Association of Single-Nucleotide Polymorphisms on FURIN and EPHA2 Genes with the Risk and Prognosis of Undifferentiated Nasopharyngeal Cancer
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Study Population
2.2. Allele and Genotype Frequencies in the Study Population
2.3. Genetic Polymorphisms and NPC Risk
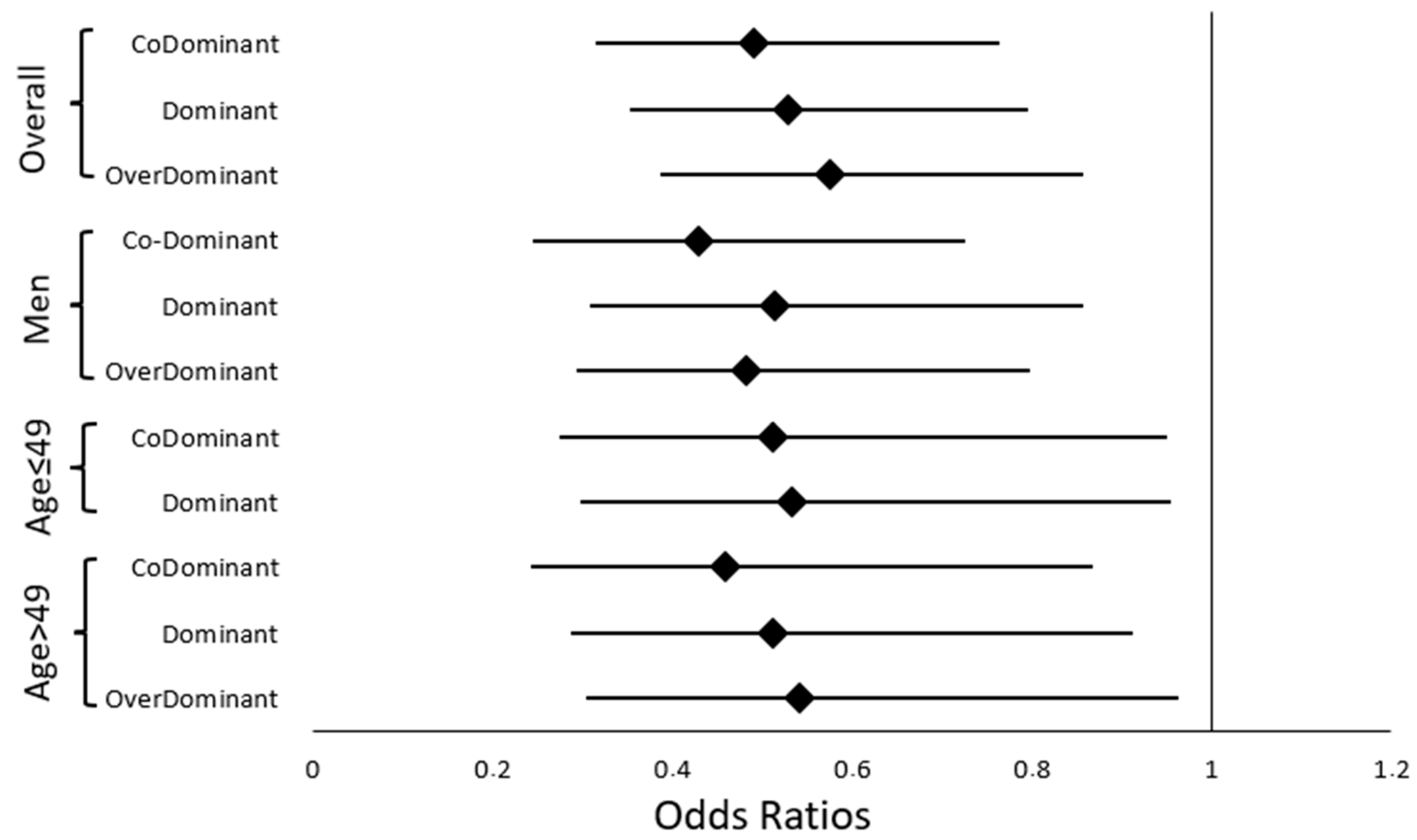
2.3.1. NPC Risk in the Overall Population
2.3.2. NPC Risk in the Gender-Stratified Population
| Men | Cases ♂ (n = 148), n (%) | Controls ♂ (n = 158) n (%) | OR (95% CI) | p-Value | OR a (95% CI) | p Value a,# | |
|---|---|---|---|---|---|---|---|
| Genetic model | rs4702 | ||||||
| CoDominant | AA | 69 (46.6) | 44 (27.8) | 1 | 1 | ||
| GG | 31 (20.9) | 34 (21.5) | 0.581 (0.314 to 1.077) | 0.084 | 0.729 (0.375 to 1.416) | 0.351 | |
| AG | 48 (32.4) | 80 (50.6) | 0.383 (0.227 to 0.644) | 0.000 | 0.429 (0.245 to 0.725) | 0.003 * | |
| Dominant | AA | 69 (46.6) | 44 (27.8) | ||||
| AG-GG | 79 (53.4) | 114 (72.2) | 0.442 (0.275 to 0.710) | 0.001 | 0.515 (0.309 to 0.858) | 0.011 * | |
| Recessive | AA-AG | 117 (79.1) | 124 (78.5) | ||||
| GG | 31 (20.9) | 34 (21.5) | 0.966 (0.558 to 1.672) | 0.903 | 1.148 (0.634 to 2.079) | 0.649 | |
| OverDominant | AA-GG | 100 (67.6) | 78 (49.4) | ||||
| AG | 48 (32.4) | 80 (50.6) | 0.468 (0.294 to 0.745) | 0.001 | 0.483 (0.293 to 0.798) | 0.005 * | |
| rs6603883 | NS | NS | |||||
| Women | Cases ♀ (n = 80), n (%) | Controls ♀ (n = 85) n (%) | |||||
| rs4702 | NS | NS | |||||
| rs6603883 | NS | NS |
2.3.3. NPC Risk in the Age-Stratified Population
| Age ≤ 49 Years | Cases (n = 116), n (%) | Controls (n = 117) n (%) | OR (95% CI) | p-Value | OR a (95% CI) | p Value a,# | |
|---|---|---|---|---|---|---|---|
| Genetic model | rs4702 | ||||||
| CoDominant | AA | 52 (44.8) | 35 (29.9) | 1 | 1 | ||
| GG | 19 (16.4) | 21 (17.9) | 0.609 (0.286 to 1.295) | 0.197 | 0.598 (0.266 to 1.344) | 0.213 | |
| AG | 45 (38.8) | 61 (52.1) | 0.497 (0.279 to 0.883) | 0.016 | 0.511 (0.275 to 0.951) | 0.034 * | |
| Dominant | AA | 52 (44.8) | 35 (29.9) | ||||
| AG-GG | 64 (55.2) | 82 (70.1) | 0.525 (0.306 to 0.901) | 0.018 | 0.534 (0.299 to 0.954) | 0.034 * | |
| Recessive | AA-AG | 97 (83.6) | 96 (82.1) | ||||
| GG | 19 (16.4) | 21 (17.9) | 0.895 (0.453 to 1.771) | 0.751 | 0.867 (0.418 to 1.801) | 0.702 | |
| OverDominant | AA-GG | 71 (61.2) | 56 (47.9) | ||||
| AG | 45 (38.8) | 61 (52.1) | 0.582 (0.346 to 0.979) | 0.041 | 0.604 (0.345 to 1.058) | 0.078 | |
| rs6603883 | NS | NS | |||||
| Age > 49 years | Cases (n = 112), n (%) | Controls (n = 126) n (%) | |||||
| Genetic model | rs4702 | ||||||
| CoDominant | AA | 51 (45.5) | 40 (31.7) | 1 | 1 | ||
| GG | 24 (21.4) | 27 (21.4) | 0.697 (0.350 to 1.388) | 0.304 | 0.629 (0.296 to 1.336) | 0.228 | |
| AG | 37 (33) | 59 (46.8) | 0.492 (0.274 to 0.881) | 0.016 | 0.459 (0.243 to 0.867) | 0.016 * | |
| Dominant | AA | 51 (45.5) | 40 (31.7) | ||||
| AG-GG | 61 (54.5) | 86 (68.3) | 0.556 (0.328 to 0.943) | 0.029 | 0.512 (0.287 to 0.912) | 0.023 * | |
| Recessive | AA-AG | 88 (78.6) | 99 (78.6) | ||||
| GG | 24 (21.4) | 27 (21.4) | 1.000 (0.538 to 1.860) | 1.000 | 0.936 (0.474 to 1.846) | 0.848 | |
| OverDominant | AA-GG | 75 (67) | 67 (53.2) | ||||
| AG | 37 (33) | 59 (46.8) | 0.560 (0.331 to 0.949) | 0.030 | 0.542 (0.305 to 0.963) | 0.037 * | |
| rs6603883 | NS | NS |
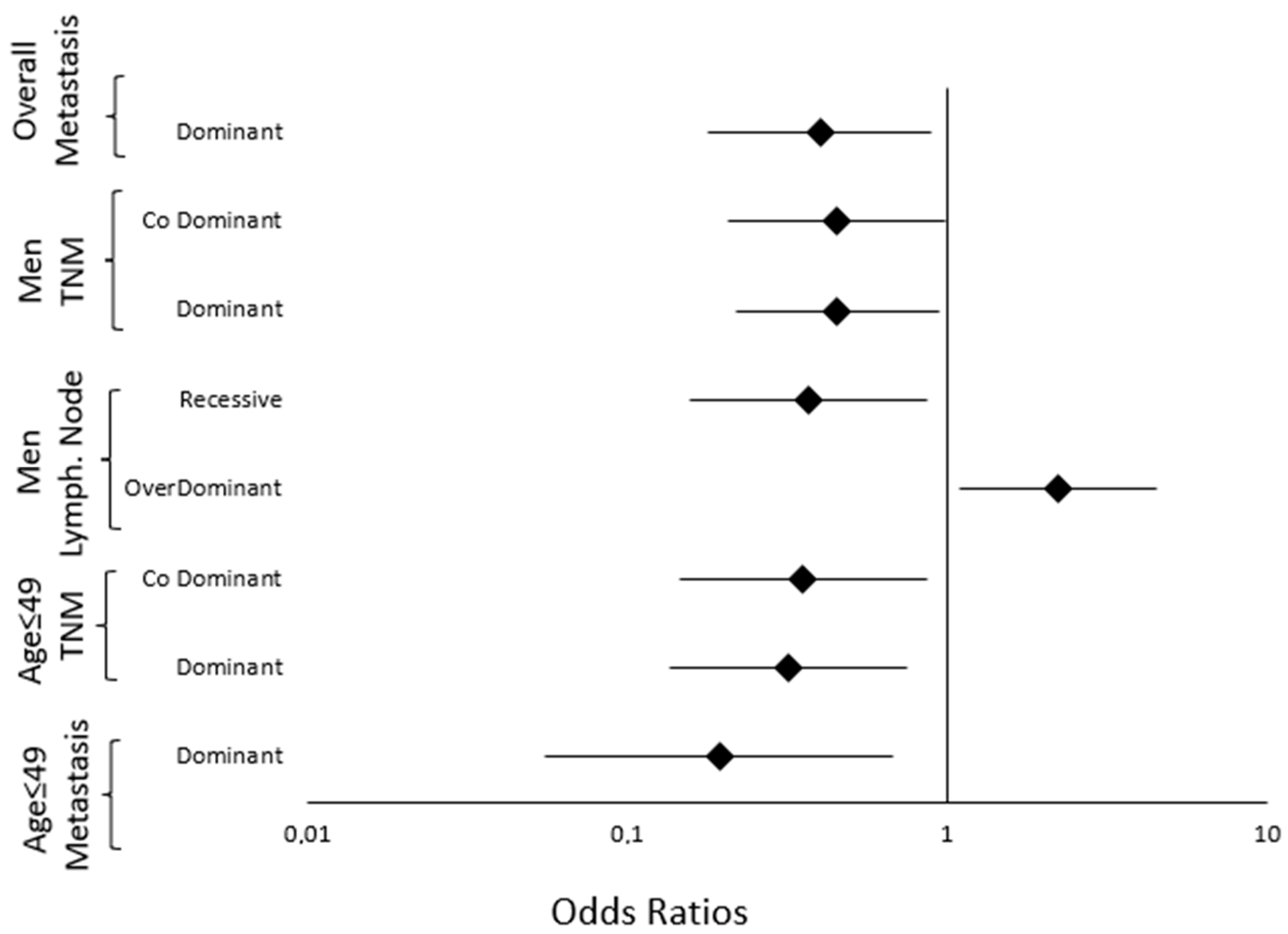
2.4. Genetic Polymorphisms and NPC Prognosis
2.4.1. Prognosis of the Overall Population
