar] [CrossRef]
- Johnston, J.B.; Ouellet, H.; Ortiz de Montellano, P.R. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010, 285, 36352–36360. [Google Scholar] [CrossRef]
- Ouellet, H.; Guan, S.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; de Montellano, P.R. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef]
- Frank, D.J.; Waddling, C.A.; La, M.; Ortiz de Montellano, P.R. Cytochrome P450 125A4, the Third Cholesterol C-26 Hydroxylase from Mycobacterium smegmatis. Biochemistry 2015, 54, 6909–6916. [Google Scholar] [CrossRef]
- García-Fernández, E.; Frank, D.J.; Galán, B.; Kells, P.M.; Podust, L.M.; García, J.L.; Ortiz de Montellano, P.R. A highly conserved mycobacterial cholesterol catabolic pathway. Environ. Microbiol. 2013, 15, 2342–2359. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.P.; Lee, N.; Choi, K.Y.; Jung, E.; Jeong, D.H.; Kim, B.G. Screening of bacterial cytochrome P450s responsible for regiospecific hydroxylation of (iso)flavonoids. Enzym. Microb. Technol. 2011, 48, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Rimal, H.; Subedi, P.; Kim, K.H.; Park, H.; Lee, J.H.; Oh, T.-J. Characterization of CYP125A13, the First Steroid C-27 Monooxygenase from Streptomyces peucetius ATCC27952. J. Microbiol. Biotechnol. 2020, 30, 1750–1759. [Google Scholar] [CrossRef]
- Doherty, D.Z.; Ghith, A.; Ho, A.; De Voss, J.J.; Bell, S.G. The bacterial cytochrome P450 (CYP) CYP125 enzymes can competitively oxidise sitosterol in the presence of cholesterol. Chem. Commun. 2023, 59, 9392–9395. [Google Scholar] [CrossRef]
- Ghith, A.; Bruning, J.B.; Bell, S.G. The oxidation of cholesterol derivatives by the CYP124 and CYP142 enzymes from Mycobacterium marinum. J. Steroid Biochem. Mol. Biol. 2023, 231, 106317. [Google Scholar] [CrossRef]
- Padayachee, T.; Lamb, D.; Nelson, D.; Syed, K. Structure–Function Analysis of the Essential Mycobacterium tuberculosis P450 Drug Target, CYP121A1. Int. J. Mol. Sci. 2024, 25, 4886. [Google Scholar] [CrossRef] [PubMed]
- Padayachee, T.; Lamb, D.C.; Nelson, D.R.; Syed, K. Structure-Function Analysis of the Biotechnologically Important Cytochrome P450 107 (CYP107) Enzyme Family. Biomolecules 2023, 13, 1733. [Google Scholar] [CrossRef]
- Wankhade, G.; Kamble, S.; Deshmukh, S.; Jena, L.; Waghmare, P.; Harinath, B. Inhibition of mycobacterial CYP125 enzyme by sesamin and [beta]-sitosterol: An in silico and in vitro study. Biomed. Biotechnol. Res. J. 2017, 1, 49. [Google Scholar]