2.4.2. Prognosis of the Gender-Stratified Population
2.4.3. Prognosis of the Age-Stratified Population
| Gender Stratification | ||||||||
| Parameters | Genetic Model | Genotype | Parameter Category n (%) | OR (95% CI) | p Value # | OR a (95% CI) | p Value a,# | |
| ♂ | Genetic Model | rs6603883 | I-II-III | IVA-IVB | ||||
| TNM stage (n = 148) | CoDominant | GG | 25 (27.5) | 26 (45.6) | 1 | |||
| AA | 21 (23) | 9 (15.8) | 0.412 (0.159 to 1.070) | 0.063 | 0.459 (0.169 to 1.249) | 0.127 | ||
| GA | 45 (49.5) | 22 (38.6) | 0.470 (0.222 to 0.994) | 0.047 | 0.452 (0.206 to 0.992) | 0.048 * | ||
| Dominant | GG | 25 (27.5) | 26 (45.6) | |||||
| GA-AA | 66 (72.5) | 31 (54.4) | 0.452 (0.225 to 0.905) | 0.025 | 0.454 (0.219 to 0.940) | 0.034 * | ||
| ♂ | rs6603883 | N0-N1 | N2-N3 | |||||
| Lymph node involvement (n = 148) | Recessive | GG-GA | 39 (69.6) | 79 (85.9) | ||||
| AA | 17 (30.4) | 13 (14.1) | 0.378 (0.167 to 0.855) | 0.019 | 0.370 (0.157 to 0.872) | 0.023 * | ||
| OverDominant | AA-GG | 37 (66.1) | 44 (47.8) | |||||
| GA | 19 (33.9) | 48 (52.2) | 2.124 (1.068 to 4.227) | 0.030 | 2.220 (1.094 to 4.507) | 0.027 * | ||
| ♀ | Genetic Model | rs4702 | N0-N1 | N2-N3 | ||||
| Lymph node involvement (n = 80) | CoDominant | AA | 16 (59.3) | 18 (34) | 1 | |||
| GG | 5 (18.5) | 7 (13.2) | 1.244 (0.329 to 4.708) | 0.747 | 0.914 (0.221 to 3.784) | 0.901 | ||
| AG | 6 (22.2) | 28 (52.8) | 4.148 (1.368 to 12.580) | 0.009 | 3.53 (1.120 to 11.133) | 0.031 * | ||
| Dominant | AA | 16 (59.3) | 18 (34) | |||||
| AG-GG | 11 (40.7) | 35 (66) | 2.828 (1.088 to 7.352) | 0.031 | 2.363 (0.869 to 6.426) | 0.092 | ||
| OverDominant | AA-GG | 21 (77.8) | 25 (47.2) | |||||
| AG | 6 (22.2) | 28 (52.8) | 3.920 (1.364 to 11.263) | 0.007 | 3.62 (1.224 to 10.693) | 0.020 * | ||
| Age Stratification | ||||||||
| Parameters | Genetic Model | Genotype | Parameter Category n (%) | OR (95% CI) | p Value # | OR b (95% CI) | p Value b,# | |
| Age ≤ 49 years | Genetic Model | rs6603883 | I-II-III | IVA-IVB | ||||
| TNM stage (n = 116) | CoDominant | GG | 22 (30.1) | 23 (53.5) | 1 | |||
| AA | 12 (16.4) | 3 (5.7) | 0.239 (0.059 to 0.964) | 0.041 | 0.198 (0.043 to 0.911) | 0.038 * | ||
| GA | 39 (53.5) | 17 (40.8) | 0.417 (0.184 to 0.943) | 0.034 | 0.355 (0.146 to 0.865) | 0.023 * | ||
| Dominant | GG | 22 (30.1) | 23 (53.5) | |||||
| GA-AA | 51 (69.9) | 20 (46.5) | 0.375 (0.172 to 0.819) | 0.013 | 0.319 (0.136 to 0.749) | 0.009 * | ||
| Age ≤ 49 years | Genetic Model | rs6603883 | M0 | M1 | ||||
| Presence of metastasis (n = 116) | Dominant | GG | 35 (34.3) | 10 (71.4) | ||||
| GA-AA | 67 (65.7) | 4 (28.6) | 0.209 (0.061 to 0.715) | 0.016 | 0.194 (0.055 to 0.682) | 0.011 * | ||
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. SNP Selection, DNA Extraction and Genotyping
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chua, M.L.K.; Wee, J.T.S.; Hui, E.P.; Chan, A.T.C. Nasopharyngeal Carcinoma. Lancet 2016, 387, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Wakisaka, N.; Kondo, S.; Endo, K.; Sugimoto, H.; Hatano, M.; Ueno, T.; Ishikawa, K.; Yoshizaki, T. Progression of Understanding for the Role of Epstein-Barr Virus and Management of Nasopharyngeal Carcinoma. Cancer Metastasis Rev. 2017, 36, 435–447. [Google Scholar] [CrossRef]
- Razak, A.R.A.; Siu, L.L.; Liu, F.-F.; Ito, E.; O’Sullivan, B.; Chan, K. Nasopharyngeal Carcinoma: The next Challenges. Eur. J. Cancer 2010, 46, 1967–1978. [Google Scholar] [CrossRef]
- Chang, E.T.; Ye, W.; Zeng, Y.-X.; Adami, H.-O. The Evolving Epidemiology of Nasopharyngeal Carcinoma. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1035–1047. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Bray, F.; Haugen, M.; Moger, T.A.; Tretli, S.; Aalen, O.O.; Grotmol, T. Age-Incidence Curves of Nasopharyngeal Carcinoma Worldwide: Bimodality in Low-Risk Populations and Aetiologic Implications. Cancer Epidemiol Biomark. Prev. 2008, 17, 2356–2365. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.T.C. Nasopharyngeal Carcinoma. Ann. Oncol. 2010, 21, vii308–vii312. [Google Scholar] [CrossRef]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein–Barr Virus Infection and Nasopharyngeal Carcinoma. Phil. Trans. R. Soc. B 2017, 372, 20160270. [Google Scholar] [CrossRef] [PubMed]
- Niedobitek, G.; Hansmann, M.L.; Herbst, H.; Young, L.S.; Dienemann, D.; Hartmann, C.A.; Finn, T.; Pitteroff, S.; Welt, A.; Anagnostopoulos, I. Epstein-Barr Virus and Carcinomas: Undifferentiated Carcinomas but Not Squamous Cell Carcinomas of the Nasopharynx Are Regularly Associated with the Virus. J. Pathol. 1991, 165, 17–24. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal Carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Damania, B.; Kenney, S.C.; Raab-Traub, N. Epstein-Barr Virus: Biology and Clinical Disease. Cell 2022, 185, 3652–3670. [Google Scholar] [CrossRef]
- Lo, K.W.; To, K.F.; Huang, D.P. Focus on Nasopharyngeal Carcinoma. Cancer Cell 2004, 5, 423–428. [Google Scholar] [CrossRef]
- Gunvén, P.; Klein, G.; Henle, G.; Henle, W.; Clifford, P. Epstein-Barr Virus in Burkitt’s Lymphoma and Nasopharyngeal Carcinoma. Antibodies to EBV Associated Membrane and Viral Capsid Antigens in Burkitt Lymphoma Patients. Nature 1970, 228, 1053–1056. [Google Scholar] [CrossRef]
- Bu, G.-L.; Xie, C.; Kang, Y.-F.; Zeng, M.-S.; Sun, C. How EBV Infects: The Tropism and Underlying Molecular Mechanism for Viral Infection. Viruses 2022, 14, 2372. [Google Scholar] [CrossRef]
- Chen, J.; Sathiyamoorthy, K.; Zhang, X.; Schaller, S.; Perez White, B.E.; Jardetzky, T.S.; Longnecker, R. Ephrin Receptor A2 Is a Functional Entry Receptor for Epstein–Barr Virus. Nat. Microbiol. 2018, 3, 172–180. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Wang, H.-B.; Zhang, A.; Chen, M.-L.; Fang, Z.-X.; Dong, X.-D.; Li, S.-B.; Du, Y.; Xiong, D.; et al. Ephrin Receptor A2 Is an Epithelial Cell Receptor for Epstein–Barr Virus Entry. Nat. Microbiol. 2018, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Longnecker, R. Epithelial Cell Infection by Epstein–Barr Virus. FEMS Microbiol. Rev. 2019, 43, 674–683. [Google Scholar] [CrossRef]
- Su, C.; Wu, L.; Chai, Y.; Qi, J.; Tan, S.; Gao, G.F.; Song, H.; Yan, J. Molecular Basis of EphA2 Recognition by gHgL from Gammaherpesviruses. Nat. Commun. 2020, 11, 5964. [Google Scholar] [CrossRef] [PubMed]
- Roebroek, A.J.; Umans, L.; Pauli, I.G.; Robertson, E.J.; van Leuven, F.; Van de Ven, W.J.; Constam, D.B. Failure of Ventral Closure and Axial Rotation in Embryos Lacking the Proprotein Convertase Furin. Development 1998, 125, 4863–4876. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef] [PubMed]