- Katariya, M.; Snee, M.; Tunnicliffe, R.; Kavanagh, M.; Boshoff, H.; Amadi Obasi, C.; Levy, C.; Munro, A.; Abell, C.; Leys, D.; et al. Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis. Chem. Weinh. Der Bergstr. Ger. 2023, 29, e202203868. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Smith, J.; Nicholls, P.; Whomsley, R.; Cariuk, P. Synthesis and biological evaluation of imidazole based compounds as cytochrome P-450 inhibitors. Drug Des. Discov. 1995, 13, 27–41. [Google Scholar]
- Urban, P.; Lautier, T.; Pompon, D.; Truan, G. Ligand Access Channels in Cytochrome P450 Enzymes: A Review. Int. J. Mol. Sci. 2018, 19, 1617. [Google Scholar] [CrossRef]
- Ouellet, H.; Kells, P.M.; Ortiz de Montellano, P.R.; Podust, L.M. Reverse type I inhibitor of Mycobacterium tuberculosis CYP125A1. Bioorg Med. Chem. Lett. 2011, 21, 332–337. [Google Scholar] [CrossRef]
- Padayachee, T.; Lamb, D.C.; Nelson, D.R.; Syed, K. Structure-Function Analysis of the Self-Sufficient CYP102 Family Provides New Insights into Their Biochemistry. Int. J. Mol. Sci. 2025, 26, 2161. [Google Scholar] [CrossRef]
- Msweli, S.M.; Padayachee, T.; Khumalo, T.; Nelson, D.R.; Lamb, D.C.; Syed, K. Structure–Function Analysis of the Steroid-Hydroxylating Cytochrome P450 109 (CYP109) Enzyme Family. Int. J. Mol. Sci. 2025, 26, 6219. [Google Scholar] [CrossRef]
- Binkowski, T.A.; Naghibzadeh, S.; Liang, J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003, 31, 3352–3355. [Google Scholar] [CrossRef]
- Schrödinger, L.D. The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC.: New York, NY, USA, 2020. [Google Scholar]
- Graham, S.E.; Peterson, J.A. How similar are P450s and what can their differences teach us? Arch. Biochem. Biophys. 1999, 369, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef] [PubMed]
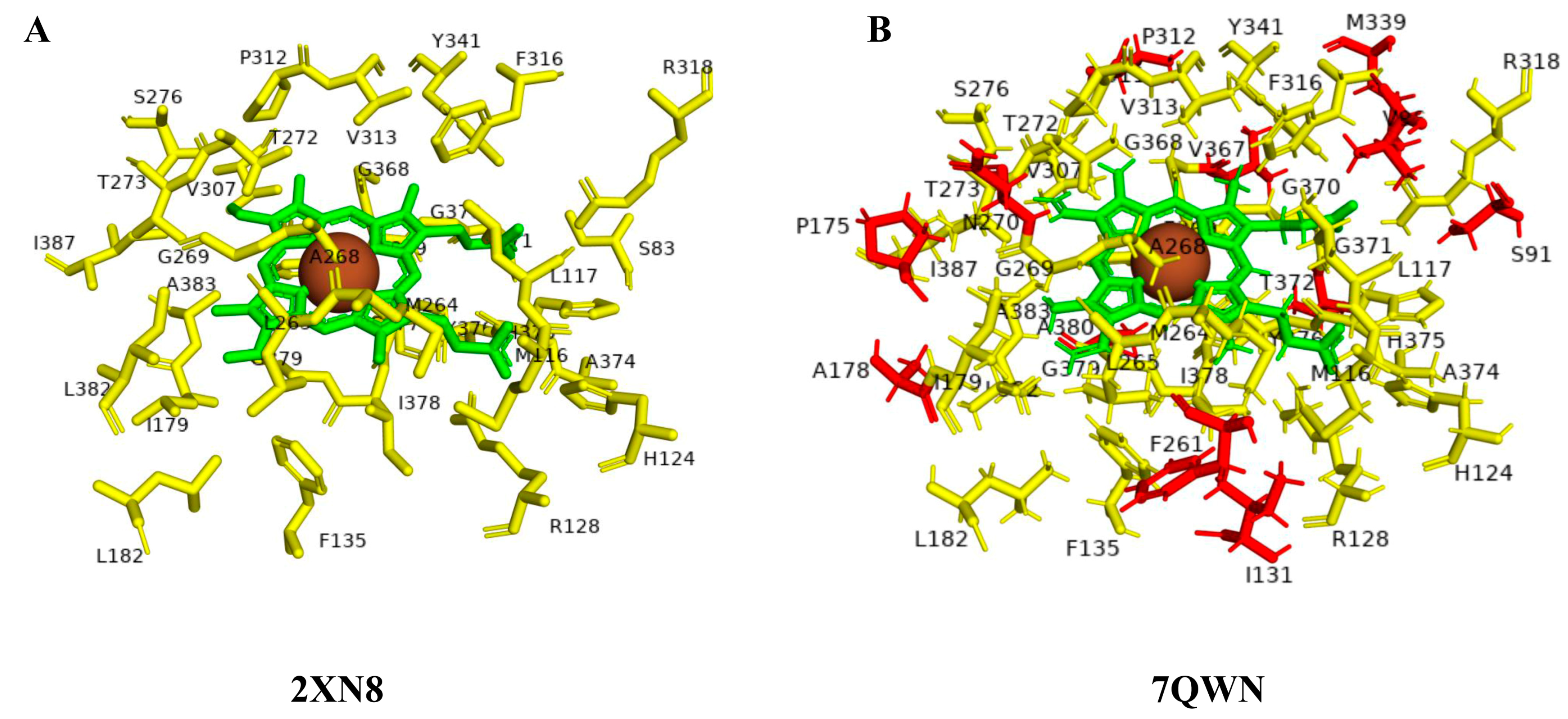

| Ligands | Species Code and P450 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mtb H37Rv CYP125A1 | Msmeg MC2 155 CYP125A3 | Msmeg MC2 155 CYP125A4 | Rjos CYP125 | Mbov ATCC BAA-935/AF2122/97 CYP125 | Mtb CDC1551 CYP125A1 | Save MA4680 CYP125A2 | Speu ATCC27952 CYP125A13 | Mmar CYP125A6 | Mmar CYP125A7 | |
| Cholesterol | √ | √ | √ | √ | √ | √ | - | √ | √ | √ |
| Cholest-4-en-3-one | √ | √ | √ | - | - | √ | - | - | √ | √ |
| 7α-hydroxycholesterol | √ | √ | √ | - | - | - | - | - | √ | - |
| 7α-hydroxy-4-cholesten-3-one | √ | X | √ | - | - | - | - | - | - | - |
| Chrysin (5,7-dihydroxyisoflavone) | - | - | - | - | - | - | √ | - | - | - |
| Cholesteryl sulfate | - | - | - | - | - | - | - | - | √ | - |
| 5β-cholestan-3α-ol | - | - | - | - | - | - | - | - | √ | √ |
| 5β-cholestan-3β-ol | - | - | - | - | - | - | - | - | √ | √ |
| cholesterol-5α,6α-epoxide | - | - | - | - | - | - | - | - | √ | √ |
| 7-ketocholesterol | - | - | - | - | - | - | - | - | √ | √ |
| 7β-hydroxycholesterol | - | - | - | - | - | - | - | - | √ | √ |
| 7-dehydrocholesterol | - | - | - | - | - | - | - | - | √ | - |