- Hallenberger, S.; Bosch, V.; Angliker, H.; Shaw, E.; Klenk, H.D.; Garten, W. Inhibition of Furin-Mediated Cleavage Activation of HIV-1 Glycoprotein Gp160. Nature 1992, 360, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Stieneke-Gröber, A.; Vey, M.; Angliker, H.; Shaw, E.; Thomas, G.; Roberts, C.; Klenk, H.D.; Garten, W. Influenza Virus Hemagglutinin with Multibasic Cleavage Site Is Activated by Furin, a Subtilisin-like Endoprotease. EMBO J. 1992, 11, 2407–2414. [Google Scholar] [CrossRef]
- Sorem, J.; Longnecker, R. Cleavage of Epstein-Barr Virus Glycoprotein B Is Required for Full Function in Cell-Cell Fusion with Both Epithelial and B Cells. J. Gen. Virol. 2009, 90, 591–595. [Google Scholar] [CrossRef]
- Bei, J.-X.; Su, W.-H.; Ng, C.-C.; Yu, K.; Chin, Y.-M.; Lou, P.-J.; Hsu, W.-L.; McKay, J.D.; Chen, C.-J.; Chang, Y.-S.; et al. A GWAS Meta-Analysis and Replication Study Identifies a Novel Locus within CLPTM1L/TERT Associated with Nasopharyngeal Carcinoma in Individuals of Chinese Ancestry. Cancer Epidemiol. Biomark. Prev. 2016, 25, 188–192. [Google Scholar] [CrossRef]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A Genome-Wide Association Study of Nasopharyngeal Carcinoma Identifies Three New Susceptibility Loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef]
- Tang, M.; Lautenberger, J.A.; Gao, X.; Sezgin, E.; Hendrickson, S.L.; Troyer, J.L.; David, V.A.; Guan, L.; Mcintosh, C.E.; Guo, X.; et al. The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the HLA Class I Antigen Recognition Groove. PLoS Genet. 2012, 8, e1003103. [Google Scholar] [CrossRef]
- Tse, K.-P.; Su, W.-H.; Chang, K.-P.; Tsang, N.-M.; Yu, C.-J.; Tang, P.; See, L.-C.; Hsueh, C.; Yang, M.-L.; Hao, S.-P.; et al. Genome-Wide Association Study Reveals Multiple Nasopharyngeal Carcinoma-Associated Loci Within the HLA Region at Chromosome 6p21.3. Am. J. Hum. Genet. 2009, 85, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.C.; Yew, P.Y.; Puah, S.M.; Krishnan, G.; Yap, L.F.; Teo, S.H.; Lim, P.V.H.; Govindaraju, S.; Ratnavelu, K.; Sam, C.K.; et al. A Genome-Wide Association Study Identifies ITGA9 Conferring Risk of Nasopharyngeal Carcinoma. J. Hum. Genet. 2009, 54, 392–397. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Coto, E.; Albaiceta, G.M.; Amado-Rodríguez, L.; García-Clemente, M.; Cuesta-Llavona, E.; Vázquez-Coto, D.; Alonso, B.; Iglesias, S.; Melón, S.; Alvarez-Argüelles, M.E.; et al. FURIN Gene Variants (Rs6224/Rs4702) as Potential Markers of Death and Cardiovascular Traits in Severe COVID-19. J. Med. Virol. 2022, 94, 3589–3595. [Google Scholar] [CrossRef]
- Hou, Y.; Liang, W.; Zhang, J.; Li, Q.; Ou, H.; Wang, Z.; Li, S.; Huang, X.; Zhao, C. Schizophrenia-Associated Rs4702 G Allele-Specific Downregulation of FURIN Expression by miR-338-3p Reduces BDNF Production. Schizophr. Res. 2018, 199, 176–180. [Google Scholar] [CrossRef]
- Yang, S.; Fu, Z.; Zhang, Y.; Fu, B.; Dong, L. The G to A Transformation of Rs4702 Polymorphism in 3’UTR of FURIN Reduced the Risk of Radiotherapy-induced Cognitive Impairment in Glioma Patients. J. Cell. Mol. Medi. 2022, 26, 684–692. [Google Scholar] [CrossRef]
- Ma, X.; Ma, Z.; Jiao, X.; Hejtmancik, J.F. Functional Non-Coding Polymorphism in an EPHA2 Promoter PAX2 Binding Site Modifies Expression and Alters the MAPK and AKT Pathways. Sci. Rep. 2017, 7, 9992. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-C. Searching for Disease-Susceptibility Loci by Testing for Hardy-Weinberg Disequilibrium in a Gene Bank of Affected Individuals. Am. J. Epidemiol. 2003, 158, 397–400. [Google Scholar] [CrossRef]
- Young, L.S.; Yap, L.F.; Murray, P.G. Epstein-Barr Virus: More than 50 Years Old and Still Providing Surprises. Nat. Rev. Cancer 2016, 16, 789–802. [Google Scholar] [CrossRef]
- Liang, C.; Kan, J.; Wang, J.; Lu, W.; Mo, X.; Zhang, B. Nasopharyngeal Carcinoma-Associated Inflammatory Cytokines: Ongoing Biomarkers. Front. Immunol. 2024, 15, 1448012. [Google Scholar] [CrossRef]
- Dobrindt, K.; Hoagland, D.A.; Seah, C.; Kassim, B.; O’Shea, C.P.; Murphy, A.; Iskhakova, M.; Fernando, M.B.; Powell, S.K.; Deans, P.J.M.; et al. Common Genetic Variation in Humans Impacts In Vitro Susceptibility to SARS-CoV-2 Infection. Stem Cell Rep. 2021, 16, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Pöhlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e5. [Google Scholar] [CrossRef] [PubMed]
- Turpeinen, H.; Seppälä, I.; Lyytikäinen, L.-P.; Raitoharju, E.; Hutri-Kähönen, N.; Levula, M.; Oksala, N.; Waldenberger, M.; Klopp, N.; Illig, T.; et al. A Genome-Wide Expression Quantitative Trait Loci Analysis of Proprotein Convertase Subtilisin/Kexin Enzymes Identifies a Novel Regulatory Gene Variant for FURIN Expression and Blood Pressure. Hum. Genet. 2015, 134, 627–636. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex Differences in Immune Responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Klein, S.L. Sex Influences Immune Responses to Viruses, and Efficacy of Prophylaxis and Treatments for Viral Diseases. Bioessays 2012, 34, 1050–1059. [Google Scholar] [CrossRef]
- Marriott, I.; Huet-Hudson, Y.M. Sexual Dimorphism in Innate Immune Responses to Infectious Organisms. Immunol. Res. 2006, 34, 177–192. [Google Scholar] [CrossRef]
- Carè, A.; Bellenghi, M.; Matarrese, P.; Gabriele, L.; Salvioli, S.; Malorni, W. Sex Disparity in Cancer: Roles of microRNAs and Related Functional Players. Cell Death Differ. 2018, 25, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Pan, J.; Hang, C.; Tan, L.; Hu, L.; Yan, Z.; Zhu, J. The Estrogen/miR-338-3p/ADAM17 Axis Enhances the Viability of Breast Cancer Cells via Suppressing NK Cell’s Function. Environ. Toxicol. 2023, 38, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Li, X.; You, B.; Shi, S.; Zhang, Q.; You, Y. MicroRNA-338 Inhibits Migration and Proliferation by Targeting Hypoxia-Induced Factor 1α in Nasopharyngeal Carcinoma. Oncol. Rep. 2015, 34, 1943–1952. [Google Scholar] [CrossRef] [PubMed]
- Bassi, D.E.; Mahloogi, H.; Al-Saleem, L.; Lopez De Cicco, R.; Ridge, J.A.; Klein-Szanto, A.J. Elevated Furin Expression in Aggressive Human Head and Neck Tumors and Tumor Cell Lines. Mol. Carcinog. 2001, 31, 224–232. [Google Scholar] [CrossRef]
- Bassi, D.E.; Lopez De Cicco, R.; Mahloogi, H.; Zucker, S.; Thomas, G.; Klein-Szanto, A.J. Furin Inhibition Results in Absent or Decreased Invasiveness and Tumorigenicity of Human Cancer Cells. Proc. Natl. Acad. Sci. USA 2001, 98, 10326–10331. [Google Scholar] [CrossRef]
- Sounni, N.E.; Baramova, E.N.; Munaut, C.; Maquoi, E.; Frankenne, F.; Foidart, J.-M.; Noël, A. Expression of Membrane Type 1 Matrix Metalloproteinase (MT1-MMP) in A2058 Melanoma Cells Is Associated with MMP-2 Activation and Increased Tumor Growth and Vascularization. Int. J. Cancer 2002, 98, 23–28. [Google Scholar] [CrossRef]
- Jaaks, P.; Bernasconi, M. The Proprotein Convertase Furin in Tumour Progression. Int. J. Cancer 2017, 141, 654–663. [Google Scholar] [CrossRef]
- Lin, C.; Qian, P.; Zhang, Y.; Liu, Z.; Dai, K.; Sun, D. Plac1 Promotes Nasopharyngeal Carcinoma Cells Proliferation, Migration and Invasion via Furin/NICD/PTEN Pathway. Tissue Cell 2021, 69, 101480. [Google Scholar] [CrossRef]