| Sitosterol | - | - | - | - | - | - | - | - | √ | |
| 5α-cholestan-3β-ol | - | - | - | - | - | - | - | - | - | √ |
| Cholesterol-5α,6α-epoxide | - | - | - | - | - | - | - | - | - | - |
| References | [20,23,24,25] | [25,26] | [25,26] | [21] | [22] | [24] | [27] | [28] | [29,30] | [29,30] |
| P450 | Confirmation | PDB Code | Number of Amino Acids in the Active Site Cavity | Common Amino Acids Among Open/Closed Conformations | Unique Amino Acids Among Open/Closed Conformations | Common Amino Acids Between Open and Closed Conformations | Unique Amino Acids in Open or Closed Confirmation |
|---|---|---|---|---|---|---|---|
| CYP125A1 | Open | 2X5L | 34 | Ser83, Met116, Leu117, His124, Arg128, Phe135, Ile179, Leu182, Met264, Leu265, Ala268, Gly269, Thr272, Thr273, Ser276, Val307, Pro312, Val313, Phe316, Arg318, Tyr341, Gly368, Phe369, Gly370, Gly371, Ala374, His375, Tyr376, Cys377, Ile378, Gly379, Leu382, Ala383, Ile387 (34) | - | Ser83, Met116, Leu117, His124, Arg128, Phe135, Ile179, Leu182, Met264, Leu265, Ala268, Gly269, Thr272, Thr273, Ser276, Val307, Pro312, Val313, Phe316, Arg318, Tyr341, Gly368, Phe369, Gly370, Gly371, Ala374, His375, Tyr376, Cys377, Ile378, Gly379, Leu382, Ala383, Ile387 (34) | |
| Open | 2XN8 | 34 | - | ||||
| Open | 3IVY | 34 | - | ||||
| Open | 3IW0 | 34 | - | ||||
| Closed | 2X5W | 34 | Met116, Leu117, His124, Arg128, Phe135, Ile179, Met264, Leu265, Ala268, Gly269, Thr272, Thr273, Ser276, Val307, Pro312, Val313, Phe316, Arg318, Tyr341, Gly368, Phe369, Gly370, Ala374, His375, Tyr376, Cys377, Ile378, Gly379, Leu382, Ala383, Ile387 (31) | Ser83, Gly371, Thr372 (3) | Ser91, Val96, Ile131, Pro175, Met339, Ala380, Ala178, Phe261, Asn270, Thr311, Thr372, Val367 (12) | ||
| Closed | 2XC3 | 34 | Ser83, Leu182, Gly371 (3) | ||||
| Closed | 3IW1 | 35 | Ser83, Leu182, Val367, Gly371 (4) | ||||
| Closed | 3IW2 | 37 | Ser91, Val96, Pro175, Met339, Val367, Ala380 (6) | ||||
| Closed | 7QKE | 45 | Ser83, Ser91, Val96, Ile131, Pro175, Ala178, Leu182, Phe261, Asn270, Thr311, Met339, Val367, Gly371, Ala380 (14) | ||||
| Closed | 7QNN | 34 | Ser83, Leu182, Gly371 (3) | ||||
| Closed | 7QWN | 42 | Ser83, Ile131, Pro175, Leu182, Phe261, Asn270, Thr311, Met339, Val367, Gly371, Ala380 (11) | ||||
| Closed | 7R1I | 36 | Ser83, Leu182, Phe261, Met339, Gly371 (5) | ||||
| Closed | 7R3U | 44 | Ser83, Val96, Ile131, Pro175, Ala178, Leu182, Phe261, Asn270, Thr311, Met339, Val367, Gly371, Ala380 (13) | ||||
| Closed | 7YXF | 42 | Ser83, Val96, Ile131, Pro175, Leu182, Asn270, Thr311, Met339, Val367, Gly371, Ala380 (11) | ||||
| Closed | 7ZLZ | 44 | Ser83, Val96, Ile131, Pro175, Ala178, Leu182, Phe261, Asn270, Thr311, Met339, Val367, Gly371, Ala380 (13) | ||||