- Yin, L.; Ding, Y.; Wang, Y.; Wang, C.; Sun, K.; Wang, L. Identification of Serum miR-501-3p and miR-338-3p as Novel Diagnostic Biomarkers for Breast Cancer and Their Target Genes Associated with Immune Infiltration. Int. J. Gen. Med. 2023, 16, 1279–1294. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Suo, J.; Wang, C.; Sun, X.; Wang, D.; He, L.; Zhang, Y.; Li, W. Downregulation of Tissue miR-338-3p Predicts Unfavorable Prognosis of Gastric Cancer. Cancer Biomark. 2017, 21, 117–122. [Google Scholar] [CrossRef]
- Lin, H.; Gao, Y.; Sun, K.; Zhang, Q.; Li, Y.; Chen, M.; Jin, F. COA3 Overexpression Promotes Non-Small Cell Lung Cancer Metastasis by Reprogramming Glucose Metabolism. Am. J. Cancer Res. 2022, 12, 3662–3678. [Google Scholar]
- Yang, Y.; Ding, T.; Cong, Y.; Luo, X.; Liu, C.; Gong, T.; Zhao, M.; Zheng, X.; Li, C.; Zhang, Y.; et al. Interferon-Induced Transmembrane Protein-1 Competitively Blocks Ephrin Receptor A2-Mediated Epstein-Barr Virus Entry into Epithelial Cells. Nat. Microbiol. 2024, 9, 1256–1270. [Google Scholar] [CrossRef]
- Wang, H.-B.; Zhang, H.; Zhang, J.-P.; Li, Y.; Zhao, B.; Feng, G.-K.; Du, Y.; Xiong, D.; Zhong, Q.; Liu, W.-L.; et al. Neuropilin 1 Is an Entry Factor That Promotes EBV Infection of Nasopharyngeal Epithelial Cells. Nat. Commun. 2015, 6, 6240. [Google Scholar] [CrossRef]
- Menges, C.W.; McCance, D.J. Constitutive Activation of the Raf–MAPK Pathway Causes Negative Feedback Inhibition of Ras–PI3K–AKT and Cellular Arrest through the EphA2 Receptor. Oncogene 2008, 27, 2934–2940. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph-Ephrin Bidirectional Signaling in Physiology and Disease. Cell 2008, 133, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Macrae, M.; Neve, R.M.; Rodriguez-Viciana, P.; Haqq, C.; Yeh, J.; Chen, C.; Gray, J.W.; McCormick, F. A Conditional Feedback Loop Regulates Ras Activity through EphA2. Cancer Cell 2005, 8, 111–118. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph Receptors and Ephrins in Cancer: Bidirectional Signalling and Beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef]
- Guo, H.; Miao, H.; Gerber, L.; Singh, J.; Denning, M.F.; Gilliam, A.C.; Wang, B. Disruption of EphA2 Receptor Tyrosine Kinase Leads to Increased Susceptibility to Carcinogenesis in Mouse Skin. Cancer Res. 2006, 66, 7050–7058. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Li, D.-Q.; Mukherjee, A.; Guo, H.; Petty, A.; Cutter, J.; Basilion, J.P.; Sedor, J.; Wu, J.; Danielpour, D.; et al. EphA2 Mediates Ligand-Dependent Inhibition and Ligand-Independent Promotion of Cell Migration and Invasion via a Reciprocal Regulatory Loop with Akt. Cancer Cell 2009, 16, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph Receptors and Ephrins in Cancer Progression. Nat. Rev. Cancer 2024, 24, 5–27. [Google Scholar] [CrossRef]
- Toracchio, L.; Carrabotta, M.; Mancarella, C.; Morrione, A.; Scotlandi, K. EphA2 in Cancer: Molecular Complexity and Therapeutic Opportunities. Int. J. Mol. Sci. 2024, 25, 12191. [Google Scholar] [CrossRef]
- Wang, X.; Zou, Z.; Deng, Z.; Liang, D.; Zhou, X.; Sun, R.; Lan, K. Male Hormones Activate EphA2 to Facilitate Kaposi’s Sarcoma-Associated Herpesvirus Infection: Implications for Gender Disparity in Kaposi’s Sarcoma. PLoS Pathog. 2017, 13, e1006580. [Google Scholar] [CrossRef]
- Fei, Z.; Hong, H.; Xu, T.; Xu, Y.; Chen, J.; Qiu, X.; Ding, J.; Feng, Y.; Huang, C.; Li, L.; et al. Analysis of Risk Characteristics for Metachronous Metastasis in Different Period of Nasopharyngeal Carcinoma. BMC Cancer 2023, 23, 165. [Google Scholar] [CrossRef]
- Su, W.-H.; Yao Shugart, Y.; Chang, K.-P.; Tsang, N.-M.; Tse, K.-P.; Chang, Y.-S. How Genome-Wide SNP-SNP Interactions Relate to Nasopharyngeal Carcinoma Susceptibility. PLoS ONE 2013, 8, e83034. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, J.; Yao, P.; Liu, X.; Chen, F.; Chen, Y.; Zhou, L.; Shen, C.; Zhou, Y.; Du, X.; et al. Spatial Transcriptomics Reveals Unique Metabolic Profile and Key Oncogenic Regulators of Cervical Squamous Cell Carcinoma. J. Transl. Med. 2024, 22, 1163. [Google Scholar] [CrossRef]
- Tang, L.-L.; Chen, Y.-P.; Mao, Y.-P.; Wang, Z.-X.; Guo, R.; Chen, L.; Tian, L.; Lin, A.-H.; Li, L.; Sun, Y.; et al. Validation of the 8th Edition of the UICC/AJCC Staging System for Nasopharyngeal Carcinoma From Endemic Areas in the Intensity-Modulated Radiotherapy Era. J. Natl. Compr. Canc. Netw. 2017, 15, 913–919. [Google Scholar] [CrossRef]
- Bursac, Z.; Gauss, C.H.; Williams, D.K.; Hosmer, D.W. Purposeful Selection of Variables in Logistic Regression. Source Code Biol. Med. 2008, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]

| Variables | Cases (n = 228), n (%) | Controls (n = 243) n (%) | OR (95% CI) | p Value # |
|---|---|---|---|---|
| Age (Mean ± SD, years) | 49.21 ± 14.70 | 49.82 ± 14.68 | 0.65 | |
| Sex | ||||
| Male | 148 (65) | 158 (64.9) | 1.00 (0.688 to 1.467) | 0.98 |
| Female | 80 (35) | 85 (35.1) | ||
| Tobacco consumption | ||||
| Yes | 123 (53.9) | 109 (44.9) | 1.440 (1.002 to 2.070) | 0.048 * |
| No | 105 (46.1) | 134 (55.1) | ||
| Alcohol consumption | ||||
| Yes | 24 (10.5) | 21 (8.6) | 1.244 (0.672 to 2.302) | 0.49 |
| No | 204 (89.5) | 222 (91.4) | ||
| BMI | ||||
| <18.5 | 8 (3.5) | 7 (2.9) | 0.93 | |
| 18.5–25 | 88 (38.6) | 95 (39.1) | ||
| >25 | 132 (57.9) | 141 (58) | ||
| Occupational Exposure | ||||
| risky occupational exposure | 134 (58.8) | 124 (51) | 1.368 (0.950 to 1.970) | 0.09 |
| non-risky occupational exposure | 94 (41.2) | 119 (49) | ||
| Repeated ENT Infections | ||||
| Yes | 89 (39) | 22 (9.1) | 6.432 (3.852 to 10.739) | 0.000 * |
| No | 139 (61) | 221 (90.9) | ||
| TNM stage | ||||
| I, II, and III | 149 (65.4) | |||
| IVA and IVB | 79 (34.6) | |||
| Local tumor invasion | ||||
| Ti, T1, and T2 | 131 (57.4) | |||
| T3 and T4 | 97 (42.6) | |||
| Lymph node involvement | ||||
| N0 and N1 | 83 (36.4) | |||
| N2 and N3 | 145 (63.6) | |||
| Presence of metastasis | ||||
| M0 | 197 (86.4) | |||
| M1 | 31 (13.6) |
| Overall Population | Cases (n = 228), n (%) | Controls (n = 243), n (%) | OR (95% CI) | p-Value | OR a (95% CI) | p Value a,# | |
|---|---|---|---|---|---|---|---|
| Genetic model | rs4702 | ||||||
| CoDominant | AA | 103 (45.2) | 75 (30.9) | 1 | |||
| GG | 43 (18.9) | 48 (19.8) | 0.652 (0.393 to 1.084) | 0.099 | 0.625 (0.361 to 1.083) | 0.094 | |
| AG | 82 (36) | 120 (49.4) | 0.498 (0.331 to 0.749) | 0.001 | 0.491 (0.315 to 0.763) | 0.002 * | |
| Dominant | AA | 103 (45.2) | 75 (30.9) | ||||
| AG-GG | 125 (54.8) | 168 (69.1) | 0.542 (0.372 to 0.79) | 0.001 | 0.529 (0.352 to 0.795) | 0.002 * | |
| Recessive | AA-AG | 185 (81.1) | 195 (80.2) | ||||
| GG | 43 (18.9) | 48 (19.8) | 0.944 (0.597 to 1.493) | 0.806 | 0.914 (0.557 to 1.499) | 0.72 | |
| OverDominant | AA-GG | 146(64) | 123 (50.6) | ||||