| Closed | 7ZQR | 36 | Ser91, Val96, Leu182, Met339, Val367 (5) | ||||
| Closed | 7ZSU | 42 | Ser83, Ser91, Val96, Ile131, Leu182, Phe261, Thr311, Met339, val367, Gly371, Ala380 (11) | ||||
| Closed | 7ZT0 | 34 | Ser83, Ile182, Gly371 (3) | ||||
| Closed | 7ZXD | 35 | Ser83, Leu182, Val367, Gly371 (4) |
| PDB Code | Compound Number | Compound Name | Compound Structure |
|---|---|---|---|
| 7R3U | 1 | 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine | 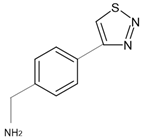 |
| 7ZQR | 2 | 4-(4-methoxyphenyl)pyridine | 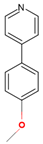 |
| 7ZSU | 3 | methyl 3-pyridin-4-ylbenzoate | 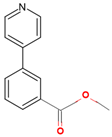 |
| 7QWN | 4 | ethyl 5-pyridin-4-yl-1~(H)-indole-2-carboxylate | 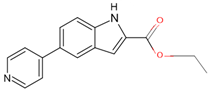 |
| 7QKE | 7 | ethyl 1-(cyclohexylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | 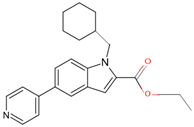 |
| 7QNN | 8 | ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | 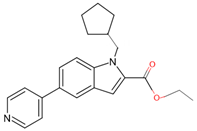 |
| 7R1I | 10 | ethyl 1-(2-piperidin-4-ylethyl)-5-pyridin-4-yl-indole-2-carboxylate | 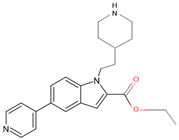 |
| 7ZLZ | 12 | ethyl 1-(2-morpholin-4-ylethyl)-5-pyridin-4-yl-indole-2-carboxylate | 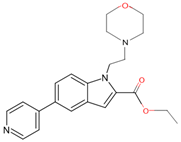 |
| 7YXF | 14 | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole | 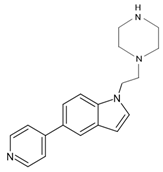 |
| 7ZT0 | 15 | 1-(2-piperazin-1-ylethyl)-5-pyridin-4-yl-indole-2-carboxamide | 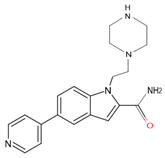 |
| 7ZXD | 19 | 1-[1-(2-piperidin-4-ylethyl)-5-pyridin-4-yl-indol-2-yl]butan-1-one |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masinga, N.; Nelson, D.R.; Syed, K. Structure–Function Analysis of Mycobacterium tuberculosis Drug Target Cytochrome P450 125 (CYP125) Enzyme Family. Int. J. Mol. Sci. 2025, 26, 8531. https://doi.org/10.3390/ijms26178531
Masinga N, Nelson DR, Syed K. Structure–Function Analysis of Mycobacterium tuberculosis Drug Target Cytochrome P450 125 (CYP125) Enzyme Family. International Journal of Molecular Sciences. 2025; 26(17):8531. https://doi.org/10.3390/ijms26178531
Chicago/Turabian StyleMasinga, Nompilo, David R. Nelson, and Khajamohiddin Syed. 2025. "Structure–Function Analysis of Mycobacterium tuberculosis Drug Target Cytochrome P450 125 (CYP125) Enzyme Family" International Journal of Molecular Sciences 26, no. 17: 8531. https://doi.org/10.3390/ijms26178531
APA StyleMasinga, N., Nelson, D. R., & Syed, K. (2025). Structure–Function Analysis of Mycobacterium tuberculosis Drug Target Cytochrome P450 125 (CYP125) Enzyme Family. International Journal of Molecular Sciences, 26(17), 8531. https://doi.org/10.3390/ijms26178531