| AG | 82 (36) | 120 (49.4) | 0.576 (0.398 to 0.833) | 0.003 | 0.576 (0.387 to 0.858) | 0.007 * | |
| rs6603883 | |||||||
| CoDominant | GG | 84 (36.8) | 89 (36.6) | 1 | |||
| AA | 40 (17.5) | 39 (16) | 1.087 (0.638 to 1.851) | 0.76 | 1.354 (0.767 to 2.390) | 0.29 | |
| GA | 104 (45.6) | 115 (47.3) | 0.958 (0.643 to 1.428) | 0.834 | 0.970 (0.629 to 1.496) | 0.89 | |
| Dominant | GG | 84 (36.8) | 89 (36.6) | ||||
| GA-AA | 144 (63.2) | 154 (63.4) | 0.991 (0.681 to 1.441) | 0.961 | 1.062 (0.708 to 1.594) | 0.77 | |
| Recessive | GG-AG | 188 (82.5) | 204 (84) | ||||
| AA | 40 (17.5) | 39 (16) | 1.113 (0.686 to 1.805) | 0.664 | 1.378 (0.825 to 2.301) | 0.22 | |
| OverDominant | GA-GG | 124 (54.4) | 128 (52.7) | ||||
| AG | 104 (45.6) | 115 (47.3) | 0.934 (0.65 to 1.341) | 0.71 | 0.879 (0.594 to 1.300) | 0.52 |
| Parameters | Genetic Model | Genotype | Parameter Category, n (%) | OR (95% CI) | p-Value | OR a (95% CI) | p Value a,# | |
|---|---|---|---|---|---|---|---|---|
| rs4702 | T1-T2 | T3-T4 | ||||||
| Local tumor invasion (n = 228) | CoDominant | AA | 53 (40.5) | 50 (51.5) | 1 | |||
| GG | 31 (23.7) | 12 (12.4) | 0.410 (0.190 to 0.886) | 0.02 | 0.382 (0.173 to 0.843) | 0.017 * | ||
| AG | 47 (35.8) | 35 (36.1) | 0.789 (0.440 to 1.415) | 0.427 | 0.846 (0.467 to 1.535) | 0.583 | ||
| Recessive | AA-AG | 100 (76.3) | 85 (87.6) | |||||
| GG | 31 (23.7) | 12 (12.4) | 0.455 (0.220 to 0.942) | 0.028 | 0.409 (0.193 to 0.869) | 0.020 * | ||
| rs6603883 | I-II-III | IVA-IVB | ||||||
| TNM stage (n = 228) | Dominant | GG | 48 (32.2) | 36 (45.6) | ||||
| GA-AA | 101 (67.8) | 43 (54.4) | 0.568 (0.324 to 0.994) | 0.048 | 0.578 (0.322 to 1.035) | 0.065 | ||
| rs6603883 | M0 | M1 | ||||||
| Presence of metastasis (n = 228) | Dominant | GG | 67 (34) | 17 (54.8) | ||||
| GA-AA | 130 (66) | 14 (45.2) | 0.424 (0.197 to 0.913) | 0.028 | 0.400 (0.180 to 0.888) | 0.024 * | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hares, S.; Hamizi, K.; Rahab, H.; Bounneche, M.H.; Aouidane, S.; Mansoura, L.; Denni, M.; Mallem, W.; Belaaloui, G. Association of Single-Nucleotide Polymorphisms on FURIN and EPHA2 Genes with the Risk and Prognosis of Undifferentiated Nasopharyngeal Cancer. Int. J. Mol. Sci. 2025, 26, 8486. https://doi.org/10.3390/ijms26178486
Hares S, Hamizi K, Rahab H, Bounneche MH, Aouidane S, Mansoura L, Denni M, Mallem W, Belaaloui G. Association of Single-Nucleotide Polymorphisms on FURIN and EPHA2 Genes with the Risk and Prognosis of Undifferentiated Nasopharyngeal Cancer. International Journal of Molecular Sciences. 2025; 26(17):8486. https://doi.org/10.3390/ijms26178486
Chicago/Turabian StyleHares, Seddam, Kamel Hamizi, Hamza Rahab, Maewa Hibatouallah Bounneche, Souhila Aouidane, Leila Mansoura, Manel Denni, Wissem Mallem, and Ghania Belaaloui. 2025. "Association of Single-Nucleotide Polymorphisms on FURIN and EPHA2 Genes with the Risk and Prognosis of Undifferentiated Nasopharyngeal Cancer" International Journal of Molecular Sciences 26, no. 17: 8486. https://doi.org/10.3390/ijms26178486
APA StyleHares, S., Hamizi, K., Rahab, H., Bounneche, M. H., Aouidane, S., Mansoura, L., Denni, M., Mallem, W., & Belaaloui, G. (2025). Association of Single-Nucleotide Polymorphisms on FURIN and EPHA2 Genes with the Risk and Prognosis of Undifferentiated Nasopharyngeal Cancer. International Journal of Molecular Sciences, 26(17), 8486. https://doi.org/10.3390/ijms26178486






